ISSN: 0973-7510
E-ISSN: 2581-690X
This study aims to determine the types of causative organism, the utility of synovial procalcitonin (PCT), C-Reactive Protein (CRP) and bacterial 16S rRNA gene-based RT-PCR and their comparison with conventional culture results in patients with clinically-suspected SA. A total of 38 patients were recruited in this cross-sectional study for performing synovial PCT and CRP assay, and bacterial gDNA quantification via RT-PCT. Records of culture results, WBC count, ESR, blood CRP, and antibiotic administration were obtained. Gross appearance and viscosity determination are significantly associated with the bacterial load. This study documents Acinetobacter radioresistens and Klebsiella pneumoniae bacteria as causative pathogens of SA in Malaysia. CRP and ESR showed a significant role in diagnosing SA. Reasons for finding no concordance between conventional culture methods and 16S rDNA RT-PCR as well as synovial PCT were comprehensively reviewed. Gross appearance and viscosity showed a significant relationship with the bacterial load. RT-PCR is useful in patients treated with antimicrobial therapy with negative culture results.RT-PCR has speed and accuracy compared to conventional culture. Awareness of Klebsiella pneumoniae and Acinetobacter radioresistens as causative bacteria should be prompted among clinicians particularly at local, regional as well as international levels. Developing guidelines for including 16S rRNA gene RT-PCR and introducing Digital PCR and next-generation sequencing to detect and identify bacterial species in diagnosing SA is recommended.
Septic Arthritis, Procalcitonin, 16S rRNA Gene, RT-PCR
Septic arthritis (SA) is an emergency condition associated with significant morbidity and mortality.1 No single laboratory test is conclusive to differentiate between septic and non-SA.2 To date, few laboratory investigations have shown sufficient sensitivity, specificity, or predictive value to be introduced into the clinical practice for the proper management of SA.3 Early diagnosis reduces morbidity and mortality. The current standard for diagnosing SA is primarily clinical-based and confirmed by the finding of positive Gram stain, synovial fluid (SF) culture, blood culture, visible pus in the SF aspirate, or response to empirical antibiotics therapy.4 Any microbial pathogen can cause SA.5 However, the main identified causative organism reported in 50% of cases of patients with SA was Staphylococcus aureus6 followed by streptococci, and together they accounted for 91% of cases.5 Preliminary inflammatory markers in patients with SA include peripheral blood white blood cell (WBC), erythrocyte sedimentation rate (ESR), and C-reactive protein (CRP). Their value does not significantly increase the confidence of SA diagnosis due to the absence of an immediate response secondary to the acute onset of inflammation.2,3,7 However, measurement of these preliminary inflammatory markers is useful in monitoring response to therapy.7,8
Procalcitonin (PCT) is a protein of 116 amino-acids with 13 kDa molecular weight. PCT is an inflammatory biomarker and mortality predictor. Its levels increase from less than 0.1 to more than 100 ng/ml in sepsis.9 PCT is specific for bacterial infections and reliable in distinguishing bacterial from viral infections.10 Unlike other inflammatory markers such as CRP, the PCT level is not altered by steroids which makes it a more accurate diagnostic marker.11 PCT has a higher diagnostic value than clinical or laboratory tests such as WBC and CRP because of its high sensitivity and specificity for diagnosing SA. It is considered as the best parameter to distinguish patients with SA from those with another joint disease, especially if the cause is systemic rather than direct joint inoculation.7 Serum PCT was found significantly increased in patients with SA in comparison with non-SA patients. The recommended cut-off value of serum PCT in diagnosing SA is 0.4 ng/ml. Discrepancy on the diagnostic value of SF PCT was reported and the use of PCT in the diagnosis of bone and joint infection was investigated by a few studies with relatively small sample sizes.12 Previous studies that evaluate the use of serum or synovial PCT level in patients with SA utilized small sample size ranging from 8 to 32 samples.11,13 Measurements of PCT level in SF is more informative than its serum level for differentiating septic from non-septic cases of arthritis.11 However, PCT level is raised in SA, but its accuracy in differentiating SA from other forms of acute arthritis is unsettled.3 Real-time quantitative PCR is the most accurate method for the detection and quantification of DNA and RNA14 and provides less handling of samples making post-PCR steps unnecessary.15
To date, studies which have addressed the utility of RT-PCR in comparison with the synovial PCT, CRP and conventional-culture results for diagnosing clinically-suspected SA are scarce and need further study as they were varied in the methodologies and outcome. Moreover, the causative organism remains unknown in a considerable number of patients with clinically-suspected SA. The aims of the present study were to determine the types of causative organism and the utility of synovial PCT, CRP and bacterial 16S rRNA gene-based RT-PCR and their accordance with conventional culture results in patients with clinically-suspected SA.
In this cross-sectional study, a total of 38 patients with clinically-suspected SA was enrolled. The synovial fluid specimens from clinically-suspected patients with SA were included. Samples with incomplete data (FBC, CRP, ESR & culture results and antibiotic administration) were excluded from analysis. The SF was stored at -20°C until the time of subsequent processing. The stored synovial aspirate was thawed at room temperature and then transferred from its sterile containers into sterile 10 ml Falcon tubes. The tubes were centrifuged at 4000 rpm for 10 min. The supernatant was collected for synovial PCT assay, CRP latex agglutination test and CRP quantitative assay, whereas the pellet was used for DNA extraction.
