ISSN: 0973-7510
E-ISSN: 2581-690X
This study aimed to investigate indigenous hydrocarbon-tolerant bacteria from a coastal environment and evaluate the bioremediation potential. Water samples were collected from high-risk oil spill areas and mixed with motor oil to find hydrocarbon-tolerant bacteria with potential uses in bioremediation. Two prospective bacterial isolates were selected for inhibition zone test, biochemical analysis, enzymatic assay, and 16S rDNA gene sequencing. The isolates were identified as Cytobacillus kochii (PQ500563) and Bacillus haikouensis (PQ395181). Phylogenetic trees constructed using the Neighbor-Joining (NJ) method confirmed the taxonomic placement with high similarity to known strains. The results showed that C. kochii degraded hydrocarbons, with an inhibition zone of 10 ± 0.55 mm, while B. haikouensis had an inhibition zone of 8 ± 1.05 mm. C. kochii had the highest dehydrogenase activity of 20.67 ± 0.22 × 10-2 μmol/min/ml, suggesting that the bacteria was very good at breaking down hydrocarbons and forming biomass. The isolate B. haikouensis had the highest catechol 2,3-dioxygenase activity of 75.90 ± 0.14 × 10-2 μmol/min/ ml, which signified being very good at breaking down aromatic chemicals. The combination of C. kochii and B. haikouensis as the consortium had a synergistic enhancement, providing significantly higher activities for key catabolic enzymes (dehydrogenase, catechol 2,3-dioxygenase, and catalase) along with increased biomass production compared to the individual pure cultures. This showed the superior efficacy of the consortium, which was driven by complementary enzymatic strengths for degrading diverse hydrocarbons in marine oil spill contexts.
Hydrocarbon-tolerant Bacteria, Dehydrogenase Enzyme, Dioxygenase Enzyme, Catabolic Enzyme, Catalase Enzyme, Cytobacillus kochii, Bacillus haikouensis
Beaches and ports worldwide are currently facing a continuous increase in critical environmental challenges that originate from substantial marine activity. Factors contributing to this deterioration include marine pollution, air and water contamination, insufficient waste management, and the unregulated application of synthetic chemicals, all of which profoundly affect coastal ecosystems.1 Maritime operations directly contribute to pollution, with oil spills, ballast water discharge, and tank cleaning procedures being substantial factors. Oil leaks transpire during cargo management, corrosion mitigation, and incidents in the form of hull fractures or collisions,2,3 as well as in shallow seas when vessels are at full capacity. Floating oil spills can damage marine ecosystems by obstructing sunlight, hindering gas exchange, and modifying water temperature.4 These alterations adversely affect plankton, the fundamental creatures in the aquatic food chain, leading to disruption of the entire ecosystem.5 All aquatic organisms often experience physical asphyxiation because of oil spills. Moreover, human health may deteriorate from the consumption of contaminated seafood and the exposure of individuals in proximity to dirty seas to toxic substances, potentially leading to numerous health complications.6 Over 1.3 million tons of petroleum hydrocarbons yearly are predicted to reach the maritime environment from both human and natural sources. Nearly 0.6 million tons of hydrocarbons from petroleum originated from natural seeps, sufficient to envelop all the world’s seas with a thickness of 20 molecules. Consequently, petroleum hydrocarbon contamination presents a significant risk to marine ecosystems, and is important to devise environmentally sustainable technology for the remediation of oil pollution in marine ecosystems.
Spill remediation methods have advanced due to the considerable ecological threats presented by oil pollution. These strategies are classified into four distinct parts, namely (1) Physical methods include the mechanical extraction of oil from the sea surface using obstacles such as booms, skimmers, and absorbents.7 Although useful in tranquil waters, these procedures lose efficiency in turbulent seas or when the oil has spread over an extensive region.8 (2) Chemical methods comprise the application of chemical dispersants to decompose oil slicks, facilitating the integration of oil into the water column. The dispersant compounds help mitigate surface pollution by distributing hydrocarbons but present considerable toxicity hazards to marine environments, affecting aquatic animals and the wider food web.9 (3) Thermal methods (in situ burning) require igniting and combusting oil on the surface, providing a rapid reaction for spill management. In situ burning emits pollutants into the environment despite being economical and expedient, increasing air pollution and posing possible risks to human health.7,8 Bioremediation is sustainable and environmentally benign process that uses microorganisms, specifically hydrocarbon-degrading bacteria, for converting hazardous oil molecules into less dangerous substances such as carbon dioxide and water.10 The method of bioremediation efficiently deconstructs complex hydrocarbons and minimizes long-term environmental impacts, serving as a cost-effective and environmentally sustainable alternative for oil spill remediation.11
Microorganisms are essential in the biodegradation of hydrocarbons, with specific bacterial species using hydrocarbons as principal sources of carbon and energy.12 A variety of hydrocarbon-degrading bacteria, including Pseudomonas, Rhodococcus, and Marinobacter, have been recognized and used in bioremediation initiatives.13 Despite significant progress in identifying hydrocarbon-degrading bacteria, the complexity of hydrocarbon molecules requires specialized enzymatic activities for effective degradation. Therefore, bacterial strains with unique or enhanced enzymatic pathways need to be identified to optimize bioremediation processes.
Enzymatic activity tests are essential for comprehending the metabolic pathways used by hydrocarbon-degrading bacteria. Essential enzymes including dehydrogenase and catechol dioxygenase are crucial in the breakdown process transforming complex hydrocarbons into simpler, less harmful substances.14 Dehydrogenase catalyzes the oxidation of hydrocarbons, while catechol dioxygenase cleaves aromatic rings.15-17 Assessing the activity of these enzymes may help to more accurately evaluate the breakdown potential of bacterial strains and the capacity to endure extreme environmental conditions. This comprehension is essential for selecting the appropriate strains for extensive bioremediation applications.
Considering the limitations, there is a growing necessity to discover indigenous, hydrocarbon-tolerant bacteria for application in marine bioremediation, particularly in areas with elevated oil spill hazards. Indigenous microorganisms are naturally adaptable to local environmental challenges and capable of initiating effective hydrocarbon breakdown under natural conditions.18 Advancements in molecular methodologies, particularly 16S rDNA sequencing, have transformed the identification and characterization of bacteria, thereby enabling accurate species determination and phylogenetic analysis.19 These methods establish a robust basis for developing customized microbial consortia intended for scalable bioremediation initiatives.
This study aimed to isolate and identify indigenous hydrocarbon-degrading bacteria from polluted maritime habitats by 16S rDNA sequencing. The breakdown capacities of the indigenous strains were assessed through enzymatic tests focusing on key enzymes, such as dehydrogenase and dioxygenase, supplemented by the clear zone diffusion method and catalase. Additionally, insights would be provided into the development of effective environmentally sustainable methods for the remediation of hydrocarbon-contaminated marine ecosystems by assessing enzymatic activities and bioremediation capacities.
