ISSN: 0973-7510
E-ISSN: 2581-690X
Global dissemination of multidrug-resistant (MDR) and extensively drug-resistant (XDR) Gram-negative bacteria (GNB) such as carbapenemase-producing Enterobacterales has resulted in reviving colistin as a final therapeutic alternative. Colistin resistance foretold a catastrophe. We aimed to detect the rates of carbapenems and colistin resistance among hospital-acquired Enterobacterales species, verify the underlying mechanisms and provide antibiogram for colistin-resistant isolates. The collected Enterobacterales isolates were tested for their antimicrobial susceptibility by the disk diffusion method and agar dilution was utilized for both imipenem and colistin. The production of ESβLs and carbapenemases was phenotypically assessed by the combined disk (CDT) and modified carbapenem inactivation (mCIM) tests, respectively. Possible attributes for colistin resistance were explored by detection of both plasmid- and efflux pump-mediated mechanisms. By multiplex PCR assay, carbapenem resistance (blaNDM-1 & blaOXA-48) and mobilized colistin-resistant-1 (mcr-1) genes were identified. A total of 160 Enterobacterales isolates were obtained of which 68.8% were MDR, 25% were XDR and 6.3% were pandrug-resistant (PDR) isolates with no statistically significant difference among Enterobacterales species (P> 0.05). Carbapenems resistance was detected in 41.3% (66/160) while colistin resistance was detected in 22% (36/160) of isolates. Proteus mirabilis expressed the highest rate of colistin resistance (100%; 16/16), followed by Enterobacter aerogenes (23.1%; 6/26), E. coli (13%; 6/46) and K.pneumoniae (11.1%; 8/72). One hundred percent (36/36) of colistin-resistant isolates proved efflux pump activity for colistin. However; only 2% (2/100) of tested Enterobacterales carried mcr-1 gene through molecular analysis. Colistin-resistant isolates exhibited variable susceptibility to the tested antimicrobial agents of which fosfomycin was the highest (94.1%). Efflux pump activity played a major role for colistin resistance among Enterobacterales species and fosfomycin could be a promising therapeutic option.
Enterobacterales, Carbapenems, Colistin, Efflux Pump & mcr-1 Gene
Antibiotic resistance is a phenomenon closely associated with both antibiotics overuse and bacterial evolution that provide worrisome prospects for all of humanity. Emergence and dissemination of MDR and XDR organisms are superbugs associated with hospital-acquired infections (HAIs) resulting in elevated mortality, therapeutic complications and economic burdens. Annually, about 1.7 million patients acquire HAIs of which 6% die.1
In the 1990s, Enterobacterales started to develop a wide range of resistance to extended-spectrum cephalosporins (ESβLs), since then carbapenems have to be utilized more as a last resort to combat MDR GNB.2 Overtime, the emergence of carbapenem resistance among Enterobacterales has become a serious health concern for both public healthcare authorities and practitioners.3 Carbapenemases production, structural mutations of outer porins and efflux pumps are key mechanisms contributing to carbapenem resistance among Enterobacterales. Of these, carbapenemase production is the commonest.4
Carbapenemases are β-lactamase enzymes that can be categorized based on need to divalent cations for their activation into metallo-carbapenemases, MβLs, (zinc-dependent class B) and non-metallo-carbapenemases (zinc-independent classes A and D) that efficiently hydrolyze all beta-lactams. Two of most prevalent carbapenemases are blaNDM (class B) and blaOXA-48 (Class D). Carbapenemases are mostly plasmid-mediated, which allows easier horizontal transfer and consequently leads to rapid spread of carbapenem resistance worldwide.4
According to the World Health Organization’s (WHO) pathogen priority list, carbapenem-resistant Enterobacterales (CRE) are classified as “critical” antibiotic-resistant pathogens that pose a tremendous threat to public health.5 Around 80% of GNB in humans belong to the Enterobacterales and are responsible for various illnesses as respiratory tract infections, urinary tract infections, bloodstream-associated infections (BSAIs), meningitis, sepsis, and other types of infections.6
Currently, serious challenges were raised and medical community can “beam back” to the pre-antimicrobial era owing to lack of effective therapeutic alternatives and limitations in novel antibiotic development.7 Due to the concerning global rise in MDR and XDR Enterobacterales, healthcare practitioners have been forced to reintroduce colistin as a final resort option to combat potentially life-threatening infections.6
Colistin (polymyxin E) is a type of bactericidal peptide with polycationic properties that exhibit strong antimicrobial activity against Enterobacterales. As it binds to lipid A component of lipopolysaccharide (LPS), consequently, Ca++ and Mg++ that form bridges between LPS are displaced leading to disruption of bacterial membrane.8 Likewise, as colistin use has increased, there has been a progressive increase in the prevalence of colistin resistance in the last few years, and the identification of underlying mechanisms is imperative.8
The intrinsic resistance to colistin among Enterobacterales was thought to be caused by chromosomal mutations in certain genes such as pmrA/B, phoP/Q and mgrB that encode regulatory proteins which govern transcription of enzymes involved in modifying the structure of LPS and thus, reducing the outer membrane’s negative charge along with weakening the electrostatic attraction of polymyxins, thereby allowing the bacteria to resist the antimicrobial effects of colistin.9
The mobilized colistin resistance gene (mcr-1) which is plasmid-encoded was first reported in E. coli isolates obtained from livestock in China. Additionally, the same gene was detected in clinical isolates of K. pneumoniae, drawing attention to the emerged colistin-resistant bacteria.10 Subsequent research on the genetic mechanisms underlying colistin resistance revealed nine other mcr genes, ranging from mcr-2 to mcr-10, in different bacterial species. Despite this, mcr-1 remains the most common globally detected gene.11 The mcr-1 gene encodes a zinc-dependant metalloenzyme which facilitates phosphoethanolamine transfer onto bacterial lipid A, thus leads to a lower binding affinity of colistin to its target site.10
Also, several studies have reported that efflux pumps are capable of reducing the susceptibility of bacteria to colistin.12 Efflux pump inhibitors (EPIs), such as carbonyl cyanide 3 chlorophenyl hydrazone CCCP, 2, 4-dinitrophenol (DNP), omeprazole and verapamil, have been investigated as potential means of reversing colistin resistance.13,14
The aim of the study was to determine the prevalence of ESβLs and carbapenems resistance among clinical Enterobacterales isolated from HAIs at Menoufia University Hospitals (MUHs), investigate the rates of colistin resistance and explore the role of both plasmid and efflux pump-mediated mechanisms. Our findings might be beneficial to elucidate the pattern of antibiotic susceptibility observed in colistin-resistant Enterobacterales isolates, thus to provide alternative treatment options for critically-ill patients, coinciding with antimicrobial stewardship of our healthcare facility.
Ethical approval and study design
The current cross-sectional study was conducted between March 2021 to August 2022 at the Faculty of Medicine Menoufia University, Medical Microbiology and Immunology Department, after obtaining approval from the Local Research Ethical Committee of Faculty of Medicine, Menoufia University (IRB No 3/2021MICR22). Informed consents were obtained from the study participants before involvement in this study.