The synovial fluid samples were observed for their characteristics such as gross appearance and viscosity. Viscosity of SF was identified by dropping it from syringe. SF with high viscosity was indicated if it formed a string about 4-6 cm long upon dropping. Highly viscous samples required predilution with normal saline before testing for PCT assay, CRP Latex agglutination test and CRP quantitative assay. Sodium hydroxide (NaOH) solution was added to the samples with high viscosity to induce liquefaction and solubility, thus allowing the ease of subsequent sample processing.4
PCT assay was done using a commercially available procalcitonin assay kit (BRAHAMS, Roche, Germany) and analyser Cobas e 411 analyser (Roche, Germany). C-reactive protein (CRP) latex reagent test was performed according to the manufacturer’s instructions (CRP direct latex kit, VEDA Laboratory, France). CRP quantitative assay was performed using QuikRead go® CRP kit (Orion Diagnostica, Finland) and QuikRead go® instrument (Orion Diagnostica, Finland).
Bacterial genomic DNA was extracted using DNeasy Blood and Tissue Kit (Qiagen Corp., Santa Clarita, CA) in accordance with the manufacturer’s instructions. The concentration and purity of the extracted DNA were determined by using NanoDropTM spectrophotometer (Thermo Scientific, USA). The DNA concentration was recorded in ng/µl and was stored at -20C˚ until use. Fluorogenic probe-based PCR targeting the 16S rRNA gene was used and aimed at determining the bacterial load in SF of patients with clinically-suspected SA. The experiment was performed using ViiA 7 Real-Time PCR System (Thermo Fisher, Scientific USA). The 2.5 times concentrated solution (3 mM Mg2+ final concentration) of the DNA-free Mastermix 16S Dye was obtained from MOLZYM, Germany. The designed universal primers amplify 160-bp fragment of any bacterial DNA in clinical sample using: universal forward primer (P891F) (5´-TGGAGCATGTGGTTTAATTCGA-3′), universal reverse primer (P1033R) (5´-TGCGGGACTTAACCCAACA-3′) and the TaqMan uniprobe (5′- CACGAGCTGACGACAGCCATGCA-3′) is labelled with the reporter dye FAM at the 5′ end, and the quencher dye MGB (Minor Groove Binder) at the 3′ end.16,17 The primers and probe for the quantification of the16S rRNA gene were supplied by Analisa Resources (M) Sdn. Bhd. Two regions of highly conserved sequences are selected as the universal primer annealing sites. The internal highly conserved sequence is used as the annealing sites of TaqMan uniprobe.
The gDNA extracted from the colonies of cultured S. aureus. was used as a standard gDNA for the quantification of the bacterial 16S rRNA. The standard curve was based on known concentrations of standard bacterial gDNA prepared by eight serial dilution of the standard gDNA. Working concentration of 1.0 to 0.00391 CFU/mL was prepared with three replicates for each dilution.
Standard control composed of pure bacterial DNA extracted from colonies of cultured Staphylococcus aureus was prepared by standard plate count method according to the standard protocol. Sterile ultrapure DNA-free water (Nacalai Tesque, Japan) was used as a negative control or no Template Control (NTC). In this study, false-positive amplification in the no Template Control (NTC) occurred which necessitated the use of the 2.5x Molzym Mastermix. DNAse I pretreatment of the reagents and adding 2 µl of the ethylene diamine tetra-acetic acid (EDTA) to the reaction mixture followed by heating at 70˚C for 10 minutes is the method used for eliminating false-positive results in the no Template Control (NTC) when using universal 16S rRNA. A volume of 23 µl of the 2.5x Molzym Mastermix was aliquoted into each well of the RT- PCR strips. For a final reaction volume of 25 μl, 2 µl of the standard DNA (positive control) or 2 µl of DNA-free water (negative control) or 2 μl of the template were added into each well containing 23 µl mastermix. The volumes for one reaction preparation were 2.5x Molzym mastermix 10.0 µl, TaqMan 16S assay 1.25 µl, MolTaq 16S enzyme 0.8 µl, DNA-free water 10.95 µl, Template 2.0 µl and total volume for one reaction 25.0 µl. Triplicates for each sample were prepared. PCR was run according to thermal cycling conditions. Thermal cycling conditions of 40 cycles was as: Hold at 95°C for ten minutes, denature at 95°C for fifteen seconds and anneal/extend at 60°C for one minute.
Precautions to avoid cross-contamination during qPCR were taken including; disinfection of the biological safety cabinet (BSC) (NUAIRE, USA), tube holder and, micropipettes followed by UV treatment for 30 minutes. Using the copies/µl unit in expressing the results of RT-PCR experiment was based on the justifications of other studies.18-21
Statistical analysis was conducted using Statistical Package for the Social Sciences (SPSS) version 22 for Windows (Chicago, IL.). Independent t test for comparing parametric and Mann-Whitney test for comparing non-parametric data were used as well as correlation analysis with chi square tests. p value of <0.05 was considered statistically significant.
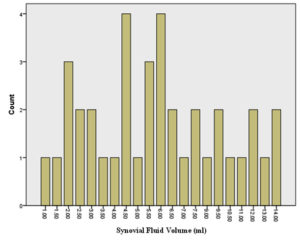
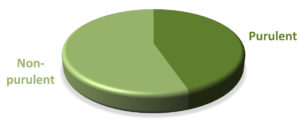
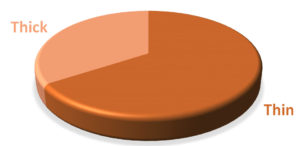
The volume of synovial fluid samples submitted to the laboratory ranged from 1 to 4 ml. Twenty-four (63.2%) synovial fluid samples were of 1-2 ml volume. Seventeen out of 38 (44.7 %) synovial fluid samples were purulent in appearance and the remaining (55.3 %) were non-purulent. With regard to the sample viscosity, 13 out of 38 (34.2 %) synovial fluid samples were thick and the remaining (65.8 %) were thin (Figure 2).