Sample collections
The sampling area was located in the Java Sea coastal waters in the Tanjung Emas port, Java, Indonesia, with coordinates of (06°57’18.51″ S, 110°25’23.12″ E). This location was selected due to the documented history of intense maritime activity near the ship refueling area, and consequently, a high potential for chronic hydrocarbon exposure, leading to being an ideal environment for isolating indigenous hydrocarbon-tolerant bacteria. At the designated area, triplicate seawater samples were collected using a sterile, wide-mouthed 500 mL bottle containing a secure cap, with each replicate obtained from a depth of 10 cm below the surface. Aseptic methods were rigorously used throughout the collection process to avoid external bacterial contamination. The collected samples were immediately kept in a cool box maintained at 4 °C and transferred to the microbiology laboratory in a suitable transport medium to preserve the integrity and prevent potential deterioration during transit.20
Bacteria isolation
The isolation process started with the serial dilution of a 1 ml marine water sample, which was mixed with 9 mL of saline solution (0.85% NaCl) in a small test tube. The mixture was vortexed, and a serial dilution was performed by adding 1 mL of the solution to 9 mL of the 0.85% NaCl solution. This process was repeated to prepare a dilution series from 10-1 to 10-8, then 0.1 mL of the last three dilutions were positioned on Petri dishes containing Zobel media agar, evenly spread using a spatula, and incubated at room temperature of 27-28 °C.21 Isolated bacteria were purified using Zobell marine agar by selecting colonies based on distinct shapes and colors. Subsequently, single colonies were purified through the streak plate method until consistent characteristics were achieved.22
Screening of hydrocarbon-tolerant bacteria
Screening for hydrocarbon tolerance was conducted by plating purified bacterial isolates on Zobell Marine Agar (HiMedia) supplemented with 1%-2% and 20% used engine oil. The screening process was performed to evaluate bacterial growth in the presence of hydrocarbons.
Hydrocarbon degradation potential test
The degradation test for hydrocarbon compounds was conducted semi-quantitatively by measuring clear zones formed. Each isolate was inoculated by depositing one loopful on Zobell marine agar using the spread plate method. A paper disc soaked in used engine oil was positioned in the center of the medium where the isolate was inoculated. The Petri dishes were incubated at room temperature for 24 hours, and then clear zones formed around the paper disc were measured using a caliper.
Characterization of bacterial isolates
The isolates were thoroughly characterized by examining the colonies based on the morphological and biochemical characteristics. This followed the procedure applied in a previous study23 performed according to Bergey’s Manual of Systematic Bacteriology.
Colony characteristics
The selected bacterial isolates were inoculated onto Zobell marine agar plates using the pour plate method and incubated at 30 °C for 24 hours to facilitate growth. Afterward, the colonies were carefully examined for various morphological traits, including color, shape, surface texture, margin, and opacity.
Motility test
Agar slants of the preserved isolates were collected and inoculated onto a cover glass using an inoculating needle. The cover glass was subsequently positioned on a concavity slide and observed under a 45x objective lens to determine the presence or absence of motile rods.24
Gram staining
After preparation of a 24-hour-old bacterial culture, a smear was applied to a glass slide and heat-fixed to adhere the cells. The slide was treated with crystal violet (HiMedia) for 2 minutes, then rinsed gently with running water. Subsequently, Gram iodine solution was applied for 2 minutes to form a complex with the crystal violet. The slide was washed with alcohol to decolorize it and then counterstained with safranin (HiMedia). After a final rinse with water, the slide was dried and examined under oil immersion.25
Endospore staining
The endospore staining procedure started with heat-fixing a bacterial smear onto a microscopic slide. Malachite green (HiMedia) was applied to the slide to specifically stain the endospores and the slide was positioned in a hot beaker for 5 minutes to facilitate the staining process. Safranin was added to counterstain the vegetative cells, then the slide was rinsed with water, blotted dry, and examined under a microscope using the oil immersion objective.26
Oxygen tolerance
Sterile nutrient agar was prepared in McCartney bottles, which were inoculated with each bacterial isolate using the stab inoculation method while the agar was in a semi-solid state. After allowing the agar to solidify, the bottles were incubated at 37 °C for 48 hours. During this incubation period, anaerobic bacteria were observed to grow at the bottom of the bottles, while aerobic bacteria developed on the surface. Specifically, facultative anaerobes showed growth from the bottom through to the top of the bottles.27
Catalase test
This test was performed using a 3% hydrogen peroxide (HiMedia) solution to evaluate the presence of catalase enzyme activity in the bacterial isolates. An overnight culture was applied to a microscopic slide, and mixed with the hydrogen peroxide using a sterilized loop. The immediate formation of bubbles both during and following the mixing process was recorded as a positive indication of catalase activity.27
Citrate utilization
Preparation and sterilization of Simmons citrate agar (HiMedia) were performed, followed by stab inoculation of all isolates into the medium and incubation at 37 °C for 18 to 24 hours. A positive result was detected by the growth of the organism on the surface of the slant and a color change from the original green to blue.28
Indole production test
Pure culture of the bacterial isolate was inoculated into a tube containing Tryptone Broth (HiMedia) and incubated at 37 °C for 48 hours. Subsequently, five drops of Kovac’s indole reagent (HiMedia) were added directly to cultured broth. A positive indole test was identified by the appearance of a red color on the top of the tube, while a negative result was determined by a yellow coloration on top of the reagent layer.29
Urea hydrolysis test
Urea agar medium was prepared and inoculated with isolated bacterial strains, then both the test and control tubes were incubated at 37 °C to allow for microbial activity. Color changes on the slants were monitored every 6 hours and daily for up to 5 days. Phenol red (HiMedia) was used as a pH indicator to detect urease activity, where a color change in the medium represented successful urea hydrolysis.25
Carbohydrate fermentation test
This test was carried out by inoculating a loopful of nutrient broth (HiMedia) culture from the test organisms into tubes containing sucrose and maltose. The inoculated tubes were incubated at 37 °C for 24 hours to allow for fermentation. Acid production was signified by a color change in the medium from reddish to yellow, while the presence of gas was shown by the formation of bubbles in the inverted Durham tubes, reflecting the metabolic activity of the organisms.30
Methyl red test
The test was conducted by inoculating a fresh bacterial culture into 5 ml of MR-VP broth (HiMedia) and incubating at 35 °C for 48 hours. After incubation, 2.5 ml of culture was transferred to a sterile tube, and five drops of Methyl Red (MR) reagent (HiMedia) were added. A positive result for the Methyl Red test was detected by the development of red coloration in the medium, signifying the production of stable acids, while a negative result was marked by a yellow coloration suggesting low acid production.31
Voges-Proskauer test
The Voges-Proskauer test was conducted using glucose phosphate broth (HiMedia), which was sterilized by autoclaving and subsequently cooled to room temperature. The 24-hour-old cultures of the selected bacterial isolates were inoculated into the broth using a sterile loop, and the tubes were incubated at 30 °C for 48 hours. Afterward, 1 ml of alpha-naphthol (HiMedia) was added to the broth and mixed thoroughly. This was followed by the addition of 0.5 ml of 40% KOH, and the mixture was shaken again. The development of a red coloration within 1 hour after reagent addition was interpreted as a positive result, representing the production of acetoin and the capacity of the organism for butylene glycol fermentation.32
Quantification and production of enzyme
Inoculum for enzyme assay microbial cultures was prepared in 250 mL sterile Erlenmeyer flasks, each containing 100 mL of Zobell Marine Broth (ZMB) (HiMedia). Each flask was supplemented with 200 ppm of anthracene as hydrocarbon substrate to simulate a contaminated environment, then aseptically inoculated with 5 mL of bacterial inoculum adjusted to an optical density (OD600) of 1.0. Cultivation was performed under aerobic conditions using a shaker incubator set at 100 rpm at room temperature (~25 °C) for 4 days. After the incubation period, microbial biomass and catabolic enzyme activity were observed.
