ISSN: 0973-7510
E-ISSN: 2581-690X
Klebsiella pneumoniae is one of the most common bacteria among all biofilm-producing as well as the beta-lactamase producing strains, which is responsible for multi-drug resistance. For better therapeutic applications, it is important to detect the biofilm production by Klebsiella pneumoniae and their antibiogram along with the ability to produce ESBL, MBL, and AmpC β-lactamases. The aim of the study was to determine the prevalence of biofilm formation and ESBL, MBL, AmpC β-lactamase phenotypes in K. pneumoniae as well as the antibiogram of all (biofilm and MBL-producing and non-producing) isolates of K. pneumoniae. Isolates of K. pneumoniae were tested for biofilm formation by the Congo-red agar method. ESBL, MBL, and AmpC β-lactamase detection were done by both screening and confirmatory tests as per CLSI guidelines. The antibiogram was obtained by the Kirby-Bauer disc diffusion method. Among the total 100 isolates of K. pneumoniae, 40% were biofilm-producing. Most of them were from urine specimens. Out of biofilm-producing isolates, ESBL – 28%, MBL- 47% and AmpC β-lactamase- 25.8% producers were observed. K. pneumoniae isolates were seen to have maximum resistance to ceftazidime and maximum sensitivity to nitrofurantoin. Study findings suggest the importance of assessment of biofilm formation for better treatment. The scenario further worsens if such biofilm-producing isolates are also MBL-positive leading to limited therapeutic options.
Biofilm Formation, Extended Spectrum Beta-lactamases, Metallo-Beta-Lactamases, Ampicillinase Cephalosporinase
A biofilm is an organized collection of microorganisms which live embedded within a self-generated matrix of extracellular polymeric substances (EPS). This biofilm is usually attached to a biotic or abiotic surface.1 The biofilm protects microorganisms from adverse effects of extreme environmental conditions. Studies have shown that biofilms can make microorganisms resistance to increase against UV radiation,2 extreme pH and temperature, high salinity, poor nutrients, and importantly to various antibiotics.3 Among Gram-negative pathogens Klebsiella pneumoniae is one of the leading bacteria responsible for biofilm formation.
Klebsiella pneumoniae possesses virulence attributes, encompassing capsule polysaccharide, lipopolysaccharide, outer membrane proteins, type 1 and type 3 fimbriae as well as factors that govern iron acquisition and nitrogen source utilization.4 These virulence elements play a critical role in enabling the pathogen to evade the host’s immune system and in facilitating the formation of robust biofilms.5 Pili along with lipopolysaccharides of biofilm-forming isolates increases the bacterial pathogenicity.6 K. pneumoniae are also known to cause biofilm-mediated infections in most hospitalized patients.4 Biofilms formed by K. pneumoniae, are bacterial communities enclosed within an extracellular matrix composed of proteins, DNA, exopolysaccharides, and lipopeptides.7 This biofilm holds the bacterial attachment to living or non-living surfaces. Also, biofilm protects the aggregate of K. pneumoniae in the biofilm from antimicrobial penetration and reduces its effects.8
K. pneumoniae has demonstrated a heightened ability to acquire resistance to antimicrobial agents compared to many other bacteria, primarily through the production of enzymes like Extended Spectrum β-Lactamase (ESBLs) and Carbapenemases.9 Furthermore, it can rapidly transfer this resistance mechanism to other bacteria. The relation between biofilm-forming and beta-lactamase-producing strains is going the extensive range to the dissemination of multi-drug resistant mechanism.10
Antimicrobial resistance (AMR) has emerged as a critical worldwide health challenge. Limited information is available regarding the relationship between biofilm formation and antimicrobial resistance in clinical isolates of K. pneumoniae. The increase in the rate of antimicrobial resistance in Enterobacterales has become a serious issue.11 Drug resistance is responsible for narrowed therapeutic options because the pathogens have developed resistance mechanisms against the commonly used antimicrobial groups like extended-spectrum beta-lactams, aminoglycosides, fluoroquinolones, and carbapenems, etc.12 ESBL-producing bacteria are not only resistant to penicillin group but also other antibiotics classes like cephalosporins, aztreonam, aminoglycosides, and fluoroquinolones.13 Carbapenems are the drug of choice in the various clinical isolates.