Collection of clinical samples & identification of bacteria
Various clinical specimens were obtained from 360 patients who were admitted to different departments and ICUs of MUHs with variable clinical types of HAIs that became evident at least 48 hours after admission such as respiratory tract infections, urinary tract infections, wound infection, burn infections and bacteremia. Enterobacterales isolates were identified through culture onto MacConkey’s, blood, nutrient and CLED media. Subsequently, the collected isolates were subjected to standard microbiological methods, which involved morphological and biochemical identification of different species.15 All the obtained species were subjected to:
Antimicrobial susceptibility test
The disk diffusion screening method (Kirby Bauer method) was applied against different antibiotic disks (Oxoid, England) as per CLSI, 2022 guidance.16 For imipenem and colistin, minimal inhibitory concentration (MIC) was determined by agar dilution method.16 The obtained Enterobacterales isolates were categorized as: MDR was defined as isolate non-susceptible to ≥ 1 agent in ≥ 3 antimicrobial categories, while XDR was used to describe an isolate non-susceptible to ≥ 1 agent in all antimicrobial categories but still sensitive to ≤ 2 categories whereas, PDR refers to non-susceptibility to all antimicrobial agents.17
Phenotypic detection of ESβLs production
ESβLs production among the isolated Enterobacterales species was detected using ceftazidime (30μg), cefotaxime (30μg), ceftriaxone (30μg) and aztreonam (30μg) disks. For zone diameter less than or equal 22 mm, 27 mm, 25mm and 27 mm, respectively to at least one disk, the isolates were considered as potential ESβLs producers (CLSI, 2022). Then, ESβLs production was confirmed by ceftazidime/clavulanic acid combined disk test (CDT). An increase of ≥5mm in the zone diameter around ceftazidime/clavulanic acid than around ceftazidime alone was interpreted as positive ESβLs production.16
Detection of carbapenemase production by phenotypic method
To confirm carbapenemase production when the tested isolate was non-susceptible to at least one of carbapenems, the modified carbapenem inactivation method (mCIM) was applied. The procedure was performed and interpreted as per CLSI, 2022 directions. A positive result was defined as an inhibition zone diameter of 6-15 mm or the presence of pinpointed colonies within a 16-18 mm zone. Whereas, a negative result was indicated by a clear zone of inhibition that measured ≥19 mm around meropenem disk.16
Demonstration of efflux pump inhibition by MIC reduction assay using CCCP as efflux pump inhibitor (EPI)
Two sets of Mueller-Hinton (MH) agar (Oxoid, England) were prepared: the first with colistin only (Sigma Aldrich; code: C4461-100MG), the second with colistin and CCCP (Sigma Aldrich; code: C2759). To prepare the stock solution of CCCP, a concentration of 5mg/mL was used, and it was dissolved in Dimethyl sulfoxide (DMSO; Sigma, 48216) (Concentration of DMSO 0.2%). The concentration of CCCP in the MH agar plates was adjusted to 10mg/L and was constantly kept whilst, that of the colistin was serially increased. A positive result indicating efflux pumps was defined as an eight-fold or greater decrease in colistin minimum inhibitory concentration (MIC) on the addition of CCCP. To calculate the mean fold change, the following formula was used: “[1/total sample size (n)] × Σ (MIC fold change × frequency of fold change)” where the ‘frequency of fold change’ is the number of times a particular MIC fold change was recorded for that species.13
Molecular characterization of carbapenem & colistin resistance
One hundred of the isolated Enterobacterales spp. (40 K. pneumoniae, 24 E. coli, 20 Enterobacter aerogenes & 16 Proteus mirabilis) were investigated to determine whether the target genes were present (blaNDM-1 and blaOXA-48 for carbapenemase production & mcr-1 for colistin resistance) by multiplex PCR assay. DNA extraction and purification for gene analysis were done in accordance with the manufacturer’s instructions by the Gene JET Kit from Thermo Scientific (Thermo Fisher Scientific, USA, K0512). Sequence of primers for detection of blaNDM-1, blaOXA-48, and mcr-1 genes are shown in Table-1.18-20
Table (1):
Sequence of primers for target genes in the study
Genes |
Primers sequence (5’-3’) |
Product size |
Reference |
|---|---|---|---|
blaNDM- 1 |
F:TTGGCGATCTGGTTTTCC R:GGTTGATCTCCTGCTTGA |
195 |
(18) |
blaOxa-48 |
F: TTGGTGGCATCGATTATCGG R:GAGCACTTCTTTTGTGATGGC |
744 |
(19) |
mcr- 1 |
F:ATGCCAGTTTCTTTCGCGTG R:TCGGCAAATTGCGCTTTTGGC |
502 |
(20) |
Statistical analysis
The data collected in this study were tabulated and statistically analyzed using a Statistical Package of Social Science (SPSS) version 29 and Epi Info 2000 programs, the Chi-square test or Fisher’s-exact test were used as tests for significance of qualitative data. While quantitative data were assessed using student T-test. The differences between groups were considered significantly different with p-values smaller than 0.05.
A total 160 non-duplicate, consecutive Enterobacterales isolates were obtained from the study participants (n= 360) with a mean age 45.8±22.6 years, of which males represented 49.2% (177/360) while females were 50.8% (183/360). Nearly, 62 (38.75%), 28 (17.5%), 25 (15.6%), 20 (12.5%), 18 (11.25%) and 7 (4.4%) isolates were recovered from urine samples, sputum, pus, surgical drains or wound swabs, bronchial aspirate, blood and burn swabs respectively. The most frequently isolated microorganisms were K. pneumoniae (72/160; 45%), followed by E. coli (46/160; 28.75%), Enterobacter aerogenes (26/160; 16.25%) and Proteus mirabilis (16/160; 10%). Among patients infected with Enterobacterales species (n=160): 70 (43.75%), 55 (34.4%) and 5 (3.1%) patients have received cephalosporins, carbapenems and colistin therapy, respectively.
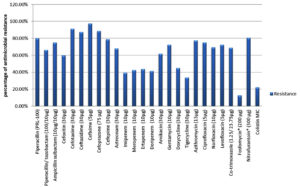
Regarding antimicrobial resistance pattern, the resistance rates reached 97.5%, 91.3%, 88.8%, 87.5%, 80.0%, 78.8%, 77.5%, 75.0%, 75.0%, 72.5%, 72.5%, 69.4%, 68.8%, 68.1%, 66.3%, 61.9%, 60.0%, 45%, 43.8%, 42.5%, 41.3%, 39.4% and 33.8% for cefixime, cefotaxime, cefoperazone, ceftazidime, piperacillin, cefepime, azithromycin, ampicillin/sulbactam, ciprofloxacin, levofloxacin, gentamycin, norfloxacin, co-trimoxazole, aztreonam, piperacillin/tazobactam, amikacin, cefoxitin, doxycycline, ertapenem, meropenem, doripenem, imipenem and tigecycline. Isolates recovered from urine samples (n=62) were resistant to nitrofurantoin (80.6%) and fosfomycin (12.9%) (Figure 1). Out of the isolated Enterobacterales spp., 68.8% (110/160), 25% (40/160) and 6.3% (10/160) were MDR, XDR and PDR respectively but with no statistically significant difference among Enterobacterales spp. (P- value >0.05).