Enzyme extraction
Cells were harvested from each culture flask to isolate enzymes included in hydrocarbon degradation. A 10 mL aliquot of bacterial culture was aseptically withdrawn from each flask and centrifuged at 5000 rpm for 10 minutes to pellet the cells. The supernatant was discarded, and the bacterial pellets were resuspended in 1 mL of 20 mM Tris-HCl buffer (HiMedia) (pH 7.4). The suspended cells were subjected to ultrasonication using an ultrasonic disintegrator at a frequency of 40 kHz for 10 minutes while maintaining the temperature at 10 °C using an ice bath to prevent protein denaturation. This step was performed to disrupt the bacterial cells and release intracellular enzymes. Following sonication, the cell lysates were centrifuged at 8000 rpm for 15 minutes at 4 °C to remove cell debris. The supernatant containing the crude enzyme extract was used immediately for subsequent enzyme activity assays. All steps were carried out under aseptic conditions to prevent contamination and ensure enzyme stability.
Enzyme activity assay
The enzymatic activities present in the supernatant were analyzed using a UV-Vis spectrophotometer. Specifically, the activity of catechol 2,3-dioxygenase (C23O) was measured at an absorbance of 260 nm using catechol (HiMedia) as the substrate. The assay mixture consisting of 2.5 mL of 0.1 M potassium phosphate buffer (HiMedia) (pH 7.5), 0.1 mL of 0.01 M catechol, and 0.1 mL of crude enzyme extract was added to a quartz cuvette with a path length of 1 cm. The enzymatic reaction was monitored by measuring the increase in absorbance at 260 nm which corresponded to the formation of cis-cis muconic acid. To assess the activity of dehydrogenase enzyme, 0.1 mL of enzyme extract was transferred to test tubes containing 2.5 mL of Tris buffer and 1 mL of a triphenyl tetrazolium chloride (TTC) (HiMedia) solution. The tubes were incubated on a shaker at room temperature for 1 hour to allow the enzymatic reaction to occur. After incubation, the reaction product, triphenyl formazan (TPF), was extracted using methanol. The concentration of TPF was quantified by measuring the absorbance at 484 nm using a UV-Vis spectrophotometer.33,34 For the measurement of catalase activity, a reaction mixture containing 2.7 mL was prepared with 0.1 ml of crude enzyme, 2.5 ml of 0.1 M potassium phosphate buffer (pH 7.0), and 0.1 ml of 0.6 mM hydrogen peroxide (HiMedia) (H2O2) as the substrate. The breakdown of hydrogen peroxide by catalase was monitored at 240 nm, showing the release of oxygen.35 Dehydrogenase, catalase, and catechol 2,3-dioxygenase enzyme activities were determined using the Beer-Lambert law by applying the molar extinction coefficients specific to each product (TPF = 1.8 × 10 tL/(mol·cm), hydrogen peroxide = 43.6 L/(mol·cm), and cis-cis muconic acid from catechol = 1.7 × 10 tL/(mol·cm)).
Molecular identification of the isolate
DNA extraction
The DNA extraction was performed using the guanidine chloride method. Samples were initially washed with 500 µl of PBS and centrifuged, followed by digestion with a solution containing WBC lysis buffer, 6 M guanidine chloride, 7.5 M ammonium acetate, and proteinase K, then incubated overnight at 37 °C. The next day, chilled chloroform was added, and samples were centrifuged, producing three layers. The supernatant was transferred to a new tube, mixed with 1 ml of cold 100% ethanol, and stored at -20 °C overnight. After a quick vortex and another centrifugation, the supernatant was discarded, and a 70% ethanol wash was applied to clean the DNA pellet, which was air-dried, dissolved in 50 µl of purified water, and stored at -20 °C.36
16S rDNA gene amplification
The isolated DNA was amplified for the universal 16S rDNA (250 bp) gene using primers F: 5′-AGAGTTTGATCCTGGCTCAG-3′ and R: 5′-CTACGGCTACCTTGTTACGA-3′. Polymerase chain reaction (PCR) amplification was carried out with the Maxime PCR PreMix Kit (i-Taq) from iNtRON Biotechnology, Korea, and a SensoQuest thermocycler, Germany. Each reaction mixture contained 2.5 units of i-Taq DNA polymerase, deoxynucleoside triphosphates (dNTPs), a 1X PCR reaction buffer, a 1X gel loading buffer, and 1 µl of DNA template. The PCR program started with an initial denaturation phase for three minutes at 94 °C, followed by 40 cycles at 94 °C for 30 seconds, 53 °C for annealing over 30 seconds, and 72 °C for a 45-second extension. After the last cycle, a final extension phase of five minutes at 72 °C completed the reaction, ensuring full strand synthesis.37
Sequencing of bacterial isolate 16S rDNA Gene
The Sanger dideoxy sequencing technology offered by Macrogen Inc. in Korea was used to purify the amplified 16S rDNA gene. This technology was also used to sequence the universal and specific forms of the amplified 16S rDNA gene.
Bioinformatics analysis
Sequence analysis
The nucleotide sequences were visualized and analyzed using Finch TV version 1.4. To identify similarities with other sequences in GenBank, a comparison was carried out using the Basic Local Alignment Search Tool (BLASTn), accessible through the NCBI website (https://blast.ncbi.nlm.nih.gov/). The 16S rDNA gene sequences were deposited in the GenBank database understanding the following accession numbers: Cytobacillus kochii (PQ500563) and Bacillus haikouensis (PQ395181).
Phylogenetic tree construction method
To initiate the analysis, 30 high-similarity nucleotide sequences were retrieved from the NCBI database using BLAST with an e-value threshold set below 0.05. These sequences were aligned through ClustalW2-BioEdit, with Gblocks subsequently used to remove divergent and poorly synchronized areas, ensuring alignment precision. Sequences were downloaded in FASTA format and processed in MEGA 10, where ClustalW was used to enhance alignment quality before phylogenetic analysis. For tree construction, the Jukes-Cantor (JC) model was applied in the Neighbor-Joining (NJ) method to assess the evolutionary relationships of the 16S rRNA and NCBI database sequences, using the Kimura 2-parameter model for calculating sequence distances. To ensure the robustness of the phylogeny, 1,000 bootstrap replicates were conducted, with nodes showing >70% values considered well-supported, and the final bootstrap test confirmed the reliability of tree associations. Alignment parameters including gap opening and extension penalties, the DNA weight matrix, and the IUB matrix were rigorously set. Before progressing, aligned sequences were manually inspected to correct any misalignments or ambiguities, enhancing data integrity for subsequent evolutionary analysis.