This research aimed to bridge the knowledge gap regarding the effect and correlation between biofilm formation in K. pneumoniae and antimicrobial resistance. Besides the prevalence of biofilm formation among K. pneumoniae clinical isolates, another objective was to study the antibiogram of Biofilm and MBL-producing and nonproducing K. pneumoniae.
A total of 100 non-repetitive K. pneumoniae isolates from clinical specimens of patients of all ages and genders admitted to Krishna Hospital and Medical Research Centre in Karad, India, were included in the study. The specimen processing and identification was done on the basis of standard methodology.14 The project was approved by the Institutional Ethics Committee and the informed consent was taken from all enrolled patients before collecting the specimen.
Biofilm detection by Congo red agar method
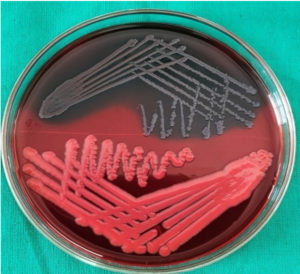
Congo red agar media was prepared by mixing brain heart infusion broth, sucrose, Congo red dye, and agar in distilled water. Isolates were inoculated on Congo red agar media and incubated at 37°C for 18-24 hrs. After incubation, the black-coloured colonies with metallic dry consistency were considered as biofilm producers and red-coloured colonies as non-producer (Figure 1).15
Antibiotic sensitivity profile
After the isolation of pure culture, organisms were tested for antimicrobial susceptibility test, which was performed by the Kirby-Bauer disc diffusion method as per the CLSI guidelines.16 For this, antibiotic discs included were nitrofurantoin (300 µg), imipenem (10 µg), ceftazidime (30 µg), cefoxitin (30 µg), ciprofloxacin (5 µg), cefotaxime (30 µg), gentamicin (10 µg), amikacin (30 µg), piperacillin/tazobactam (100/10 µg), fosfomycin (200 µg), co-trimoxazole (1.25/23.75 µg), meropenem (10 µg), tigecycline (15 µg). The zone of inhibition was interpreted as per the CLSI (2021) standards.
Phenotypic detection of drug resistance
The isolates were further screened for phenotypic detection of enzymes like ESBL, MBL, and AmpC β-lactamase. For the screening of ESBL production ceftazidime and ceftazidime plus clavulanic acid discs were used and a difference of 5mm zone size between the two was considered significant.17 MBL production was detected with imipenem and imipenem EDTA discs with respect to 7 mm inhibition zone size18 difference and for the AmpC β-lactamase detection ≥4 mm inhibition zone difference was considered significant between cefoxitin and cefoxitin plus cloxacillin discs.19
Among 100 isolates of K. pneumoniae maximum biofilm-producing isolates were from urine 12 (12%) followed by ETT 9 (9%), pus 7 (7%), blood 8 (8%), and least in sputum 3 (3%) and CSF 1 (1%) (Table 1).