Figure 1. Antimicrobial resistance pattern of Enterobacterales isolates
*Fosfomycin and nitrofurantoin were tested against urine samples according to guidance of CLSI 2022
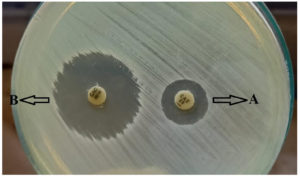
About 97.5% (156/160) of Enterobacterales isolates were ESβLs producers by disk diffusion screening test. Meanwhile, the CDT revealed only 68.1% (109/160) of them as positive ESβLs producers with a significance statistical difference between the two tests (P-value ˂ 0.05) (Table 2 & Figure 2).
Table (2):
Comparison between phenotypic screening and confirmatory tests for ESβLs and carbapenemase production among the isolated Enterobacterales species
| Phenotypic tests For ESβL detection | K. pneumoniae (72) | E. coli (46) | E. aerogenes (26) | P. mirabilis (16) | Total Enterobacterales | P value ** | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| +ve | -ve | +ve | -ve | +ve | -ve | +ve | -ve | +ve | -ve | |||
| Disk diffusion screening | 70 (97.2%) | 2 (2.8%) | 44 (95.7%) | 2 (4.3%) | 26 (100%) | 0 (0.0%) | 16 (100%) | 0 (0.0%) | 156 (97.5%) | 4 (2.5%) | .625 | |
| CDT | 54 (75%) | 18 (25%) | 29 (63%) | 17 (37%) | 16 (61.5%) | 10 (38.5%) | 10 (62.5%) | 6 (37.5%) | 109 (68.1%) | 51 (31.9%) | .413 | |
| P value * | 0.0002 | 0.0002 | 0.0007 | 0.018 | <.0001 | |||||||
| Phenotypic tests for carbapenem resistance | K. pneumoniae (72) | E. coli (46) | E. aerogenes (26) | P. mirabilis (16) | Total Enterobacterales (160) | P value ** | ||||||
| R | S | R | S | R | S | R | S | R | S | .055 | ||
| Disk diffusion | 36 (50%) | 36 (50%) | 22 (47.8%) | 24 (52.2%) | 20 (76.9%) | 6 (23.1%) | 12 (75%) | 4 (25%) | 90 (56.2%) | 70 (43.8%) | ||
| Agar dilution (Imipenem MIC) | 28 (38.9%) | 44 (61.1%) | 17 (37%) | 29 (63%) | 15 (57.7%) | 11 (42.3%) | 6 (37.5%) | 10 (62.5%) | 66 (41.3%) | 94 (58.7%) | .32 | |
| P value *** | .179 | .291 | .139 | .033 | .007 | |||||||
| +ve | -ve | +ve | -ve | +ve | -ve | +ve | -ve | +ve | -ve | .225 | ||
| mCIM | 26 (36.1%) | 46 (63.9%) | 17 (37%) | 29 (63%) | 15 (57.7%) | 11 (42.3%) | 6 (37.5%) | 10 (62.5%) | 64 (40%) | 96 (60%) | ||
*Comparison between disk diffusion screening method and CDT by Fisher’s exact test
**Comparison between different Enterobacterales species for each test.
***Comparison between disk diffusion and agar dilution as screening for carbapenem resistance by Chi-square test.
Figure 2. Combined disk test (CDT) for ESβLs production. Letter A represents ceftazidime alone. Letter B represents ceftazidime/clavulanate. There was an increase of inhibitory zone diameter ≥5 mm around ceftazidime/clavulanate (CAC) than ceftazidime alone.
Nearly, 56.2% (90/160) of Enterobacterales isolates were carbapenem resistant by disk diffusion method compared to only 41.3% (66/160) by imipenem agar dilution method with a significance statistical difference (P- value ˂ 0.05). Confirmatory test for carbapenemase detection (mCIM), detected 40% (64/160) of Enterobacterales spp. as carbapenemase producers (Table 2 & Figure 3).
Figure 3. Detection of carbapenemase production by modified carbapenem inactivation test. Letter A represents the reference strain E. coli ATCC 25922 which is meropenem sensitive while letter B represents meropenem disk (10μg). Isolate 3 showed zone of inhibition less than 15mm and was considered as positive carbapenemase producer. Isolates 5 showed a zone of inhibition ≥19 mm that indicated a negative result.
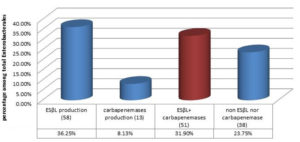
About 31.9% (51/160) of the isolated Enterobacterales spp. were ESβLs/carbapenemases co-producers, a statistically significant association was noticed between ESβLs and carbapenemases production among the tested isolates (P- value=0.01) (Figure 4).
Figure 4. Association between ESβLs production (by CDT) and carbapenemases production (by mCIM) among Enterobacterales isolates (X2 =6.567 and P- value =.0104)
Colistin resistance among Enterobacterales isolates by agar dilution method reached 22.2% (36/160). Proteus mirabilis isolates exhibited the the highest degree of colistin resistance (100%; 16/16), followed by Enterobacter aerogenes (23.1%; 6/26), E. coli (13%; 6/46) and the least resistance was for K.pneumoniae (11.1%; 8/72). Approximately 72.2% (26/36), 16.7% (6/36), and 11.1% (4/36) of the isolates showed MIC values at 64µg/ml, 16µg/ml, and 8µg/ml for colistin, respectively (Table 3).
Table (3):
Colistin MIC reduction assay when using CCCP as efflux pump inhibitor among colistin-resistant isolates (n=36)
| Isolates (n=36) | MIC of colistin (µg/ml) | MIC of colistin + CCCP (Fold change) | Conclusion effect | Mean fold change of colistin MIC ± SD | Mean fold change of colistin +CCCP MIC | P value |
|---|---|---|---|---|---|---|
| Klebsiella pneumoniae (n=8) | ||||||
| 6 isolates | 64 | 0.125 (512) | Reverse | 50 ± 25.9 | 0.125 ± 0 | .00096 |
| 2 isolates | 8 | 0.125 (64) | Reverse | |||
| E. coli (n=6) | ||||||
| 2 isolates | 8 | 0.125 (64) | Reverse | 29.3 ±27.09 | 0.125 ± 0 | .04592 |
| 2 isolates | 16 | 0.125 (128) | Reverse | |||
| 2 isolates | 64 | 0.125 (512) | Reverse | |||
| Enterobacter aerogenes (n=6) | ||||||
| 4 isolates | 64 | 0.125 (512) | Reverse | 48 ± 24.78 | 0.104± .032 | <.001 |
| 2 isolates | 16 | 0.125 (128) | Reverse | |||
| Proteus mirabilis (n=16) | ||||||
| 2 isolates | 16 | 0.125 (128) | Reverse | 58 ± 16.4 | 0.56 ± .84 | < .00001 |
| 4 isolates | 64 | 2 (32) | Reverse | |||
| 2 isolates | 64 | 0.0625 (1024) | Reverse | |||
| 8 isolates | 64 | 0.125 (512) | Reverse | |||
Notably, when using CCCP as efflux pump inhibitor, it could drop the mean fold of colistin MIC for K. pneumoniae from 50 to 0.125, from 29.3 to 0.125 for E. coli, from 48 to 0.104 for Enterobacter aerogenes and from 58 to 0.56 for Proteus mirabilis (Table 3).