Phylogenetic tree visualization
The resulting NJ tree was visualized using MEGA 10, followed by the examination of the topology, including branch lengths and bootstrap values. An appropriate outgroup species was used to root the tree, providing context for the evolutionary relationships. The final tree figures were exported in Newick format and visualized with FigTree to create publication-quality images.
Statistical analysis
All enzyme activity and biomass production assays were performed in triplicate, with data presented as mean ± standard deviation (SD). One-way analysis of variance (ANOVA) was used to compare the mean values for each measured parameter among the three experimental groups, including C. kochii pure culture, B. haikouensis pure culture, and the consortium. When ANOVA showed a significant difference (p <0.05), Tukey’s HSD post-hoc test was performed for pairwise comparisons to identify specific group means that differed significantly. Analyses were conducted using SPSS version 30, and a p-value <0.05 was considered statistically significant.
Sampling area description
Seawater samples were collected from small boats in areas with a high potential for oil spills due to intense marine activity. These were gathered using a horizontal bottle and immediately transferred into pre-sterilized 500 mL dark Schott Duran bottles to prevent light exposure, then stored at 4 °C in a refrigerator to maintain the integrity for analysis.
The combination of continuous ship traffic and the potential for oil spills produced an environment vulnerable to pollution. Consequently, the sampling area served as an ideal location for investigating hydrocarbon-tolerant bacteria. These environments showed a tendency to host bacteria with high hydrocarbon-degrading capabilities, offering valuable insights into the potential for bioremediation.
Hydrocarbon-tolerant bacteria isolation
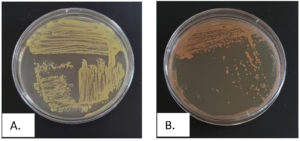
The microbial diversity was investigated by directly plating triplicate marine water samples onto Zobell Marine Agar, which generated an initial 28 distinct bacterial isolates after purification into pure cultures. To specifically isolate hydrocarbon-tolerant bacteria, aliquots from the same triplicate marine water samples passed through serial dilution up to 10-8. Further aliquots from the dilutions were plated onto Zobell Marine Agar supplemented with different used engine oil concentrations of 1%-2% and 20%. This targeted method for hydrocarbon tolerance provided two isolates designated B1 and B2, which showed robust growth under the selective conditions. The characterization and purification results for the two hydrocarbon-tolerant isolates are presented in Figure 1 and Table 1.
Table (1):
Morphological and biochemical characterization of isolates
| Characteristic | Bacteria isolates | |
|---|---|---|
| B 1 | B 2 | |
| Color | Yellow | Orange-red |
| Shape | Rod | Rod |
| Surface | Flat | Convex |
| Edge | Entire | Entire |
| Gram staining | + | + |
| Motility | Yes | Yes |
| Endospore Staining | Positive | Positive |
| Oxygen tolerance | Aerobic | Aerobic |
| Sucrose | – | – |
| Maltose | – | + |
| Indole | – | + |
| Methyl red | + | + |
| Urease | – | + |
| Voges-Proskauer | + | + |
| Citrate utilization | – | + |
| Catalase | + | + |
Figure 1. Streak plate purification results of hydrocarbon-tolerant bacteria, A. Isolate B1, B. Isolate B2
Evaluation of hydrocarbon degradation capacity using the disk diffusion method
The formation of a clear or inhibition zone around a paper disk on agar media serves as a crucial parameter for identifying hydrocarbon degradation ability of each bacterial isolate. The clear zone suggests that the bacteria can use hydrocarbons as a carbon source, an essential characteristic for bioremediation applications. To assess this potential, each bacterial isolate was inoculated onto Zobell marine agar, followed by disk diffusion testing known as a widely accepted microbiological method for evaluating the antimicrobial and biodegradation properties of microorganisms. The disk diffusion method provides a visual representation of hydrocarbon degradation potential, as larger clear zones majorly represent greater enzyme activity and a stronger capacity for hydrocarbon utilization. The assessment presented in Table 2 showed the capability of each isolate to degrade hydrocarbons, offering insight into the potential for use in environmental clean-up efforts.
Table (2):
Hydrocarbon degradation potential test using zone of inhibition methods
No. |
Isolate code |
Diameter (mm) |
|---|---|---|
1 |
B 1 |
10 ± 0.55 |
2 |
B 2 |
8 ± 1.05 |
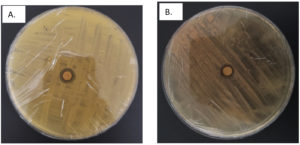
Both isolates B1 and B2 had positive hydrocarbon degradation activity (Figure 2), as shown by the formation of a clear zone. The clear zone of isolate B1 had a wider result, being 2 mm wider than isolate B2 which had a width of 8 mm. This variation in clear zone size directly correlates with the bacterial enzymatic capacity to degrade hydrocarbons. A larger clear zone around isolate B1 suggests a higher concentration of hydrocarbon-degrading enzymes, which enhances the bioremediation potential. This observation is consistent with previous results that clear zones reflect extracellular enzyme activity included in the degradation of complex compounds.38
Figure 2. Hydrocarbon degradation potential assessment results for (A) Isolate B1 and (B) Isolate B2
Evaluation of hydrocarbon degradation potential through catabolic enzyme assay and catalase enzyme
Two bacterial isolates and the consortium were further assessed to quantify biomass and catabolic enzyme activity, specifically dehydrogenase and catechol 2,3-dioxygenase, using spectrophotometry. Biomass and enzyme quantification was carried out through the shake flask method using Zobell marine broth supplemented with anthracene as hydrocarbon substrate, where biomass was measured at OD600 nm. Dehydrogenase, catechol 2,3-dioxygenase, and catalase enzymes were measured at OD 484 nm, 260 nm, and 240 nm with TTC, catechol, and hydrogen peroxide as the substrates, respectively.