Table (1):
Distribution of biofilm-producing and non-producing K. pneumoniae among various clinical specimens
| Clinical Specimens | Biofilm producers | Biofilm non-producers | ||
|---|---|---|---|---|
| Number (n) | % | Number (n) | % | |
| Urine | 12 | 12 | 13 | 13 |
| ETT | 9 | 9 | 13 | 13 |
| Pus | 7 | 7 | 15 | 15 |
| Blood | 8 | 8 | 10 | 10 |
| Sputum | 3 | 3 | 5 | 5 |
| CVP tip | 0 | 0 | 2 | 2 |
| Ascitic fluid | 0 | 0 | 1 | 1 |
| CSF | 1 | 1 | 0 | 0 |
| Pleural fluid | 0 | 0 | 1 | 1 |
| Total | 40 | 40 | 60 | 60 |
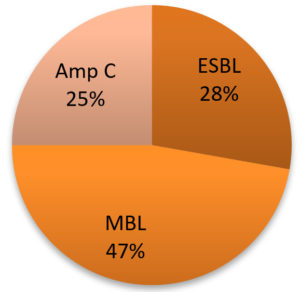
Among the biofilm-producing strains of K. pneumoniae, 25% were AmpC β-lactamase-producing, 28% were ESBL-producing and 47% were MBL-producing isolates (Figure 2).
When the biofilm-producing strains of Klebsiella pneumoniae were tested against antimicrobial agents, they showed maximum sensitivity to nitrofurantoin 23 (57.5%), meropenem 22 (55%) and resistance to ceftazidime 36 (90%), ciprofloxacin, cefoxitin, tigecycline 31 (77.5%) each (Table 2).
Table (2):
Antimicrobial profile of biofilm producing strains of K. pneumoniae
Antibiotics |
Sensitive |
Sensitive % |
Resistant |
Resistant % |
|---|---|---|---|---|
Ceftazidime |
4 |
10 |
36 |
90 |
Imipenem |
10 |
25 |
30 |
75 |
Ciprofloxacin |
9 |
22.5 |
31 |
77.5 |
Nitrofurantoin |
23 |
57.5 |
17 |
42.5 |
Meropenem |
22 |
55 |
18 |
45 |
Cefotaxime |
12 |
3 |
28 |
70 |
Cefoxitin |
9 |
22.5 |
31 |
77.5 |
Co-trimoxazole |
17 |
42.5 |
23 |
57.5 |
Gentamicin |
17 |
42.5 |
23 |
57.5 |
Piperacillin/ Tazobactam |
10 |
25 |
30 |
75 |
Amikacin |
14 |
35 |
26 |
65 |
Tigecycline |
9 |
22.5 |
31 |
77.5 |
Fosfomycin |
21 |
52.5 |
19 |
47.5 |
Antimicrobial sensitivity pattern of Biofilm and MBL-producing K. pneumoniae isolates showed maximum sensitivity to nitrofurantoin 10 (58.82%), fosfomycin 9 (52.94%) and resistance to ceftazidime, 16 (94.11%) (Table 3).
Table (3):
Antimicrobial profile of biofilm and MBL-producing strains
Antibiotics |
Sensitive |
Sensitive % |
Resistant |
Resistant % |
|---|---|---|---|---|
Ceftazidime |
1 |
5.88 |
16 |
94.11 |
Ciprofloxacin |
3 |
17.64 |
14 |
82.35 |
Nitrofurantoin |
10 |
58.82 |
7 |
41.17 |
Cefotaxime |
4 |
23.52 |
13 |
76.47 |
Cefoxitin |
2 |
11.76 |
15 |
88.23 |
Co-Trimoxazole |
4 |
23.52 |
13 |
76.47 |
Gentamicin |
7 |
41.17 |
10 |
58.82 |
Piperacillin/Tazobactam |
4 |
23.52 |
13 |
76.47 |
Amikacin |
4 |
23.52 |
13 |
76.47 |
Tigecycline |
4 |
23.52 |
13 |
76.47 |
Fosfomycin |
9 |
52.94 |
8 |
47.05 |
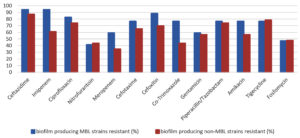
When biofilm and MBL-producing strains were analyzed, their antimicrobial pattern showed maximum resistance to ceftazidime (94.11%) and imipenem (94.11%). The biofilm-producing non-MBL isolates showed resistance to ceftazidime (86.95%) and imipenem (60.86%). The MBL-positive isolates were comparatively more resistant than MBL-negative isolates among the biofilm-producing Klebisella pneumoniae isolates (Figure 3).