No statistically significant correlation between the blood and synovial CRP. CRP Latex agglutination test of the SF showed positive results in 26/38 (71 %) samples. Significant association was seen between the results of latex agglutination test and the quantitative assay of the CRP in SF. The synovial CRP level ranged from 10.0 to 1466 and blood CRP level ranged from 10.0 to 304 mg/L (Table 1).
Table (1):
Comparison of synovial CRP assay with semi-quantitative latex agglutination test and blood CRP assay (n=38).
| Variable | Synovial CRP assay (mg/L) Median (IQR) | z statisticsa | p valuea |
|---|---|---|---|
| Synovial latex agglutination test | |||
| Positive (n=26) | 48.0 (77.1) | 2.84- | 0.004 |
| Negative (n=12) | 0.0 (21.7) | ||
| Blood CRP (mg/L) | |||
| Undetectable (n=14) | 82.40 (120) | -0.408752 | 0.68 |
| Detectable (n=24) | 21.50 (86) | ||
a Mann-Whitney test
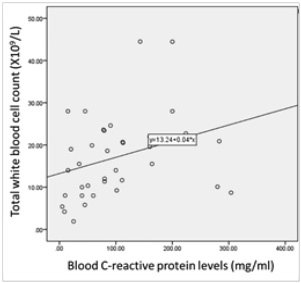
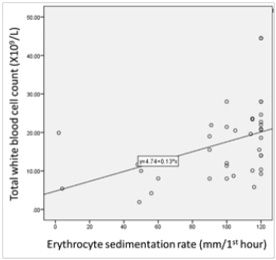
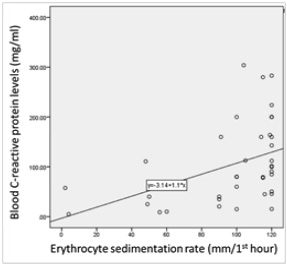
Significant correlations among the total white blood cell count, C-reactive protein and erythrocyte sedimentation rate (ESR) were found (Figure 1).
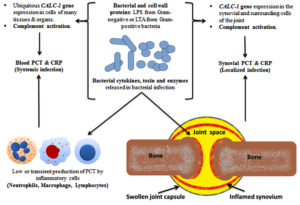
Figure 1. Pathogenesis of SA with PCT and CRP production: The bacterial-induced ubiquitous CALC-1 gene expression in parenchymal cells of many different tissues and organs, in the presence of severe systemic bacterial sepsis, might indicate the pivotal role of PCT as a detrimental marker of sepsis survival and outcomes. The released endotoxin (lipopolysaccharide, LPS) from Gram-negative bacteria or lipoteichoic acid (LTA) from Gram-positive bacteria and proinflammatory cytokines are the main stimuli for CALC-1 gene expression. The bacterial endotoxin and proinflammatory cytokines (e.g., interleukin-1b, tumour necrosis factor-α, interleukin-6) stimulate the secretion of PCT from parenchymal cells which in turn, reinforce immune cells production of cytokines that augment the high PCT levels in patients with systemic or local infection. PCT is produced by inflammatory cells including monocytes, lymphocytes, and neutrophils and accumulates in the synovial fluid
A total of 35 samples (92.1%) showed positive 16S rDNA RT-PCR. The range of bacterial DNA amount in the synovial fluid was 0.001- 3.074 copies/µl. Standard curve had an efficiency of 90% and R2: 0.957 which passes the standard curve criteria. On comparing sample viscosity and RT-PCR results, thick synovial fluid showed statistically higher bacterial 16S rDNA gene quantity compared with thin fluid. Purulent synovial fluid showed statistically higher bacterial 16S rDNA gene quantity compared to fluid with Non-purulent appearance. Synovial PCT level was detectable in twenty (52.6%) samples and found to be below the detectable level (less than 0.025 ng/ml) in 18 (47.4%) samples. The synovial PCT level was ranged from 0.025 to 2.52 ng/ml. The synovial PCT levels were not statistically different according to the synovial fluid consistency, gross appearance, culture results and the presence or absence of evidence of systemic infection. The synovial PCT levels were not statistically different according to the synovial fluid consistency, gross appearance, and culture results (Table 2).
Table (2):
Comparison among synovial procalcitonin levels and bacterial 16S rRNA gene quantity by RT-PCR (copies/µl) with synovial fluid characteristics
| Synovial fluid characteristics | Synovial procalcitonin (ng/ml) Median (IQR) | z statisticsa | p valuea | Synovial Absolute 16S rRNA gene (copies/µl) Median (IQR) | z statisticsa | p valuea |
|---|---|---|---|---|---|---|
| Synovial fluid consistency (viscosity) | ||||||
| Thin (n= 25) | 0.024 (0.275) | -0.433 | 0.693 | 0.002 (2.99) | -2.175 | 0.030 |
| Thick (n=13) | 0.024 (0.175) | 2.099 (4.17) | ||||
| Synovial fluid gross appearance | ||||||
| Purulent(n=17) | 0.146 (0.275) | -1.699 | 0.121 | 0.002 (2.74) | -2.031 | 0.042 |
| Non-purulent(n=21) | 0.190 (0.092) | 1.139 (3.69) | ||||
a Mann-Whitney test
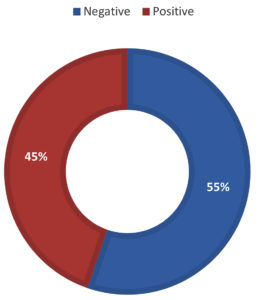
No statistically significant differences in the 16S rRNA gene quantity with the positive culture results (n=38). All culture-positive samples 17/38 (44.7 %) were found to be RT-PCR positive. Three of the culture-negative samples (14.3 %) were found to be RT-PCR negative. No false negative 16S rDNA RT-PCR in consideration of the culture results. Seventeen of 38 (45%) samples gave positive culture, whereas 21 of 38 (55%) samples were negative (Figure 1). Staphylococcus aureus was isolated in 9 of 17 (52.9%) samples, of which three were methicillin-resistant S. aureus (MRSA). Burkholderia pseudomallei, Pseudomonas aeruginosa, Acinetobacter radioresistens and Klebsiella pneumoniae each was found in one sample (5.8%). Four of 17 (23.5%) samples showed mixed growth of three or more organisms which were not specifically identified. No statistically significant differences in PCT assay results compared to the synovial fluid culture results (n=38) (Table 3).