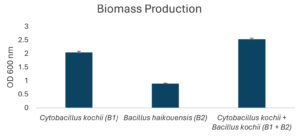
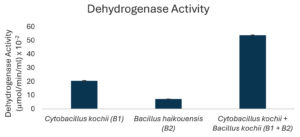
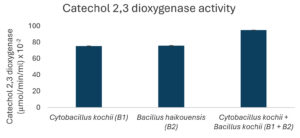
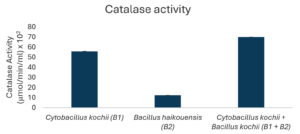
The potential to degrade hydrocarbons by the two bacterial isolates C. kochii and B. haikouensis, along with the consortium was assessed through the quantification of biomass production and key catabolic enzyme activities using anthracene as a model polycyclic aromatic hydrocarbon (PAH). Biomass production (Figure 3), measured at OD600 nm, was highest in the consortium (2.54 ± 0.04), followed by C. kochii (2.05 ± 0.04), with B. haikouensis showing the lowest (0.90 ± 0.02). Dehydrogenase activity (Figure 4) followed this trend, with the consortium presenting markedly higher activity (54.00 ± 0.17 x 10-2 µmol/min/ml) compared to C. kochii (20.67 ± 0.22 x 10-2 µmol/min/ml) and B. haikouensis (7.33 ± 0.12 x 10-2 µmol/min/ml). The two pure cultures showed similar high levels of catechol 2,3-dioxygenase (C23O) activity (Figure 5) (C. kochii: 75.35 ± 0.20 x 10-2 µmol/min/ml; B. haikouensis: 75.90 ± 0.14 x 10-2 µmol/min/ml), but the consortium had the greatest activity (95.10 ± 0.10 x 10-2). Catalase activity (Figure 6) was highest in the consortium (69.98 ± 0.11 x 10² µmol/min/ml), followed by C. kochii (55.75 ± 0.11 x 10² µmol/min/ml), while B. haikouensis had the lowest activity (12.39 ± 0.08 x 10² µmol/min/ml). For all measured parameters, including biomass production, dehydrogenase activity, catechol 2,3-dioxygenase activity, and catalase activity, the one-way ANOVA showed statistically significant differences (p <0.05) among the mean values of the three experimental groups (C. kochii pure culture, B. haikouensis pure culture, and their consortium). Post hoc Tukey’s HSD test showed that the consortium had significantly higher activity than both C. kochii (p <0.05) and B. haikouensis (p <0.05), while the difference between the two pure cultures was statistically significant (p <0.05).
Molecular identification of hydrocarbon-tolerant bacteria
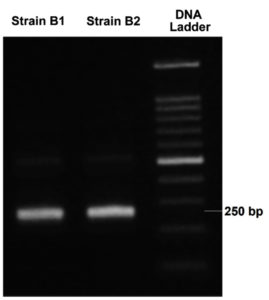
The genomic DNA of isolates B1 and B2 was extracted, followed by amplification using PCR. Subsequently, the PCR products were visualized through agarose gel electrophoresis, as shown in Figure 7.
Sequencing results were analyzed through NCBI’s BLAST tool, allowing for comparison with global DNA databases. Isolate B1 shared 98.87% sequence similarity with C. kochii, while B2 showed 100% similarity to B. haikouensis. Values above 97% majorly signified species-level matches, and those greater than 95% represented genus-level identification.39
Phylogenetic tree visualization
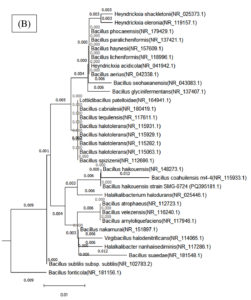
In this study, the NJ method and the Kimura 2-parameter model were used to build an evolutionary tree based on 30 bacterial sequences from the NCBI database for a phylogenetic analysis. Sequence alignment was carried out with ClustalW in MEGA 10, trimming ambiguous areas to enhance accuracy. The resulting phylogenetic tree provided insight into evolutionary relationships among the bacteria, supported by using an outgroup species for accurate distance estimations (Figure 8).
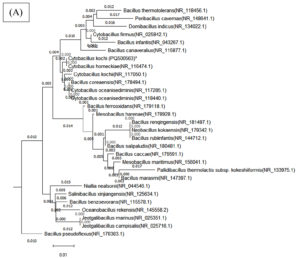
Figure 8. Phylogenetic tree of both bacterial isolates (A) Cytobacillus kochii (PQ500563) and (B) Bacillus haikouensis (PQ395181), which was constructed using Nj method by MEGA (10) software
A 98% bootstrap value supported the grouping of closely related species in Clade A, which contained C. kochii. This high value represents strong evolutionary relationships in Clade A, where shorter branch lengths suggest recent divergence events among the species. Clade B, which included B. haikouensis, had a 95% bootstrap value and comprised species less closely related to the counterparts in Clade A despite sharing a common ancestor. The longer branch lengths in Clade B reflected a higher degree of evolutionary divergence, suggesting an earlier split from the lineage of Clade A.
The robustness of the tree was confirmed through 1000 bootstrap replicates, with nodes showing over 70% bootstrap support considered reliable. Bootstrap values above 90% for deeper nodes in the tree increased confidence in the wider evolutionary separations across clades. Based on more recent divergences shown in Clade A, this pattern implied that the divergence between Clades A and B was an ancient occurrence. Clade C, where species from all over the world clustered together, showed unexpected relationships, signifying horizontal gene transfer or convergent evolution.
The quest for effective and sustainable solutions to marine hydrocarbon pollution necessitates a thorough understanding of the microorganisms capable of degrading recalcitrant compounds.40,41 This study attempted to provide solutions by starting with the isolation of indigenous marine bacteria, which led to an assessment of the individual and synergistic bioremediation potential against a model PAH, known as anthracene.
Recovery of 28 distinct bacterial isolates from marine water samples through plating on Zobell Marine Agar signified the inherent microbial diversity in the sampled marine environment. Subsequent purification ensured the establishment of pure cultures, a prerequisite for accurate downstream characterization. The critical screening for hydrocarbon tolerance included exposing these isolates to Zobell Marine Agar supplemented with challenging used engine oil concentrations of 1%-2% and 20%. The used engine oil was a complex amalgam of aliphatic, aromatic, and PAH serving as a stringent selective pressure.42 The production of only two isolates, designated B1 and B2, capable of growing under the demanding conditions showed the significant tolerance and intrinsic potential to use these complex hydrocarbons as carbon and energy sources. The rigorous selection process effectively narrowed the field to the most promising candidates for detailed investigation into the bioremediation capabilities in oil-contaminated marine environments. The successful purification of the two resilient isolates was visually documented, as presented in Figure 1.
Further morphological and biochemical characterization (Table 1) provided crucial insights into the physiological and metabolic attributes of B1 and B2. The two isolates shared several fundamental characteristics conducive to survival and activity in challenging environments. The characteristics include being rod-shaped, significantly capable of forming endospores, Gram-positive, and motile. The formation of endospores is a well-recognized survival strategy, enabling bacteria to endure harsh conditions such as nutrient scarcity, desiccation, or the presence of toxic xenobiotics often prevalent in contaminated areas, and to resume metabolic activity when conditions improve.43 The aerobic nature implies a dependence on oxygen for metabolic processes, which is a common trait for efficient hydrocarbon degradation pathways primarily including oxidative mechanisms. Moreover, catalase-positive status of both B1 and B2 suggests the presence of enzymatic defenses against hydrogen peroxide, a reactive oxygen species (ROS) generated during aerobic metabolism and often increased by the stress of pollutant degradation.44 Shared positive results for methyl red and Voges-Proskauer tests, as well as the inability to ferment sucrose provided additional common metabolic fingerprints, suggesting some overlap in the central metabolic pathways.