K. pneumoniae isolation in patients can raise serious concern because globally it is one of the most important causes of MDR infections.19 Also in the case of HAIs outbreaks or longer stays in the hospitals due to MDR; it results in inflated healthcare costs.20
In this study, we investigated the biofilm-producing capabilities of Klebsiella pneumoniae isolated from a range of clinical specimens. Our findings show some of the K. pneumoniae isolates were biofilm-producing strains. The formation of biofilms by nosocomial opportunistic pathogens like K. pneumoniae on host-tissue surfaces represents a pivotal step in the progression of infection. In this study, 40% of isolates were biofilm producers. Similar findings are reported by Madhavi S. Hullur et al.21 stated that 54% K. pneumoniae isolates were identified as biofilm producers. In another study, Rakesh Prasad Sah et al.22 reported that 57.4% K. pneumoniae strains were biofilm producers. Among biofilm-producing strains of K. pneumoniae the higher reported findings were 77.55% by Rabina Dumaru et al.10 The variation in biofilm-forming ability among each isolate can be attributed to a multitude of influencing factors, including the physiochemical properties of K. pneumoniae, the nature of the surface to which the biofilm adheres as well as the environmental factors like temperature and pH.23 The findings in other studies were 60% by Gedif Meseret Abebe et al.,24 65.46% by Roshani Nhuchhen Pradhan et al.25 etc. The occurrence of the lowest biofilm-producing strains of K. pneumoniae was reported as 24.3% by Sabina Fatima et al.26
Because of the ability to produce β- lactamase enzymes and biofilms, Klebsiella spp. is resistant to a wide range of antibiotics,27 which is also observed in our study. Among the 40 biofilm-producing K. pneumoniae isolates 28% were ESBL producers, 47% MBL producers and 25% AmpC β-lactamases producers. These findings are slightly comparable with Susmita Kuinkel et al.27 where they observed 17.9% ESBL producers, 33.3% MBL producers, and 20.5% AmpC β-lactamases producers among biofilm-positive strains. In the study of Rabina Dumaru et al.10 30.61% were ESBL and 26.53% were MBL producers,10 respectively among biofilm producers.
Comparatively slightly higher prevalence of ESBL, MBL and AmpC β-lactamases production in biofilm-producing K. pneumoniae in present study is justifiable as half (49.05%) of these biofilm producers were from various ICUs where patients usually have an invasive equipment for bacteria to form biofilm on.
Biofilm-producing strains have the tendency of ESBL, MBL, and AmpC β-lactamases production which is responsible for multi-drug resistance factors worldwide.28
Overall, 40% of isolates were detected as biofilm producers in our study. When these biofilm-producing strains were further got tested with antimicrobials, they showed multiple antibiotic resistance patterns. In the present study, maximum isolates were resistant to cephalosporin -ceftazidime (90%), carbapenem – imipenem (75%), and fluoroquinolone- ciprofloxacin (77.5%). On the other hand, Hera Nirwati et al.8 found in their study that most of the biofilm-producer isolates were resistant to cefazolin (65.29%), cefuroxime (58.74%) and ceftriaxone (56.64%) respectively. Klebsiella with resistance of ceftazidime (66.5%), cefepime (59.39%), piperacillin/Tazobactam (57.87%), and ofloxacin (57.87%) were observed in the study of Rabina Dumaru et al.10
Biofilm producing MBL-positive isolates showed more resistance to aminoglycoside, cephalosporins, quinolones, and carbapenem group of antimicrobials compared to MBL negative biofilm producing isolates.