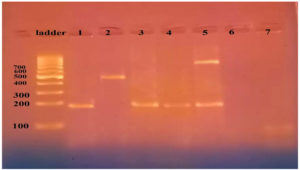
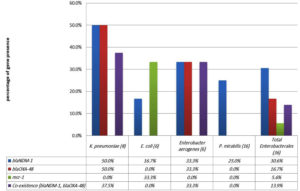
As regards distribution of carbapenemase genes among 100 Enterobacterales isolates, 35% (35/100) and 20% (20/100) were positive for blaNDM-1 and blaOXA-48, respectively. Additionally, 16% (16/100) of them revealed co-existence of the two genes. The highest prevalence of blaNDM-1 was detected among E. coli (45.8%; 11/24) followed by K. pneumoniae (35%; 14/40), Enterobacter aerogenes (30%; 6/20) and finally Proteus mirabilis isolates (25%; 4/16) (P> 0.05). E. coli and K. pneumoniae showed equal frequency for blaOXA-48 gene (25% for each) followed by Enterobacter aerogenes (20%; 4/20) without significant difference as well (P> 0.05). As a genetic determinant of colistin resistance, mcr-1 gene was only identified in 2% (2/100) of the tested isolates (only in E .coli) and none of the remaining species (98/100; 98%) harbored the gene (Figure 5 &
Table 4).
Table (4):
Distribution of blaNDM-1, blaOXA-48 and mcr-1 genes by multiplex PCR among Enterobacterales isolates
| Target genes | Enterobacterales isolates (n=100) | ||||||
|---|---|---|---|---|---|---|---|
| K.pneumoniae (40) | E. coli (24) | E.aerogenes (20) | P. mirabilis (16) | Total (100) | |||
| +ve | +ve | +ve | +ve | +ve | -ve | ||
| Single gene | |||||||
| blaNDM-1 | 6 (15%) | 7 (29.2%) | 2 (10%) | 4 (25%) | 19 (19%) | 81 (81%) | |
| blaOXA-48 | 2 (5%) | 2 (8.3%) | – | – | 4 (4%) | 96 (96%) | |
| mcr-1 | 0 | 2 (8.3%) | – | – | 2 (2%) | 98 (98%) | |
| Co-existence | |||||||
| blaNDM-1& blaOXA-48 | 8 (20%) | 4 (16.7%) | 4 (20%) | – | 16 (16%) | 84 (84%) | |
| Total | P- value | ||||||
| blaNDM-1 | 14 (35%) | 11 (45.8%) | 6 (30%) | 4 (25%) | 35 (35%) | 65 (65%) | .660 |
| blaOXA-48 | 10 (25%) | 6 (25%) | 4 (20%) | – | 20 (20%) | 80 (80%) | .311 |
| mcr-1 | – | 2 (8.3% ) | – | – | 2 (2%) | 98 (98%) | .091 |
Figure 5. Agarose gel electrophoresis for the multiplex PCR-amplified products of Enterobacterales isolates. Ladder is DNA molecular size marker (100 bp). Lanes 1, 3 and 4, were positive for blaNDM-1 (195bp), Lane 2 was positive to mcr-1 (502), lane 5 was positive for blaNDM-1 & blaOXA-48 (744). Lanes 6 & 7 were negative
Among colistin-resistant (n=36) Enterobacterales, 11/36 (30.6%) and 6/36 (16.7%) isolates were respectively positive for blaNDM-1 and blaOXA-48. Moreover, 5/36 (13.9%) isolates displayed co-existence of the two carbapenemase genes (Figure 6).
Figure 6. Genetic profile for blaNDM-1, blaOXA-48 and mcr-1 among colistin-resistant Enterobacterales species
The characteristics of colistin-resistant isolates, both phenotypic and genotypic, are detailed in table 5. Out of 36 colistin-resistant isolates, 17 isolates were recovered from urine, 6 from wound, 4 from burn, 4 from bronchial aspirates, 3 from blood and 2 from sputum specimens. The highest percentage of colistin-resistant isolates (47.2%; 17/36) was from ICUs. 77.8% (28/36) and 61.1% (22/36) were respectively ESβLs and carbapenemase producers. Furthermore, 33.3% (12/36) of colistin-resistant isolates were MDR, 38.9% (14/36) were XDR and 27.8% (10/36) displayed non susceptibility to all the tested antibiotics and were reported as PDR cases.