Despite the similarities, distinct differences in colony morphology (B1: yellow, flat; B2: orange-red, convex) as well as in the biochemical profiles showed varied ecological adaptations and specialized degradation capabilities. Isolate B2 presented the unique ability to ferment maltose, produce indole, show urease activity, and use citrate as a carbon source, while B1 was negative for these tests. The stated differences suggest that both isolates adapted to hydrocarbon-rich environments may possess distinct enzymatic repertoires and metabolic pathways. For example, urease activity in B2 could confer a competitive advantage in nitrogen-limited marine environments by allowing the utilization of urea as a nitrogen source.45 Citrate utilization by B2 points to a broader metabolic versatility compared to B1. These distinct biochemical profiles strongly suggested that B1 and B2 might occupy slightly different ecological or functional niches in a contaminated environment and could play different, potentially complementary roles in the degradation of complex hydrocarbon mixtures. The early indication of differentiation was crucial in conceiving the potential for synergistic action in a microbial consortium for more comprehensive bioremediation.
C. kochii (B1) showed a striking capacity for robust biomass production (OD 600 nm = 2.05) in the presence of anthracene, suggesting a strong ability to tolerate and actively use the PAH or the initial breakdown products for substantial growth and proliferation. This vigorous growth was intrinsically connected to the significantly higher dehydrogenase activity (20.67 x 10-2 µmol/min/ml) compared to B. haikouensis (B2). Dehydrogenase enzymes are important, broad-spectrum indicators of overall metabolic activity and play a crucial role in the initial oxidative steps of hydrocarbon catabolism by facilitating electron transfer reactions.46 The significant dehydrogenase activity in C. kochii and the great biomass accumulation represent a high metabolic flux and an efficient, possibly less specific system for initiating the oxidative breakdown of the anthracene structure. The activity positions C. kochii as an effective primary degrader capable of making the complex PAH structure more accessible for further, more specialized enzymatic actions. The ability to form a substantial clear zone (10 ± 0.55 mm) on used engine oil supports the role performed as an active initial degrader. Moreover, the identification of B1 as C. kochii correlates the results with known characteristics of the species,47 including endospore formation (Table 1) and the inherently high catalase activity (55.75 x 10² µmol/min/ml). These traits show significant environmental resilience, crucial for survival and sustained activity in harsh, hydrocarbon-contaminated marine environments where oxidative stress is a prevalent challenge during active degradation.
B. haikouensis had lower biomass production (OD 600 nm = 0.90) and markedly lower dehydrogenase activity (7.33 x 10-2 µmol/min/ml). This suggests a less vigorous initial action on the parent anthracene molecule or a slower total metabolism under the conditions. However, the most substantial result for B2 was the catechol 2,3-dioxygenase (C23O) activity (75.9 x 10-2 µmol/min/ml), which was comparable to and slightly higher than the activity of C. kochii (75.35 x 10-2 µmol/min/ml). C23O is an indispensable enzyme in the meta-cleavage pathway of catechols, which are common and critical intermediates formed during the aerobic degradation of many aromatic compounds, including PAHs such as anthracene.48 The high C23O activity in B. haikouensis strongly points towards a specialized metabolic strategy despite showing modest growth and lower initial oxidative capacity, as signified by dehydrogenase levels. B. haikouensis isolate appears to channel more of the metabolic resources into producing specific, high-activity enzymes required for subsequent more recalcitrant steps of PAH degradation, such as aromatic ring cleavage, rather than prioritizing rapid biomass accumulation on the parent PAH. This specialization is essential for the complete breakdown and detoxification of complex aromatic pollutants. The identification of B. haikouensis is particularly relevant considering that the species is recognized for halotolerance and resilience in extreme environments, which are traits highly beneficial for application in saline marine bioremediation scenarios.49,50 The inherent hardiness, combined with the specialized C23O enzymatic machinery observed in this study, positions B. haikouensis as a potent candidate for tackling the more persistent aromatic fractions of oil pollution. The moderate clear zone formation (8 ± 1.05 mm) on used engine oil provides more evidence of the degradative capabilities.
Catalase activity profiles further clarified the distinct physiological responses of the two isolates to anthracene metabolism. C. kochii had substantially higher catalase activity (55.75 x 10² µmol/min/ml) compared to B. haikouensis (12.39 x 10² µmol/min/ml). Aerobic metabolism of hydrocarbons, particularly PAHs including multiple oxidative steps, inherently generates ROS,44 which necessitates robust antioxidant defense mechanisms. The significantly higher catalase levels in C. kochii suggested experiences of greater oxidative stress which could be a direct consequence of the vigorous metabolic activity and rapid initial processing of anthracene, leading to a higher intracellular accumulation of ROS. The enhanced catalase production reflected a well-developed adaptive defense system crucial for mitigating ROS-induced cellular damage. However, the lower catalase activity in B. haikouensis might represent a lower total metabolic rate when dealing with anthracene, a more channeled and specific degradation pathway that generates fewer ROS. This might signify a reliance on other, unmeasured antioxidant mechanisms or a more efficient ROS management system beyond catalase only. The provided interpretation corresponded with the specialization in later-stage degradation, potentially encountering different types or levels of oxidative stress compared to a primary attacker.
The consortium achieved the highest biomass production (OD 600 nm = 2.54), greater than the biomass generated by both isolates and even exceeding the additive potential. This enhanced growth suggests improved nutrient utilization efficiency, possible cross-feeding of growth-promoting metabolites, or the effective removal of inhibitory intermediates when the two strains co-exist and co-metabolize. Moreover, dehydrogenase activity in the consortium increased to 54 x 10-2 µmol/min/ml, which appeared substantially greater than the sum of the individual activities and nearly 2.6 times higher than the value obtained for the more active C. kochii only. The dramatic increase shows a highly stimulated total metabolic activity in the consortium, suggesting a more aggressive, efficient, and potentially broader initial action on the anthracene substrate.
C23O activity peaked in the consortium (95.10 x 10-2 µmol/min/ml), approximately 1.26 times greater than the already high levels observed in the individual isolates. This shows that the consortium is more vigorous in the initial breakdown stages reflected by dehydrogenase activity and proficient in processing the subsequent aromatic intermediates. The enhanced C23O activity is crucial for preventing the accumulation of potentially toxic catechol intermediates and driving the degradation pathway toward complete mineralization, a key goal of bioremediation. Consistent with the enhanced total metabolic activity and potential for increased ROS generation, catalase activity was highest in the consortium (69.98 x 10² µmol/min/ml). The observed catalase activity signifies that while the combined metabolic efforts may generate more ROS, the consortium is collectively better equipped to neutralize the oxidative stress, potentially contributing to improved growth, stability, and degradative performance.