Biofilms are the bacterial cells that have ability to self-produce a polymeric matrix which is capable to adhering a layer on living or non-living surface. The ability of microorganisms to produce biofilm leads to high prevalence of multidrug resistance which in turn leads to treatment failure. Hence, it is crucial to emphasize both antimicrobial therapy and anti-biofilm strategies, as they collectively offer the most effective treatment options. The resistance pattern in biofilm producing isolates is further aggravated if the isolates are Metallo-Beta-Lactamase producing strains.
In the present study, we observed high prevalence (40%) of biofilm formation among Klebsiella pneumoniae isolates. Most of the biofilm producers were from urine and catheter tips. Among these biofilm producing strains prevalence of ESBL, MBL and AmpC β-lactamases. β-lactamases was seen in 28%, 47%, and 25% of the isolates, respectively. K. pneumoniae as well as biofilm producing K. pneumoniae strains had higher resistance to ceftazidime and higher sensitivity to nitrofurantoin antibiotic.
Significant proportions of the study isolates (40%) were positive for biofilm formation suggesting importance of biofilm assessment routinely. The scenario further worsens if such biofilm producing isolates are also MBL-positive making limited therapeutic options. The study suggests that it is very important to screen biofilm production and phenotype detection in urinary and catheter related Klebsiella pneumoniae infections.
ACKNOWLEDGMENTS
The authors are grateful to the Krishna Hospital and Medical Research Center and the Department of Microbiology, Krishna Institute of Medical Sciences, KVV DU, Karad, for providing the necessary research facilities to carry out the project.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
The study was approved by the Institutional Ethics Committee, Krishna Institute of Medical Sciences, KVV DU, Karad, with Protocol number 051/2021-2022.
INFORMED CONSENT
Written informed consent was obtained from the participants before enrolling in the study.
- Yin W, Wang Y, Liu L, He J. Biofilms: the microbial “protective clothing” in extreme environments. Int J Mol Sci. 2019;20(14):3423.
Crossref - de Carvalho CC. Biofilms: Microbial strategies for surviving UV exposure. Adv Exp Med Biol. 2017:996:233-239.
Crossref - Hathroubi S, Mekni MA, Domenico P, Nguyen D, Jacques M. Biofilms: microbial shelters against antibiotics. Microbial Drug Resist. 2017;23(2):147-156.
Crossref - Ragupathi NK, Sethuvel DP, Dwarakanathan HT, et al. The influence of biofilm formation on carbapenemresistance in clinical Klebsiella pneumoniae infections: phenotype vs genome-wide analysis. bioRxiv. 2020.
Crossref - Murphy CN, Clegg S. Klebsiella pneumoniae and type 3 fimbriae: nosocomial infection, regulation and biofilm formation. Future Microbiol. 2012;7(8):991-1002.
Crossref - Vuotto C, Longo F, Balice MP, Donelli G, Varaldo PE. Antibiotic Resistance Related to Biofilm Formation in Klebsiella pneumoniae. Pathogens. 2014;3(3):743-758.
Crossref - Donlan RM. Biofilms: microbial life on surfaces. Emerg Infect Dis. 2002;8(9):881.
Crossref - Nirwati H, Sinanjung K, Fahrunissa F, et al. Biofilm formation and antibiotic resistance of Klebsiella pneumoniae isolated from clinical samples in a tertiary care hospital, Klaten, Indonesia. BMC Proc. 2019;13(Suppl 11):20.
Crossref - Podschun R, Ullmann U. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev. 1998;11(4):589-603.
Crossref - Dumaru R, Baral R, Shrestha LB. Study of biofilm formation and antibiotic resistance pattern of gram-negative Bacilli among the clinical isolates at BPKIHS, Dharan. BMC Res Notes. 2019;12(1):1-6.
Crossref - Gniadkowski M. Evolution and epidemiology of extended-spectrumbeta-lactamases (ESBLs) and ESBL-producing microorganisms. Clin Microbiol Infect. 2001;7(11):597-608.