Table (3):
Comparison of the synovial fluid culture results with procalcitonin assay results and bacterial 16S rRNA gene quantity by RT-PCR (n=38).
| Parameters Synovial fluid culture results | Procalcitonin assay | Bacterial 16S rRNA gene quantity | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Result | Frequency (%) | Rate value | c2 | p – valuea | Frequency (%) | Rate value | c2 | p-valuea | |
| Negative | Detectable | 11 | 47.6 | 18 | 85.7 | ||||
| Non-detectable | 10 | 52.4 | 0.001 | 0.973 | 3 | 14.3 | 2.637 | 0.104 | |
| Positive | Detectable | 9 | 47.1 | 17 | 100 | ||||
| Non-detectable | 8 | 52.9 | 0 | 0 | |||||
a Chi square test
(g)
Figure 2. Distribution of patients according to the synovial fluid volume (A), gross appearance of the synovial fluid (B), viscosity of the synovial fluid (C) and culture result of the synovial fluid (D) and Correlations among the total white blood cell count, C-reactive protein and erythrocyte sedimentation rate (ESR) levels among patients (n=38) (E to G). The volume of synovial fluid samples submitted to the laboratory ranged from 1 to 4 ml. twenty-four (63.2%) synovial fluid samples were of 1-2 ml volume. Gross appearance analysis of SF relied on the visual inspection to determine appearance and viscosity. Seventeen out of 38 (44.7 %) synovial fluid samples were purulent in appearance and the remaining (55.3 %) were non-purulent. With regard to the sample viscosity, 13 out of 38 (34.2 %) synovial fluid samples were thick and the remaining 25 (65.8 %) were thin. The synovial fluid volume is statistically different in accordance to synovial fluid viscosity (Synovial fluid volume 4.11(3.0) and 7.46(3.4), respectively, with p=0.005). Significant fair direct positive correlation between white blood cell count and blood C-reactive protein levels in the blood (Spearman’s correlation, r = 0.422, p value= <0.001) (E) and erythrocyte sedimentation rate (Spearman’s correlation, r = 0.510, p value= <0.001) (F). A significant fair direct positive correlation between blood C-reactive protein levels and ESR (Spearman’s correlation, r = 0.466, p value= <0.001) (G)
Distinguishing SA from other inflammatory joint conditions is based on clinical and routine laboratory investigations representing a diagnostic challenge.4 The SF volume in normal joints varies from a few drops in small joints to several millilitres in larger joints. SF volume in the normal knee joint is from 0.2 to 2 ml. Pathological changes in the joint are reflected in the volume, cellular composition and particulate load within the SF. In the present study, 63.2% of the synovial fluid samples volume were of 1-2 ml (Figure 1) which might reflect the inflammatory nature of the SF samples rather than being an exudate of non-inflammatory origin with larger volume. The volume of SF is of little relevance for diagnosing SA.22
The bacterial-induced ubiquitous CALC-1 gene expression in parenchymal cells of many different tissues and organs, in the presence of severe systemic bacterial sepsis, might indicate the pivotal role of PCT as a detrimental marker of sepsis survival and outcomes. The released endotoxin (lipopolysaccharide, LPS) from Gram-negative bacteria or lipoteichoic acid (LTA) from Gram-positive bacteria and proinflammatory cytokines are the main stimuli for CALC-1 gene expression. The bacterial endotoxin and proinflammatory cytokines (e.g., interleukin-1b, tumour necrosis factor-a, interleukin-6) stimulate the secretion of PCT from parenchymal cells which in turn, reinforce immune cells production of cytokines that augment the high PCT levels in patients with systemic or local infection. PCT is produced by inflammatory cells including monocytes, lymphocytes, and neutrophils and accumulates in the synovial fluid (Figure 2). A significant correlation between white blood cell (WBC) count and both erythrocyte sedimentation rate (ESR) (r = 0.510, p value= <0.001) and blood C-reactive protein (CRP) levels (r = 0.422, p value= <0.001) was demonstrated. These findings confirm the role of the blood CRP in supporting the diagnosis of clinically suspected SA. In this study, a significant correlation between CRP and ESR (r = 0.466, p value= <0.001) was found, thus suggesting the role for both CRP and ESR in assisting the diagnosis of clinically-suspected SA. Other studies found that CRP is a better independent predictor of SA than ESR.11,23 The lack of correlation between blood and synovial CRP found might be due to its deposition in areas of tissue necrosis of the affected joints by SA.23 This finding suggests that CRP from synovial fluid does not offer a diagnostic advantage in the detection of joint infection. In this study, a significant relationship between synovial CRP using latex agglutination test for semi-quantitative antigen concentration determination with the quantitative synovial CRP assay (p = 0.004) (Table 1) was found. Thus, the present study is the first to reveal the utility of CRP assessment using latex agglutination test in SF from patients with clinically-suspected SA and its relationship with the quantitative synovial CRP assay.