The results strongly support a “division of labor” model for the synergistic degradation of anthracene by the consortium. C. kochii that has the high dehydrogenase activity and robust growth may initiate the oxidation of anthracene and efficient conversion into simpler, partially oxidized intermediates. Additionally, C. kochii tends to play a role in degrading more labile components during exposure of the consortium to a complex mixture such as crude oil. B. haikouensis which has a specialized and further enhanced C23O pathway in the consortium efficiently cleaves the aromatic rings of the intermediates, tackling the more recalcitrant parts of the PAH molecule. The cooperative metabolism, where one strain processes the products or byproducts generated by the other, prevents the accumulation of potentially toxic intermediates, and facilitates a more complete and rapid PAH mineralization. The synergistic interactions, including metabolic cross-feeding and detoxification, are critical for the effective bioremediation of complex hydrocarbon mixtures often found in polluted marine environments and offer a significant advantage over the application of single strains. This corresponds with broader ecological principles where microbial communities often achieve more complex tasks than individual species, as shown in other studies reporting that certain consortia outperform pure cultures in hydrocarbon degradation.51,52
In conclusion, this study identified two indigenous marine bacteria, C. kochii and B. haikouensis, from hydrocarbon-polluted waters and evaluated the PAH degradation potential. Individually, C. kochii showed robust biomass (OD 600 nm = 2.05) as well as high dehydrogenase (20.67 x 10-2 µmol/min/ml) and catalase (55.75 x 10² µmol/min/ml) activities, signifying vigorous initial hydrocarbon oxidation and stress resilience. B. haikouensis presented superior catechol 2,3-dioxygenase (C23O) activity (75.9 x 10-2 µmol/min/ml), specializing in aromatic ring cleavage. The combination of the C. kochii and B. haikouensis consortium had significant synergistic enhancements, achieving the highest biomass (OD 2.54) and markedly increased dehydrogenase (54 x 10-2 µmol/min/ml), C23O (95.10 x 10-2 µmol/min/ml), and catalase (69.98 x 102 µmol/min/ml) activities compared to individual strains. This observation supported a “division of labor” model, where C. kochii initiated broad hydrocarbon oxidation and B. haikouensis processed aromatic intermediates, leading to more comprehensive PAH mineralization. The results showed the potential of the targeted indigenous microbial consortium for developing effective, sustainable bioremediation strategies for marine hydrocarbon contamination. Synergy was shown with anthracene, but optimal conditions and long-term stability for broader applications were unknown. More investigations should be conducted to optimize the ratio of C. kochii and B. haikouensis in the consortium for maximal synergistic activity against various hydrocarbon mixtures. Studies on the stability of the consortium over time and under varying environmental conditions, such as temperature, salinity, and nutrient levels, would be crucial for practical application.
ACKNOWLEDGMENTS
The authors would like to extend their sincere gratitude to all those who contributed to the successful completion of this research. Their invaluable support and assistance have been greatly appreciated.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
KK performed data collection, resources, and managed the laboratory. MTJ performed funding acquisition and supervised the experiments. MBA carried out the experiment and analyzed the data. MO analyzed and interpretating the molecular 16S rDNA data.MBA drafted the manuscript. MO and MTJ reviewed and edited the manuscript. All authors read and approved the final manuscript for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
Not applicable.
- Tornero V, Hanke G. Chemical contaminants entering the marine environment from sea-based sources: A review with a focus on European seas. Marine Pollution Bulletin. 2016;112(1-2):17-38.
Crossref - Kurniawan ST, Imron MF, Roziqin A, et al. Cases of oil spills in the Indonesian coastal area: Ecological impacts, health risk assessment, and mitigation strategies. Regional Studies in Marine Science. 2024;79:103835.
Crossref - Bogalecka M. Collision and Contact – Analysis of Accidents at Sea. Trans Nav. 2024;18(1):75-85.
Crossref - Fuad MAZ. Oil Spill Trajectory Simulation and Environmental Sensitivity Index Mapping: A Case Study of Tanjung Priok, Jakarta. J Environ Eng Sustain Technol. 2021;8(2):47-54.
Crossref - Akbar RTM, Devi SN, Kurniawan ID, Ulfa RA, Darniwa AV. Diversity of Plankton in the Waters of Sanghyang Kenit Rajamandala Cave, Indonesia. Depik. 2023;12(3):291-301.
Crossref - D’andrea M, Reddy K. Crude Oil Spill Exposure and Human Health Risks. Journal of occupational and environmental medicine/American College of Occupational and Environmental Medicine. 2014;56:1029-1041.
Crossref - Sharma V, Ramish A, Sahu O. Oil Spill Recovery Techniques in Petroleum Industry: A Review on Treatment Process. J Oil Gas Petrochemical Sci. 2021;3:1-5.
Crossref - Obi EO, Kamgba FA, Obi DA. Techniques of Oil Spill Response in the Sea. IOSR J Appl Phys. 2014;6(1):36-41.
Crossref - Osborne OE, Willie MMC, O’Hara PD. The Effects of Oil Spill Dispersant Use on Marine Birds: A Review of Scientific Literature and Identification of Information Gaps. Environ Rev. 2022;31(2):243-255.
Crossref - Ayilara MS, Babalola OO. Bioremediation of Environmental Wastes: The Role of Microorganisms. Front Agron. 2023;5.
Crossref - Sonune N. Microbes: A Potential Tool for Bioremediation. In: Kumar V, Prasad R, Kumar M, eds. Rhizobiont in Bioremediation of Hazardous Waste. 2021:391-407.
Crossref - Agnez-Lima LF, Vainstein MH, Zhang X. Editorial: Microbial Hydrocarbon Degradation and Bioremediation: From Genes to Pathways. Front Microbiol. 2024;15:1416516.
Crossref - Das N, Das A, Das S, et al. Petroleum Hydrocarbon Catabolic Pathways as Targets for Metabolic Engineering Strategies for Enhanced Bioremediation of Crude-Oil-Contaminated Environments. Fermentation. 2023;9(2):196.
Crossref - Ghosal D, Ghosh S, Dutta TK, Ahn Y. Current State of Knowledge in Microbial Degradation of Polycyclic Aromatic Hydrocarbons (PAHs): A Review. Front Microbiol. 2016;7:1369.
Crossref - Alonso-Gutierrez J, Teramoto M, Yamazoe A, Harayama S, Figueras A, Novoa B. Alkane-Degrading Properties of Dietzia Sp. H0B, a Key Player in the Prestige Oil Spill Biodegradation (NW Spain). J Appl Microbiol. 2011;111(4):800-810.
Crossref - Ge X, Campbell RE, Van de Rijn I, Tanner ME. Covalent Adduct Formation with a Mutated Enzyme: Evidence for a Thioester Intermediate in the Reaction Catalyzed by UDP-Glucose Dehydrogenase. J Am Chem Soc. 1998;120(26):6613-6614.
Crossref - Kita A, Kita S, Fujisawa I, et al. An Archetypical Extradiol-Cleaving Catecholic Dioxygenase: The Crystal Structure of Catechol 2,3-Dioxygenase (Metapyrocatechase) from Pseudomonas Putida Mt-2. Structure. 1999;7(1):25-34.
Crossref - Gao M, Tan F, Shen Y, Peng Y. Rapid Detection Method of Bacterial Pathogens in Surface Waters and a New Risk Indicator for Water Pathogenic Pollution. Sci Rep. 2024;14(1):1614.
Crossref - Nelson JO, Kumon T, Yamashita YM. rDNA Magnification Is a Unique Feature of Germline Stem Cells. Proc Natl Acad Sci U S A. 2023;120(47).
Crossref - Soto-Varela ZE, Rosado-Porto D, Bolivar-Anillo HJ, et al. Preliminary Microbiological Coastal Water Quality Determination along the Department of Atlantico (Colombia): Relationships with Beach Characteristics. J Mar Sci Eng. 2021;9(2):122.