Table (5):
Phenotypic and genotypic characteristics of colistin-resistant isolates (n=36)
| Isolates (n=36) | Specimens | Departments | Colistin MIC (µg/ml) | Efflux pump activity | mcr-1 | blaNDM-1 | blaOXA-48 | CDT | mCIM | Pheno-types |
|---|---|---|---|---|---|---|---|---|---|---|
| K.pneumoniae1 | Urine | ICU | 64 | + | – | + | + | + | + | PDR |
| K.pneumoniae 2 | Urine | ICU | 64 | + | – | – | + | + | + | XDR |
| K.pneumoniae 3 | Wound | Surgery | 64 | + | – | – | – | + | + | XDR |
| K.pneumoniae 4 | Burn swab | Burn unit | 8 | + | – | + | – | + | + | PDR |
| K.pneumoniae 5 | Urine | Urology | 8 | + | – | + | + | + | + | XDR |
| K.pneumoniae 6 | Urine | Paediatrics | 64 | + | – | + | + | + | + | PDR |
| K.pneumoniae 7 | Bronchial aspirate | ICU | 64 | + | – | – | – | + | + | PDR |
| K.pneumoniae 8 | Blood | ICU | 64 | + | – | – | – | + | + | XDR |
| E.coli 1 | Urine | ICU | 8 | + | + | + | – | + | + | XDR |
| E.coli 2 | Urine | ICU | 16 | + | + | – | – | + | + | XDR |
| E.coli 3 | Urine | Internal medicine | 64 | + | – | – | – | + | – | PDR |
| E.coli 4 | Burn swab | Burn unit | 8 | + | – | – | – | + | + | PDR |
| E.coli 5 | Urine | Urology | 64 | + | – | – | – | + | + | PDR |
| E.coli 6 | Urine | ICU | 16 | + | – | – | – | + | – | PDR |
| E.aerogenes 1 | Sputum | Chest | 64 | + | – | + | + | + | + | PDR |
| E.aerogenes 2 | Bronchial aspirate | ICU | 16 | + | – | – | – | – | – | MDR |
| E.aerogenes 3 | Wound | ICU | 16 | + | – | – | – | + | + | XDR |
| E.aerogenes 4 | Bronchial aspirate | ICU | 64 | + | – | – | – | + | – | PDR |
| E.aerogenes 5 | Bronchial aspirate | ICU | 64 | + | – | + | + | – | + | MDR |
| E.aerogenes 6 | Sputum | Chest | 64 | + | – | – | – | + | + | XDR |
| P. mirabilis 1 | Urine | Urology | 16 | + | – | – | – | + | – | XDR |
| P. mirabilis 2 | Urine | Urology | 64 | + | – | – | – | – | – | MDR |
| P. mirabilis 3 | Blood | Urology | 64 | + | – | – | – | – | – | MDR |
| P. mirabilis 4 | Wound | Surgery | 64 | + | – | – | – | – | – | MDR |
| P. mirabilis 5 | Wound | Urology | 64 | + | – | – | – | + | – | XDR |
| P. mirabilis 6 | Burn swab | Burn unit | 64 | + | – | – | – | + | + | XDR |
| P. mirabilis 7 | Urine | Paediatrics | 64 | + | – | + | – | + | + | MDR |
| P. mirabilis 8 | Urine | ICU | 64 | + | – | + | – | + | + | MDR |
| P. mirabilis 9 | Urine | ICU | 16 | + | – | – | – | + | – | XDR |
| P. mirabilis 10 | Urine | ICU | 64 | + | – | – | – | – | – | MDR |
| P. mirabilis 11 | Blood | Paediatrics | 64 | + | – | – | – | – | – | MDR |
| P. mirabilis 12 | Wound | Surgery | 64 | + | – | – | – | – | – | MDR |
| P. mirabilis 13 | Wound | ICU | 64 | + | – | – | – | + | – | XDR |
| P. mirabilis 14 | Burn swab | Burn unit | 64 | + | – | + | – | + | + | XDR |
| P. mirabilis 15 | Urine | ICU | 64 | + | – | + | – | + | + | MDR |
| P. mirabilis 16 | Urine | ICU | 64 | + | – | – | – | + | + | MDR |
| Total | 36 (100%) | 2 (5.6%) | 11 (30.6%) | 6 (16.7%) | 28 (77.8%) | 22 (61.1%) | ||||
| Total MDR=12 (33.3%) Total XDR= 14 (38.9%) Total PDR=10 (27.8%) |
||||||||||
The antimicrobial susceptibility pattern of colistin-resistant Enterobacterales by disk diffusion is showed in table 6. The susceptibility rates reached 94.1% and 50%, respectively for fosfomycin and piperacillin/tazobactam, 38.9% for all of cefoxitin, aztreonam, imipenem and meropenem, 33.3% for each of ampicillin/sulbactam, amikacin, doripenem and norfloxacin. Much lower susceptibility rates were observed for piperacillin, cefepime, gentamycin, ertapenem and levofloxacin (27.8% for each). Only 22.2% of colistin-resistant isolates were susceptible to tigecycline and ciprofloxacin and about 16.7% were susceptible to each of co-trimoxazole and ceftazidime. The lowest susceptibility was for doxycycline, nitrofurantoin (11.1% for each) and azithromycin (5.6%). Notably, all (100%) isolates were resistant to cefotaxime, cefixime and cefoperazone.
Table (6):
Susceptibility profile of colistin-resistant Enterobacterales isolates (n=36)
| Antimicrobial agents | Colistin-resistant Enterobacterales (n=36) | X2 | P value | ||
|---|---|---|---|---|---|
| S (%) | R (%) | ||||
| Fosfomycin (200μg)* | 16 (94.1%) | 1 (5.9%) | 0.26 | ||
| Piperacillin/ tazobactam (100/10μg) | 18 (50.0%) | 18 (50.0%) | 6.517 | .011 | |
| Cefoxitin (30μg) | 14 (38.9%) | 22 (61.1%) | 0.0004 | .984 | |
| Aztreonam (30μg) | 14 (38.9%) | 22 (61.1%) | 3.871 | .049 | |
| Imipenem (10μg) | 14 (38.9%) | 22 (61.1%) | 4.815 | .028 | |
| Meropenem (10μg) | 14 (38.9%) | 22 (61.1%) | 2.783 | .095 | |
| Ampicillin/ sulbactam (10μg/10μg) | 12 (33.3%) | 24 (66.7%) | 5.308 | .011 | |
| Amikacin (30μg) | 12 (33.3%) | 24 (66.7%) | 0.084 | .772 | |
| Norfloxacin (10μg) | 12 (33.3%) | 24 (66.7%) | 2.376 | .123 | |
| Doripenem (10μg) | 12 (33.3%) | 24 (66.7%) | 5.887 | .015 | |
| Levofloxacin (5μg) | 10 (27.8%) | 26 (72.2%) | .733 | .392 | |
| Cefepime (30μg) | 10 (27.8%) | 26 (72.2%) | .733 | .392 | |
| Piperacillin (PRL-100) | 10 (27.8%) | 26 (72.2%) | 8.193 | .004 | |
| Ertapenem (10μg) | 10 (27.8%) | 26 (72.2%) | 8.065 | .005 | |
| Gentamycin (10μg) | 10 (27.8%) | 26 (72.2%) | 0.048 | .827 | |
| Ciprofloxacin (5μg) | 8 (22.2%) | 28 (77.8%) | 0.357 | .55 | |
| Tigecycline (30μg) | 8 (22.2%) | 28 (77.8%) | 32.385 | < 0.001 | |
| Ceftazidime (30μg) | 6 (16.7%) | 30 (83.3%) | .755 | .385 | |
| Cotrimoxazole (1.25/23.75 μg) | 6 (16.7%) | 30 (83.3%) | 1.611 | .204 | |
| Doxycycline (30μg) | 4 (11.1%) | 32 (88.9%) | < 0.00001 | ||
| Nitrofurantoin (300 μg)* | 2 (11.8%) | 15 (88.2%) | 0.712 | ||
| Azithromycin (15μg) | 2 (5.6%) | 34 (94.4%) | 0.25 | ||
| Cefotaxime (30μg) | 0 0.0% | 36 (100%) |
1 | ||
| Cefixime (5μg) | 0 0.0% | 36 (100%) |
1 | ||
| Cefoperazone (75 μg) | 0 0.0% | 36 (100%) |
1 | ||
*Among colistin-resistant isolates (n=36), 17 isolates were recovered urine samples and tested against fosfomycin and nitrofurantoin according to guidance of CLSI 2022.
Indiscriminate use of antibiotics resulted in worldwide dissemination of drug-resistant organisms of which carbapenem-resistant Enterobacterales are of major concern. This limits physicians’ therapeutic options, particularly in developing nations where infectious diseases are abound and microbes exist in their most resistant phenotypes.21
The lack of new antibiotics led to reintroduction of old antimicrobials to treat severe infections. In that regard, colistin has gained clinical value owing to its activity against MDR GNB. However, there has been a significant increase in the frequency of colistin resistance in recent years.22 Hence, we conducted this study to survey carbapenems and colistin resistance rates and the related mechanisms among Enterobacterales species isolated from patients with HAIs at MUHs, Egypt and assessed the antimicrobial susceptibility patterns of the recovered colistin-resistant isolates to provide alternative treatment lines.