Normal SF is highly viscous and its viscosity is dependent on the concentration of synovium-derived hyaluronic acid. Infectious fluid is of low viscosity and proportional to the degree of local inflammation.24 The finding of statistically higher (2.099, p=0.042) bacterial 16S rRNA gene quantity in the thick synovial fluid compared with thin fluid (p=0.002) (Table 2) might be attributed to the lower concentration and molecular weight of synovial hyaluronan and the addition of various serum-derived proteins substantially increase the viscosity of SF as a result of inflammatory process in the joint affected with SA.25 The finding of twenty-five (65.8 %) thin and less viscous SF seems to be attributed to the dilution of the SF with plasma from leaky synovial blood vessels, and the reduction of production of the hyluronans by the synoviocytes. Moreover, the production and release of degrading digestive enzymes into the fluid leads to the reduction of viscosity and poor clot formation.14 Depolymerization of the hyaluronic acid primarily by lysosomal enzymes of neutrophils, synovial lining cells, and cartilage cells cause destruction of the joint cartilage within several days.24 However, measurement of SF viscosity was reported to be of limited clinical utility.26 In this study, SF with higher viscosity (thick) was found to be significantly (p=0.005) of smaller volume compared to the volume of the less viscous (thin) SF samples (Table 2). The smaller volume of the more viscous SF might reflect the technical difficulties in the aspiration of the SF or might be due to the higher protein and cellular contents of the inflammatory SF in patients with SA. The thick viscosity of the synovial fluid was more consistent with the clinical suspicion of SA which might indicate a usefulness of the SF viscosity assessment for the appropriate diagnosis of SA.
The statistically higher bacterial 16S rRNA gene quantity (1.139) found in seventeen (44.7 %) SF samples with purulent gross appearance compared with non-purulent fluid (0.002, p=0.030) (Table 2) might indicate the utility of the gross appearance determination in the diagnosis work up for patients with clinically-suspected SA. However, the gross appearance of SF is useful in differentiating inflammatory from non-inflammatory synovial fluid and is related to its cellularity.15,27 In another study, the purulent appearance of the SF was reported in (74%) of SA cases, and all non-SA cases have non-purulent appearance.13 It was reported that the gross appearance analysis is 94% sensitive and 58% specific for differentiating inflammatory and non-inflammatory causes of acute arthritis with positive predictive value of 0.69 and negative predictive value of 0.91.13 The gross appearance and viscosity showed significant association with the bacterial load measured by 16S rRNA gene RT-PCR assay.
The finding of positive synovial fluid culture results with the clinical suspicion of SA in 17 (44.7 %) out of the total 38 synovial fluid samples (Table 3) might indicate the limitation of SF culture in diagnosing SA and the need of an efficient diagnostic tool for proper diagnosis of SA. Prior antibiotic administration by all patients with clinically suspected SA for at least 24-48 before the SF aspiration might be another reason for the finding of negative culture results. Other reasons for the possible false negative cultures in patients with clinically-suspected SA include; mishandling of fluids, inappropriate culture media, fastidious organisms, samples of small volume, transient bacteremia, and nonbacterial infection.16 Non-purulent arthritis (NPA) is also called reactive arthritis or Reiter syndrome is defined as arthritis without purulent drainage or associated abscess with infection in another part of the body, most often sepsis or urinary tract infection. The mechanism of NPA is unclear but speculatively toxin-mediated.28 In this study, the mechanism of NPA is explained, at least in part, by documenting the presence of bacterial DNA in the SF via RT-PCR in samples with culture negative result.
The finding of Staphylococcus aureus including MERSA as the causative bacteria in 9 of the 17 samples (52.9%) was in accordance with previously published data on the causative pathogens in patients with SA.3,6,7 Moreover, each of the Burkholderia pseudomallei, Pseudomonas aeruginosa, Acinetobacter radioresistens, and Klebsiella pneumoniae was isolated from the studied patients with SA. B. pseudomallei is a Gram-negative, aerobic, motile, non-spore-forming bacillus with complex bacterial genomes, causes melioidosis disease which is endemic in South-east Asia and northern Australia.11 B. pseudomallei as a causative organism of septic arthritis/osteomyelitis in 20 (12.7 %) of patients presented with melioidosis was reported in a North-Eastern state of Malaysia.26 Klebsiella oxytoca is a member of the family Enterobacteriaceae and known to be responsible for nosocomial infections. The first cases of SA caused by K. oxytoca was reported in 2010. A. radioresistens and K. pneumoniae have not been reported as causative bacteria of SA in Malaysia. Infections with K. pneumoniae are increasingly reported and cause life-threatening community-acquired infections even in patients with competent immune response.29 The first cases of SA caused by K. pneumoniae was reported in 2013 from Taiwan.30 A. radioresistens is Gram-negative, aerobic, and non-spore-forming, coccobacilli causing opportunistic infections in critically-ill patients with impaired immunity due to having serious underlying predisposing conditions or are on treatments affecting the immune response. These opportunistic infections include pneumonia, skin and soft-tissue infections, wound infections, urinary tract infections, meningitis, and bloodstream infections. Community-acquired infections by A. radioresistens have been reported mainly from South-east Asia and tropical Australia. This organism is frequently multi drug-resistant and is capable of causing morbidity and mortality in patients with severe underlying immune deficiency. It is grown easily on simple culture media, forming domed, smooth, pigmented pale yellow or grey colonies of about two mm diameter.31 Mixed bacterial infection has been reported in 10 % of intravenous drug users.7 In this study, cases with mixed infections have been categorized as ‘positive culture’, even though specific identification of each bacterium was not performed.