Crossref - Purwaningsih S, Sutisna E, Nugroho AA. Characterization, diversity, and effectiveness phosphate solubilizing bacteria from the soil and rhizosphere on the growth of Glycine max L. in greenhouse. IOP Conf Ser Earth Environ. Sci. 2022;976:012030.
Crossref - Liu Y, Wang J, Zhao R, et al. Bacterial isolation and genome analysis of a novel Klebsiella quasipneumoniae phage in Southwest China’s karst area. Virol J. 2024;21(1):56.
Crossref - Sharmin S, Towhid Hossain MT, Anwar MN. Isolation and characterization of a protease-producing bacterium Bacillus amovivorus and optimization of some factors of culture conditions for protease production. J Biol Sci. 2005;5(3):358-362.
Crossref - Legesse DY. Optimization and partial characterization of Bacillus protease isolated from soil and agro-industrial wastes. Int J Nutr Food Sci. 2017;6(1):31-38.
Crossref - Harley JP, Prescott LM. Laboratory exercises in microbiology. 5th Ed. The McGraw-Hill Companies. 2002.
- Olutiola PO, Famurewa O, Sontang HG. An introduction to general microbiology: a practical approach. Ca. Heidelberg verlagsanstaltund Dreuckerei GMbh., Heidelberg, Germany. 1991.
- Fawole MO, Oso BA. Laboratory Manual of Microbiology. Rev. ed. Ibadan: Spectrum Books. 2001.
- MacWilliams MP. Citrate Test Protocol. Washington, DC: American Society for Microbiology. 2009.
- MacWilliams MP. Indole Test Protocol. Washington, DC: American Society for Microbiology. 2012.
- Kowser J, Aziz MG, Uddin MB. Isolation and characterization of Acetobacter aceti from rotten papaya. J Bangladesh Agric Univ. 2016;13(2):199.
Crossref - McDevitt S. Methyl Red and Voges-Proskauer Test Protocols. Washington, DC: American Society for Microbiology. 2009.
- Han M, Luo W, Gu Q, Yu X. Isolation and characterization of a keratinolytic protease from a feather-degrading bacterium Pseudomonas aeruginosa C11. Afr J Microbiol Res. 2012;6(9):2211-2221.
Crossref - Ghaly AE, Mahmoud NS. Optimum conditions for measuring dehydrogenase activity of Aspergillus niger using TTC. Am J Biochem Biotechnol. 2006;2(4):186-194.
Crossref - Burdock T, Brooks M, Ghaly A, Dave D. Effect of assay conditions on the measurement of dehydrogenase activity of Streptomyces venezuelae using triphenyl tetrazolium chloride. Adv Biosci Biotechnol. 2011;2(4):214-225.
Crossref - Szczepanczyk M, Paul L, Ruzgas T, Bjorklund S. Comparison of oxygen electrode chronoamperometry and spectrophotometry for determination of catalase activity. Oxygen. 2023;3(1):77-89.
Crossref - Damayanti KI, Mulyanib NS, Aminin ALN. Freeze-thaw system for thermostable b-galactosidase isolation from Gedong Songo Geobacillus sp. isolate. J Kimia Sains Dan Aplikasi. 2020;23(11):383-389.
Crossref - Estiningtyas R. Diversity of polyethylene degrading bacteria in Ambon mangrove forest. Ber Biol. 2022;21:221-230.
Crossref - Jabiri S, Legrifi I, Benhammou M, et al. Screening of rhizobacterial isolates from apple rhizosphere for their biocontrol and plant growth promotion activity. Appl Microbiol. 2023;3(3):948-967.
Crossref - Johnson JS, Spakowicz DJ, Hong BY, et al. Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nat Commun. 2019;10:5029.
Crossref - Alori ET, Gabasawa AI, Elenwo CE, Agbeyegbe OO. Bioremediation techniques as affected by limiting factors in soil environment. Frontiers in Soil Science. 2022;2:937186.
Crossref - Bekele GK, Gebrie SA, Mekonen E, Fida TT, Woldesemayat AA, Abda EM, Tafesse M, Assefa F. Isolation and Characterization of Diesel-Degrading Bacteria from Hydrocarbon-Contaminated Sites, Flower Farms, and Soda Lakes. Int J Microbiol. 2022:1-12.
Crossref - Ishaya S, Usman S, Nweke OD, et al. Degradation of used engine oil by alcaligenes sp. strain isolated from oil contaminated site: Isolation, identification, and optimization of the growth parameters. Case Studies in Chemical and Environmental Engineering. 2023;8:100516.
Crossref - Riley EP, Schwarz C, Derman AI, Lopez-Garrido J. Milestones in Bacillus subtilis sporulation research. Microbial Cell. 2021;8(1):1-16.
Crossref - Aragaw TA, Bogale FM, Gessesse A. Adaptive Response of Thermophiles to Redox Stress and Their Role in the Process of dye Degradation From Textile Industry Wastewater. Front Physiol. 2022;13:908370.
Crossref - Pei P, Aslam M, Wang H, et al. Diversity and ecological function of urease-producing bacteria in the cultivation environment of Gracilariopsis lemaneiformis. Microb Ecol. 2024;87(1):35.
Crossref - Pi YR, Bao MT. Investigation of kinetics in bioaugmentation of crude oil via high-throughput sequencing: Enzymatic activities, bacterial community composition and functions. Pet Sci. 2022;19(4):1905-1914.
Crossref - Al-Sharidah A, Richardt A, Golecki JR, Dierstein R, Tadros MH. Isolation and characterization of two hydrocarbon-degrading Bacillus subtilis strains from oil contaminated soil of Kuwait. Microbiol Res. 2000;155(3):157-164.
Crossref - Davoodi SM, Miri S, Brar SK, Martel R. Continuous fixed-bed column studies to remove polycyclic aromatic hydrocarbons by degrading enzymes immobilized on polyimide aerogels. Journal of Water Process Engineering. 2023;53:103597.
Crossref - Kumar P, Fulekar MH, Hiranmai RY, Kumar R, Kumar R. 16S rRNA molecular profiling of heavy metal tolerant bacterial communities isolated from soil contaminated by electronic waste. Folia Microbiol (Praha). 2020;65(6):995-1007.
Crossref - Li J, Yang G, Lu Q, Zhao Y, Zhou S. Bacillus haikouensis sp. nov., a facultatively anaerobic halotolerant bacterium isolated from a paddy soil. Antonie van Leeuwenhoek. 2014;106(4):789-794.
Crossref - Eio EJ, Kawai M, Niwa C, Ito M, Yamamoto S, Toda T. Biodegradation of bisphenol A by an algal-bacterial system. Environ Sci Poll Res. 2015;22(19):15145-15153.
Crossref - Chuah LF, Chew KW, Bokhari A, Mubashir M, Show PL. Biodegradation of crude oil in seawater by using a consortium of symbiotic bacteria. Environ Res. 2022;213:113721.
Crossref
© The Author(s) 2025. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.