In this study, 160 Enterobacterales isolates were tested against different antimicrobial agents. Even, among members of the same antibiotic class, different levels of susceptibility were detected. Collectively, 68.8%, 25%, and 6.3% of the tested species were respectively MDR, XDR, and PDR isolates. This came in agreement with a previous study that reported 65.7% of hospital-acquired Enterobacterales as MDROs.23 A study conducted in Saudi Arabia revealed 57.3% and 3.5% of Enterobacterales clinical isolates as MDR and XDR organisms, respectively, but no PDRs were detected.24 A higher prevalence of MDR hospital isolates (81.0%) was reported by other study.25 Literature review summarized MDR rates among Enterobacterales in Egypt, from 30% to 70%.26 The elevated rates of MDR infections observed in this study is ultimately attributed to the critically-ill status for most of the studied patients, empirical use of broad-spectrum antibiotics and poor hygienic conditions in developing countries.26
ESβLs production poses a great challenge in the management of Enterobacterales infections and is one of the major mechanisms for emergence of MDROs. Accordingly, the magnitude of ESβLs reached 68.1% in our study and K. pneumoniae proved the highest frequency (54/72; 75%). Varying rates for ESβLs-producing Enterobacterales were reported in the Northeast of Iran (50.8 %),27 and in Saudi Arabia (51.4%).24 Lower reports were declared in Brazil (21.3%),27 Mexico (30.7%),28 and Zimbabwe (14%).29 On the contrary, much higher rates were observed in Ethiopia (70.9%),29 Germany (83.6%),30 Congo (92%),31 and Cambodia (93.4%).32 One potential reason for the high incidence of ESβLs could be the selective pressure resulting from the widespread use of beta-lactam antibiotics as the primary treatment option for bacterial infections caused by Enterobacterales in African countries.29
Emergence of CRE has evolved into a formidable threat to community and they keep escalating trends stably during the later years. By 2022, carbapenem resistance accounted for 61.1% in Egypt.33 In USA, the prevalence of CRE colonization varied widely from 1%–30.4%.34 In the current study, 66/160 (41.3%) isolates were carbapenem resistant and 64/160 (40%) were carbapenemases producers by the mCIM method. Carbapenem resistance rate was relatively higher than other studies that reported carbapenem resistance range from 20–30% among Enterobacterales.35 Makharita et al. in Egypt recorded 36.1% of Enterobacterales as carbapenem resistant,36 which was lower than that reported in Sindh province of Pakistan (59%).37
Class B (blaNDM-1) and D (blaOXA-48) carbapenemase genes were identified in 35% and 20%, respectively, of the tested Enterobacterales isolates. The metallo-beta-lactamase blaNDM-1 gene proved the highest prevalence among E. coli isolates (45.8%). Several studies have addressed blaNDM-1 as the predominant carbapenemase gene in CRE.22,38 Ongoing higher rates were detected by Wang et al.39 who found blaNDM-1 in 75% of E. coli isolates and Chaudhary et al.40 recognized blaOXA-48 in 32.6% of MDR Enterobacterales species.
Our results revealed 31.9% of Enterobacterales species as ESβLs/carbapenemase co-producers. Tayh et al.41 reported 20% resistance to imipenem among ESβL-producing Enterobacterales isolates and Qadi et al.1 found that 43.9% and 68.3% of ESβLs- producing Enterobacterales were respectively non-susceptible to imipenem and meropenem. The co-expression of ESβLs and carbapenemases β-lactamases has exacerbated the emergence of XDR clinical strains, which are challenging to manage and pose a significant threat due to the potential for clonal spread of these genes. It is critical to establish guidelines to prevent the misuse and overuse of antibiotics, particularly carbapenems beside infection control policies and vigilant surveillance on a routine basis.41
Colistin is a key drug for MDR GNB, so its resistance is a significant concern, owing to shortage of alternative therapies.1 Our study found 36/160 (22.2%) Enterobacterales isolates were resistant to colistin. Proteus mirabilis exhibited the highest colistin resistance (16/16; 100%) which may be due to the modification of LPS of outer membrane.42 The frequency of colistin resistance among other Enterobacterales species was 23.1% (6/26) for Enterobacter aerogenes, 13% (6/46) for E. coli, and 11.1% (8/72) for K. pneumoniae. In another publication by Mahmoud et al.43 in Egypt, colistin resistance appeared in 42.9% of K. pneumoniae and E. coli isolates. In Greek, a report by Meletis et al.44 examined 718 clinical Enterobacterales isolates of which 57 (7.9%) isolates were colistin resistant. In Italy, colistin resistance reached 24.7% and was as high as in Enterobacter spp. (47%) and K. pneumoniae (43%).45 In Gaza, 41% of Enterobacterales isolates were classified as colistin resistant and the Proteus group exhibited the highest resistance to colistin, with a rate of 63.2%, followed by Serratia (57.1%). In contrast, Klebsiella isolates had the lowest resistance rate, with only 31.6% exhibiting resistance to colistin.1
According to the current results, genotypic surveys for plasmid-encoded genes, has identified mcr-1 in only two E. coli isolates (2%; 2/100) and none of other species expressed this gene. Such finding came in line with Ejaz et al. who detected only 2.6% of MDR GNB harbouring mcr-1 gene.46 Other studies done in Pakistan and Iran, reported mcr-1 in 3% and 3.2% of clinical isolates, respectively.47,48 About emergence of colistin resistance gene (mcr-1) among colistin-resistant K. pneumoniae in Jordan, Gharaibeh et al. declared that only 1.1% of the tested isolates had mcr-1 gene.49 In Egypt, Zaki et al.50 detected mcr-1 in two isolates (one E. coli strain & one K.pneumoniae strains). Also, Ibrahim et al. reported mcr-1 gene in 7.1% of K.pneumoniae isolates recovered from urinary tract infection of ICU-admitted 70 years male patient.51
Mobilized colistin resistance mcr-1 gene revealed higher frequencies in Greece and Italy (43% and 20.8%, respectively)44,45 with higher predominance in E. coli compared to other species.52,53 Nevertheless, mcr-1 gene is a particularly concerning public health issue, because it can be transmitted more easily across diverse bacteria by horizontal gene transfer than chromosomal colistin resistance genes.54 Among Enterobacterales E. coli tops the list in rapid acquisition/transfer of resistance traits by horizontal gene transfer.55 The mcr-1 has likely been emerged and accelerated by the use of colistin on farms in China and Southeast Asia,10 and subsequently spread to other countries.52
Efflux pumps allow bacteria to move antimicrobials agents out of cells leading to antimicrobial resistance. Efflux pump inhibitors (EPIs) inhibit efflux and could reverse antimicrobial resistance.12 Our results demonstrated that 100% (36/36) of colistin-resistant isolates proved efflux pump activity against colistin when using CCCP as EPI. The Role of CCCP as EPI to rescue colistin susceptibility was studied by Baron and Rolain.13 who reported that, CCCP was found to be effective in reversing colistin resistance in all investigated strains, and demonstrated ability to restore colistin susceptibility. Ni et al.56 suggested that, this effect may be attributed to renewing the negative charges of outer membrane. Park and Ko57 proposed that enhanced colistin activity in these cells could be related to a decrease in ATP synthesis caused by CCCP action.