The finding of no concordance between conventional culture methods and 16S rDNA RT-PCR (Table 3) disagrees with the conclusion of other studies.13,32 The possible reasons for discrepancy in terms of finding positive RT-PCR and negative culture might be attributed to a number of reasons; presence of contaminants, insufficient number of bacteria in the samples to reach the level of detectability by culture as the quality and quantity of samples are important to ensure recovery of pathogens. Other reasons include prior treatment with antimicrobial therapy,33 low bacterial concentration within infectious site particularly after the initiation of antibiotic treatment33 and the presence of fastidious growing bacteria such as Granulicatella adiacens.
RT-PCR detects bacterial DNA from both viable and nonviable bacteria. The presence of non-viable anaerobic bacteria leads to negative culture results.16 This might explain the discrepancies between culture and RT-PCR. Nevertheless, false-positive results can also occur due to amplicon contamination.34 Leukocyte DNA may result in false negative PCR by competing with the small amount of bacterial DNA and inhibits bacteria-specific PCR within the sample.35 Therefore, a newly available reagent kit to remove human DNA before the Qiagen bacterial gDNA extraction is recommended. Proteinase K, is insufficient for complete cell wall lysis owing to its inhibition by SF components or to the overwhelming abundance of synovial proteins.17 Thermal and enzymatic inductions of cell lysis in the sample processing protocol allow more effective extraction of microbial DNA from the difficult-to-lyse cell walls such as Gram-positive organisms. RT-PCR is 40-fold more sensitive for total bacterial load detection compared to culture methods.19 In this study, all culture positive samples were also positive by RT-PCR. In order to achieve an adequate signal to noise ratio during PCR assay and to avoid false positive results, different methods of decontaminating the PCR reagents and eliminating contaminating DNA were used. Untreated commercial PCR master mix can be contaminated at a level of about 10 copies per well.36 DNA decontamination methods can lead to loss of sensitivity when using universal 16S rDNA PCR assay. The enzymatic treatment methods for decontaminating PCR reagents include the use of deoxyribonuclease (DNAse) treatment and restriction endonuclease digestion. The physical and chemical methods include, ultraviolet irradiation and 8-methoxypsoralen treatment in combination with UV irradiation.36 The DNAse I eliminate up to 100 copies of contaminant DNA without causing PCR inhibition.37 DNAse I pretreatment of the reagents with or without addition of ethylene diamine tetra-acetic acid (EDTA) followed by heating is reported as the most effective method of eliminating false-positive results in the NTC when using universal 16S rRNA.36 In this study, the decontamination procedure by using DNAse I lead to poor efficiency of PCR that might be attributed to the DNAse-induced degradation of the Taq polymerases38 or to long heating step in the decontamination procedure. The high affinity of Taq DNA polymerase for DNA makes it impossible to completely eliminate contaminating DNA. The use of universal 16S rDNA PCR with the TaqMan system necessitates the use of ultrapure Taq DNA polymerase such as Molzym Mastermix, ultrapure reagents, and DNA-free plastic.38 The finding that RT-PCR was positive for the presence of bacteria in 35 samples while SF culture was only positive in 17 samples suggests that the TaqMan PCR assay has analytical and clinical utility with speed and accuracy compared to culture for SA. RT-PCR is a useful tool for molecular diagnosis even in patients treated with antimicrobial therapy.39,40
The value of RT-PCR is enhanced when coupled with another inflammatory biomarker such as PCT.16 PCT concentration strongly correlates with bacterial load and severity of infections, thus making it beneficial for use to guide antibiotic therapy, support therapeutic decision making, decrease the unnecessarily prolonged use of antibiotics in non-bacterial disease, and allow early diagnosis of bacterial SA. PCT as a biomarker for differentiating SA from non-SA has been previously evaluated with conflicting results.10 In localized infection a specific, cut-off value is needed to increase the sensitivity of the test. 11 The proposed reasons for the lack of statistical difference in the synovial PCT levels according to the synovial fluid gross appearance, viscosity, conventional culture results and 16S rDNA RT-PCR include the facts that; PCT is secreted at lower levels in body fluids other than serum,12 its levels are generally low during infection in biological fluids38 and it lacks sensitivity in local infections and increase only if the infection involves surrounding tissues or the presence of systemic infection. Other reasons include; delayed processing of frozen-stored samples for longer than 24 hrs while the half life of PCT in the synovial fluid is about 20 to 24 hrs39 and its level is elevated when the cause of SA is systemic rather than local.11,12 In addition, higher PCT level was reported in patients with Gram-negative than those with Gram-positive blood-born infection.41 The role of PCT in arthritic destruction is uncertain.7 Low or transient production of PCT by inflammatory cells including monocytes, lymphocytes, and neutrophils and its accumulation in the synovial fluid thereafter is typically evident in the follow-up measurements.10,12 PCT levels showed no significant difference in patients with SA compared to those without evidences of SA.9 However, the previously reported significantly higher levels of PCT in the SF of patients with SA and the consideration of PCT assay in SF as positive indicators of SA might attribute to the freshness of the SF samples used in the experiments.41 PCT has been reported to be stable in samples stored at -20˚C for a long period.34
There is no current globally accepted reference standard for the diagnosis of SA. The gross appearance and viscosity showed significant relationship with the bacterial load measured by 16S rRNA gene RT-PCR assay in SF samples of patients with clinically-suspected SA. RT-PCR is a useful tool for molecular diagnosis of SA even in patients treated with antimicrobial therapy, detecting bacteria in samples with negative SF culture results, and has clinical utility with speed and accuracy compared to culture for SA. The awareness of K. pneumoniae and A. radioresistens as causative bacteria in patients with SA regardless of their immune status should be prompted among clinicians. Synovial CRP assessment using latex agglutination is useful and found to be significantly associated with the quantitative synovial CRP assay. Locally-developed guideline involving 16S rRNA gene RT-PCR assay for diagnosing SA should be established to improve the diagnostic performance. Precautions and procedural modification to reduce the problem of amplification of background DNA contamination is essential. Identification of the individual bacteria using techniques with lower detection limit such as Digital PCR (d-PCR) technology and next-generation sequencing (NGS) to identify unculturable bacteria including fastidious bacteria or new pathogen in SF samples is recommended.