Crossref - Namratha KG, Sreeshma P, Subbannayya K, Dinesh PV, Champa H. Characterization and antibiogram of Klebsiella spp. isolated from clinical specimen in a rural teaching hospital. Sch J App Med Sci. 2015;3(2E):878-883.
- Nepal K, Pant ND, Neupane B, et al. Extended spectrum beta-lactamase and metallo beta-lactamase production among Escherichia coli and Klebsiella pneumoniae isolated from different clinical samples in a tertiary care hospital in Kathmandu, Nepal. Ann Clin Microbiol Antimicrob. 2017;16(1):62.
Crossref - Mackie TJ, collee J.G. McCartney. Mackie & McCartney practical medical microbiology 14th edition. New York: Churchill Livingstone. 2006.
- Rajmane A, Joshi PA, Shikhare V, et al. A study of biofilm production and antimicrobial susceptibility pattern among urinary isolates. Indian J Microbiol Res. 2021;8(4):268-273.
Crossref - Clinical and Laboratory standard institute performance standard for Antimicrobial Susceptibility testing, CLSI document M100-E31.E2021
- Segar LA, Kumar SH, Joseph NM, Sivaraman UM. Prevalence of extended spectrum beta-lactamases among Enterobacteriaceae and their anti-biogram pattern from various clinical samples. Asian J Pharma Clin. Res. 2015;8(5).
- Ali FS, Authman SH, Al Marjani MF. Imipenem Resistance and Biofilm Formation In Klebsiella pneumoniae from some Hospitals in Baghdad City. J Pharm Sci Res. 2019;11(1):233-238.
- Singh P. The incidence of AmpC β-lactamases producing Klebsiella pneumoniae subspecies pneumoniae. International Journal of Research in Medical Sciences. 2018;6(4):1169.
- Kidd TJ, Mills G, Sa-Pessoa J, et al. A Klebsiella pneumoniae antibiotic resistance mechanism that subdues host defences and promotes virulence. EMBO Mol Med. 2017;9(4):430-447.
Crossref - Hullur MS, Natarajan A, Sreeramulu P. Phenotypic Characterization of Virulence Factors and Antibiogram of Klebsiella pneumoniae Isolates from Various Clinical Samples – A Cross Sectional Study. J Pure Appl Microbiol. 2022;16(3):1783-1791.
Crossref - Sah RP. Phenotypic and genotypic study of klebsiella species with special reference to virulence factors and antimicrobial resistance pattern.
Crossref - Abebe GM. Detection of Biofilm Formation and Antibiotic Resistance in KlebsiellaOxytoca and Klebsiella pneumoniae from Animal Origin Foods. Int J Microbiol Biotechnol. 2020;5(3):120-130.
Crossref - Pradhan RN, Madhup SK, Pant SP. Antibiogram and Biofilm Formation Among Carbapenem Resistant Klebsiella pneumoniae. Tribhuvan University Journal of Microbiology. 2019;6:63-69.
Crossref - Fatima S, Prasanthi K, Nagamani K. Comparative evaluation of biofilm production in multidrug resistant and sensitive Gram negative clinical isolates. Int J Curr Microbiol App Sci. 2015;4(6):918-926.
- Kuinkel S, Acharya J, Dhungel B, et al. Biofilm formation and phenotypic detection of ESBL, MBL, KPC and AmpC β-lactamases enzymes and their coexistence in Klebsiella spp. isolated at the National Reference Laboratory, Kathmandu, Nepal. Microbiol Res. 2021;12(3):683-697.
Crossref - Shahriar A, Faijanur-Rob-Siddiquee M, Ahmed H, Mahmud AR. Comparative and Correlative Analysis of Biofilm Formation and Antimicrobial Resistance Traits towards Extended Spectrum β-Lactamase (ESBL) and Metallo-β-Lactamase (MBL) Producing Pathogenic Bacteria among the Clinical Isolates. J Microbiol Immunol. 2021;3(5):001-009.
© The Author(s) 2024. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.