The characteristics of colistin-resistant isolates, both phenotypic and genotypic, were also analyzed and revealed that 30.6% (11/36) and 16.7% (6/36) of them harbored blaNDM-1 and blaOXA-48 respectively. Zafer et al. found 9/40 and 7/40 of colistin-resistant isolates were positive for blaOXA-48 and blaNDM-1 genes, respectively.22 In Jordan,50 19% and 11.5% of colistin-resistant Enterobacterales isolates were positive for blaOXA-48 and blaNDM-1, respectively. The occurrence of colistin resistance in conjunction with carbapenemase genes poses significant risks in the use of carbapenems and colistin to combat infections.58
Our colistin-resistant strains were most frequently isolated from urine specimens (17/36; 47.2%) and 47.2% of colistin-resistant strains were obtained from ICUs’ samples. Zafer et al. found colistin-resistant Klebsiella and E. coli among cancer patients were highly recovered from blood specimens (60%),22 this could be owing to the cancer patients’ neutropenic state, which favours GNB bloodstream infection treated with colistin. Panigrahi et al. noticed that 31.4% of colistin-resistant GNB were isolated from respiratory samples followed by 25% from blood samples among ICU patients.59 Sorour et al. detected that ICUs were the highest frequent site (66.7%) for isolation of colistin-resistant GNB compared to other hospital departments.60
Also, we assessed the susceptibility profile of colistin-resistant Enterobacterales to provide antimicrobial stewardship team with data required for implementation of antibiotic policy in our healthcare facility. Colistin-resistant isolates showed a considerable high susceptibility to fosfomycin (94.1%), piperacillin/ tazobactam (50%), aztreonam, imipenem, meropenem and cefoxitin (38.9% for each of them). On other hand, lower susceptibility was observed against other antimicrobial agents ranging from 33.3% to 5.6%. Surprisingly, absolute non susceptibility was detected for each of cefotaxime, cefixime and cefoperazone. These findings were remarkably similar to those of Gharaibeh et al., who documented significant resistance to ceftazidime, tobramycin, and imipenem and average susceptibility to fosfomycin among colistin-resistant Enterobacterales.49
Both colistin and fosfomycin are considered a salvage treatment for MDR and XDR CRE.61 It is worth noting that fosfomycin kept activity against strains of mcr-1 gene carrying colistin-resistant Enterobacterales. As shown in our study, fosfomycin susceptibility among colistin-resistant Enterobacterales isolated from urinary tract infections was 94.1% (16/17). Other study showed that 83.3% of strains carrying mcr-1 gene showed fosfomycin susceptibility.62 Beyond UTI as the main focus of fosfomycin prescription, fosfomycin also showed excellent diffusion to various tissues. Thus, it should be considered for managing various other types of infectious caused by MDR, XDR Enterobacterales.63
Carbapenem and colistin resistance reached alarming rates in Egypt. The high prevalence of MDR and XDR among Enterobacterales isolates was concerning and their higher rate among colistin-resistant Enterobacterales adds another layer of concern to this escalating problem. Efflux pump is a major contributor to the emerged colistin-resistant Enterobacterales. Plasmid-borne colistin resistance is now spreading all over the world. The coexistence of class D blaOXA-48 and class B blaNDM-1 carbapenemases genes was notable among CRE isolates. Fosfomycin achieved excellent activity against colistin-resistant isolates. Immediate action to monitor the usage of antimicrobials, especially colistin, is a must.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
AFM, SAMA and ABM designed and contributed to all aspects of the study. AFL provided and analyzed clinical information of the studied participants. MEE and RME performed the experiments. AME interpreted the clinical and laboratory data, and wrote the manuscript. All authors reviewed, edited and approved the final manuscript for publication.
FUNDING
None.
DATA AVAILABILITY
The datasets generated and/or analysed during the current study are available from the corresponding author on reasonable request.
ETHICS STATEMENT
This study was approved by the Institutional Ethics Committee, Faculty of Medicine, Menoufia University, Egypt, with reference number 3/2021MICR22.
INFORMED CONSENT
Written informed consent was obtained from the participants before enrolling in the study.
- Qadi M, Alhato S, Khayyat R, Elmanama AA. Colistin resistance among Enterobacteriaceae isolated from clinical samples in Gaza Strip. Can J Infect Dis Med Microbiol. 2021;1-6.
Crossref - Castanheira M, Simner PJ, Bradford PA. Extended-spectrum β-lactamases: An update on their characteristics, epidemiology and detection. JAC-Antimicrob Resist. 2021;3(3):dlab092.
Crossref - Fadlallah M, Salman A, Salem-Sokhn E. Updates on the status of carbapenem-resistant Enterobacterales in Lebanon. International Journal of Microbiology. 2023;2023:1-10.
Crossref - Suay-Garcia, Perez-Gracia. Present and future of carbapenem-resistant Enterobacteriaceae (CRE) infections. Antibiotics. 2019;8(3):122.
Crossref - World Health Organization. WHO publishes list of bacteria for which new antibiotics are urgently needed. https://www.who.int/news/item/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed. 2017.
- Binsker U, Kasbohrer A, Hammerl JA. Global Colistin use: A review of the emergence of resistant Enterobacterales and the impact on their genetic basis. FEMS Microbiol Rev. 2021;46(1):fuab049.
Crossref - Uddin TM, Chakraborty AJ, Khusro A, et al. Antibiotic resistance in microbes: History, mechanisms, therapeutic strategies and future prospects. Journal of Infection and Public Health. 2021;14(12):1750-1766.
Crossref - Srisakul S, Wannigama DL, Higgins PG, et al. Overcoming addition of phosphoethanolamine to lipid A mediated colistin resistance in Acinetobacter baumannii clinical isolates with colistin-sulbactam combination therapy. Sci Rep. 2022;12(1):11390.
Crossref - Olaitan AO, Morand S, Rolain J-M. Mechanisms of polymyxin resistance: Acquired and intrinsic resistance in bacteria. Front Microbiol. 2014;5:643.
Crossref - Sun J, Zhang H, Liu Y-H, Feng Y. Towards understanding MCR-like Colistin resistance. Trends Microbiol. 2018;26(9):794-808.
Crossref - Savin M, Bierbaum G, Blau K, et al. Colistin-resistant Enterobacteriaceae isolated from process waters and wastewater from German poultry and pig Slaughterhouses. Front Microbiol. 2020;11:575391.
Crossref - Telke AA, Olaitan AO, Morand S, Rolain J-M. SoxRS induces colistin hetero-resistance in Enterobacter asburiae and Enterobacter cloacae by regulating the acrab-tolc efflux pump. J Antimicrob Chemother. 2017;72(10):2715-2721.