ACKNOWLEDGMENTS
The authors would like to thank the staff members in the Medical Microbiology and Parasitology Laboratory at Hospital Universiti Sains Malaysia for the samples collection and storage. We are grateful to all the respondents who participated in this study. Thanks are extended to Dr Aniza Abd Aziz for supporting in the statistical analysis of this study data.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
RAHA, UYHA and AH conceptualized and designed the study. TMBTMS, AFBS and SBI contributes to the admin, technical and material support. RAHA performed data acquisition, data analysis and interpretation. UYHA performed statistical analysis. RAHA and AH drafted the manuscript. UYHA critically revised the manuscript. AH supervised and approved the final manuscript for publication.
FUNDING
None.
DATA AVAILABILITY
All data generated or analysed during this study are included in this manuscript.
ETHICS STATEMENT
Ethical approval was obtained from Jawatankuasa Etika Penyelidikan (Manusia) of Universiti Sains Malaysia (JEPeM) (Ref. No, USM/JEPeM/16040163) and the Ministry of Health (MOH) Research and Ethics Committee (MREC) (Ref. No. 12KKM/NIHSEC/P15-1616).
- John J, Chandran L. Arthritis in Children and Adolescents. Pediatr Rev. 2011;32(11):470-479.
Crossref - Horowitz DL, Katzap E. Approach to Septic Arthritis. American Family Physician. 2011;84(6):653-660. PMID: 21916390.
- Mathews CJ, Weston VC, Jones A, Field M, Coakley G. Bacterial septic arthritis in adults. The Lancet. 2010;375(9717):846-855.
Crossref - Yang S, Ramachandran P, Hardick A, et al. Rapid PCR-Based Diagnosis of Septic Arthritis by Early Gram-Type Classification and Pathogen Identification.
J Clin Microbiol. 2008;46(4):1386-1390.
Crossref - Margaretten ME, Kohlwes J, Moore D, Bent S. Does This Adult Patient Have Septic Arthritis? JAMA. 2007;297(13):1478-1488.
Crossref - Helito CP, Zanon BB, de Souza Miyahara H, et al. Clinical and epidemiological differences between septic arthritis of the knee and hip caused by oxacillin-sensitive and -resistant s. aureus. Clinics. 2015;70(1):30-33.
Crossref - Martinot M, Sordet C, Soubrier M, et al. Diagnostic value of serum and synovial procalcitonin in acute arthritis: a prospective study of 42 patients. Clin Exp Rheumatol. 2005;23(3):303-310.
- Lynn MM, Mathews CJ. Advances in the management of bacterial septic arthritis. International Journal of Clinical Rheumatology. 2012;7(3):335-342.
Crossref - Maruna P, Nedelnikova K, Gurlich R. Physiology and genetics of procalcitonin. Physiol Res. 2000;49(Suppl 1):57-61. PMID: 10984072.
- Schuetz P, Albrich W, Mueller B. Procalcitonin for diagnosis of infection and guide to antibiotic decisions: past, present and future. BMC Med. 2011;9(1):107.
Crossref - Talebi-Taher M, Shirani F, Nikanjam N, Shekarabi M. Septic versus inflammatory arthritis: discriminating the ability of serum inflammatory markers. Rheumatol Int. 2013;33(2):319-324.
Crossref - Shen CJ, Wu MS, Lin KH, et al. The use of procalcitonin in the diagnosis of bone and joint infection: a systemic review and meta-analysis. Eur J Clin Microbiol Infect Dis. 2013;32(6):807-814.
Crossref - Gadsby NJ, Onen A, Phillips SA, et al. Evaluation of Real-Time 16S rDNA PCR and Pyrosequencing for Routine Identification of Bacteria in Joint Fluid and Tissue Specimens. OJMM. 2011;1(1):1-6.
Crossref - Kaltenboeck B, Wang C. Advances in Real-Time PCR: Application to Clinical Laboratory Diagnostics. In: Advances in Clinical Chemistry. 2005: 40: 219-259.
- Murphy J, Bustin SA. Reliability of real-time reverse- transcription PCR in clinical diagnostics: gold standard or substandard? Expert Review of Molecular Diagnostics. 2009;9(2):187-197.
Crossref - Rowther FB, Rodrigues CS, Deshmukh MS, et al. Prospective Comparison of Eubacterial PCR and Measurement of Procalcitonin Levels with Blood Culture for Diagnosing Septicemia in Intensive Care Unit Patients. J Clin Microbiol. 2009;47(9):2964-2969.
Crossref - Yang S, Lin S, Kelen GD, et al. Quantitative Multiprobe PCR Assay for Simultaneous Detection and Identification to Species Level of Bacterial Pathogens. J Clin Microbiol. 2002;40(9):3449-3454.
Crossref - Větrovský T, Baldrian P. The Variability of the 16S rRNA Gene in Bacterial Genomes and Its Consequences for Bacterial Community Analyses PLoS ONE. 2013;8(2):e57923.
Crossref - Nadkarni MA, Martin FE, Jacques NA, Hunter N. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology. 2002;148(1):257-266.
Crossref - Sutton S. Accuracy of Plate Counts. Journal Of Validation Technolgy. 2011;17(3):42-46. http://www.microbiologynetwork.com/content/file/JVT_2011_v17n3_Accuracy-of-Plate-Count.pdf
- de Bruin OM, Birnboim HC. A method for assessing efficiency of bacterial cell disruption and DNA release. BMC Microbiol. 2016;16(1):197.
Crossref - Punzi L, Oliviero F. Arthrocentesis and Synovial Fluid Analysis in Clinical Practice. Ann NY Acad Sci. 2009;1154(1):152-158.
Crossref - Singhal R, Perry DC, Khan FN, et al. The use of CRP within a clinical prediction algorithm for the differentiation of septic arthritis and transient synovitis in children. The Journal of Bone and Joint Surgery British. 2011;93(11):1556-1561.
Crossref - Brannan SR, Jerrard DA. Synovial fluid analysis. The Journal of Emergency Medicine. 2006;30(3):331-339.
Crossref - Tamer TM. Hyaluronan and synovial joint: function, distribution and healing. Interdisciplinary Toxicology. 2013;6(3):111-125.
Crossref - Pascual E, Jovaní V. Synovial fluid analysis. Best Practice & Research Clinical Rheumatology. 2005;19(3):371-386.
Crossref - Zueter A, Yean CY, Abumarzouq M, Rahman ZA, Deris ZZ, Harun A. The epidemiology and clinical spectrum of melioidosis in a teaching hospital in a North-Eastern state of Malaysia: a fifteen-year review. BMC Infect Dis. 2016;16(1):333.
Crossref - Pal B, Jones P, Baildam AD. Acute inflammatory (non- purulent) arthritis concomitant with a developing breast abscess. Scand J Rheumatol. 1999;28(2):123- 124.
Crossref - Kishibe S, Okubo Y, Morino S, et al. Pediatric hypervirulent Klebsiella pneumoniae septic arthritis: hv K. pneumoniae septic arthritis. Pediatrics International. 2016;58(5):382-385.
Crossref - Ménard A, Harambat J, Pereyre S, Pontailler JR, Mégraud F, Richer O. First Report of Septic Arthritis Caused by Klebsiella oxytoca. J Clin Microbiol. 2010;48(8):3021-3023.
Crossref - Visca P, Seifert H, Towner KJ. Acinetobacter infection – an emerging threat to human health. IUBMB Life. 2011;63(12):1048-1054.
Crossref - Clifford RJ, Milillo M, Prestwood J, et al. Detection of Bacterial 16S rRNA and Identification of Four Clinically Important Bacteria by Real-Time PCR. PLoS ONE. 2012;7(11):e48558.
Crossref - Mencacci A, Leli C, Cardaccia A, et al. Comparison of conventional culture with SeptiFast real-time PCR for microbial pathogen detection in clinical specimens other than blood. Journal of Medical Microbiology. 2011;60(12):1774-1778.
Crossref - Simon TD, Van Yserloo B, Nelson K, et al. Use of quantitative 16S rRNA PCR to determine bacterial load does not augment conventional cerebrospinal fluid (CSF) cultures among children undergoing treatment for CSF shunt infection. Diagnostic Microbiology and Infectious Disease. 2014;78(2):188-195.
Crossref - Gaibani P, Mariconti M, Bua G, et al. Development of a Broad-Range 23S rDNA Real-Time PCR Assay for the Detection and Quantification of Pathogenic Bacteria in Human Whole Blood and Plasma Specimens. BioMed Research International. 2013;
1-8.
Crossref - Muhla H, Kochemb A-J, Disquea C, et al. Activity and DNA contamination of commercial polymerase chain reaction reagents for the universal 16S rDNA real-time polymerase chain reaction detection of bacterial pathogens in blood. Diagn Microbiol Infect Dis. 2010;66(1):41-9.
Crossref - Silkie SS, Tolcher MP, Nelson KL. Reagent decontamination to eliminate false-positives in Escherichia coli qPCR. Journal of Microbiological Methods. 2008;72(3):275-282.
Crossref - Corless CE, Guiver M, Borrow R, Edwards-Jones V, Kaczmarski EB, Fox AJ. Contamination and Sensitivity Issues with a Real-Time Universal 16S rRNA PCR. J Clin Microbiol. 2000;38(5):1747-1752.
Crossref - Christ-Carin M, Muller B. Procalcitonin in bacterial infections – hype, hope, more or less? Swiss Med Wkly. 2005;135(31-32):451-460.
Crossref - Wang C, Zhong D, Liao Q, Kong L, Liu A, Xiao H. Procalcitonin levels in fresh serum and fresh synovial fluid for the differential diagnosis of knee septic arthritis from rheumatoid arthritis, osteoarthritis and gouty arthritis. Experimental and Therapeutic Medicine. 2014;8(4):1075-1080.
Crossref - Streit G, Alber D, Toubin MM, Toussirot E, Wendling D. Procalcitonin, C-reactive protein, and complement-3a assays in synovial fluid for diagnosing septic arthritis: Preliminary results. Joint Bone Spine. 2008;75(2):238-239.
Crossref
© The Author(s) 2023. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.