Crossref - Baron SA, Rolain J-M. Efflux pump inhibitor CCCP to rescue colistin susceptibility in MCR-1 plasmid-mediated colistin-resistant strains and gram-negative bacteria. J Antimicrob Chemother. 2018;73(7):1862-1871.
Crossref - Osei Sekyere J, Amoako DG. Carbonyl cyanide M-chlorophenylhydrazine (CCCP) reverses resistance to Colistin, but not to carbapenems and tigecycline in multidrug-resistant Enterobacteriaceae. Front Microbiol. 2017;8:228.
Crossref - Tille PM. Evolve Resources for Bailey and Scott’s Diagnostic Microbiology, 15th Edition;2018. http://evolve.elsevier.com/Tille/micro/
- Clinical and Laboratory Standards Institute (CLSI): Performance standards susceptibility testing. for antimicrobial 2022 32nd Ed. CLSI supplement M100.Wayne, PA, USA.
- Magiorakos A-P, Srinivasan A, Carey RB, et al. Multidrug-resistant, extensively drug-resistant and Pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268-281.
Crossref - Zheng F, Sun J, Cheng C, Rui Y. The establishment of a duplex real-time PCR assay for rapid and simultaneous detection of blaNDM and blaKPC genes in bacteria. Ann Clin Microbiol Antimicrob. 2013;12(1):30.
Crossref - Poirel L, Heritier C, Tolun V, Nordmann P. Emergence of Oxacillinase-Mediated Resistance to Imipenem in Klebsiella pneumoniae. Antimicrob Agents Chemother. 2004;48(1):15-22.
Crossref - Lescat M, Poirel L, Nordmann P. Rapid multiplex polymerase chain reaction for detection of MCR-1 to MCR-5 Genes. Diagn Microbiol Infect Dis. 2018;92(4):267-269.
Crossref - Furqan W, Ali S, Usman J, et al. Assessing colistin resistance by phenotypic and molecular methods in carbapenem-resistant Enterobacterales in a tertiary care hospital in Pakistan. Infect Drug Resist. 2022;15:5899-5904.
Crossref - Zafer MM, El-Mahallawy HA, Abdulhak A, Amin MA, Al-Agamy MH, Radwan HH. Emergence of colistin resistance in multidrug-resistant Klebsiella pneumoniae and Escherichia coli strains isolated from cancer patients. Ann Clin Microbiol Antimicrob. 2019;18(1):40.
Crossref - Mirzaei B, Babaei R, Bazgir ZN, Goli HR, Keshavarzi S, Amiri E. Prevalence of Enterobacteriaceae spp. and its multidrug-resistant rates in clinical isolates: A two-center cross-sectional study. Mol Biol Rep. 2021;48(1):665-675.
Crossref - Alkofide H, Alhammad A, Alruwaili A, et al. Multidrug-resistant and extensively drug-resistant Enterobacteriaceae: Prevalence, treatments, and outcomes: A retrospective cohort study. 2020.
Crossref - Bandy A, Tantry B. ESBL activity, MDR, and carbapenem resistance among predominant Enterobacterales isolated in 2019. Antibiotics. 2021;10(6):744.
Crossref - El-Kholy A, El-Mahallawy HA, Elsharnouby N, Abdel Aziz M, Helmy AM, Kotb R. Landscape of multidrug-resistant gram-negative infections in Egypt: Survey and literature review. Infect Drug Resist. 2021;14:1905-1920.
Crossref - Rizi KS, Mosavat A, Youssefi M, et al. High prevalence of blaCMY AMPC beta-lactamase in ESBL co-producing Escherichia coli and Klebsiella spp. clinical isolates in the northeast of Iran. J Glob Antimicrob Resist. 2020;22:477-482.
Crossref - Nogueira K da, Conte D, Maia FV, Dalla-Costa LM. Distribution of extended-spectrum β-lactamase types in a brazilian tertiary hospital. Rev Soc Bras Med Trop. 2015;48(2):162-169.
Crossref - Onduru OG, Mkakosya RS, Aboud S, Rumisha SF. Genetic determinants of resistance among ESBL-producing Enterobacteriaceae in community and hospital settings in east, Central, and Southern Africa: A systematic review and meta-analysis of prevalence. Can J Infect Dis Med Microbiol. 2021;5153237.
Crossref - Muller-Schulte E, Tuo MN, Akoua-Koffi C, Schaumburg F, Becker SL. High prevalence of ESBL-producing Klebsiella pneumoniae in clinical samples from central Cote d’Ivoire. Int J Infect Dis. 2020;91:207-209.
Crossref - Moges F, Eshetie S, Abebe W, et al. High prevalence of extended-spectrum beta-lactamase-producing Gram-negative pathogens from patients attending Felege Hiwot Comprehensive Specialized Hospital, Bahir Dar, Amhara Region. PLOS ONE. 2019;14(4):e0215177.
Crossref - Vlieghe ER, Huang T-D, Phe T, et al. Prevalence and distribution of beta-lactamase coding genes in third-generation cephalosporin-resistant Enterobacteriaceae from bloodstream infections in Cambodia. Eur J Clin Microbiol Infect Dis. 2015;34(6):1223-1229.
Crossref - Gandor NH, Amr GE-S, Eldin Algammal SM, Ahmed AA. Characterization of carbapenem-resistant K. pneumoniae isolated from intensive care units of Zagazig University Hospitals. Antibiotics. 2022;11(8):1108.
Crossref - Chen H-Y, Jean S-S, Lee Y-L, et al. Carbapenem-resistant Enterobacterales in long-term care facilities: A global and narrative review. Front Cell Infect Microbiol. 2021;11.
Crossref - Mustafai MM, Hafeez M, Munawar S, et al. Prevalence of carbapenemase and extended-spectrum β-lactamase producing Enterobacteriaceae: A cross-sectional study. Antibiotics. 2023;12(1):148.
Crossref - Makharita RR, El-kholy I, Hetta HF, et al. antibiogram and genetic characterization of carbapenem-resistant gram-negative pathogens incriminated in healthcare-associated infections. Infect Drug Resist. 2020;13:3991-4002.
Crossref - Apanga PA, Ahmed J, Tanner W, et al. Carbapenem-resistant Enterobacteriaceae in sink drains of 40 healthcare facilities in Sindh, Pakistan: A cross-sectional study. PLOS ONE. 2022;17(2):e0263297.
Crossref - Wang Q, Wang X, Wang J, et al. Phenotypic and genotypic characterization of carbapenem-resistantent Erobacteriaceae: Data from a longitudinal large-scale CRE study in China (2012-2016). Clin Infect Dis. 2018;67(Suppl 2):S196-S205.
Crossref - Chaudhary M, Payasi A. Prevalence, genotyping of Escherichia coli and Pseudomonas aeruginosa clinical isolates for oxacillinase resistance and mapping susceptibility behaviour. J Microb Biochem Technol. 2014;06(02):063-067.
Crossref - Tayh G, Al Laham N, Ben Yahia H, Ben Sallem R, Elottol AE, Ben Slama K. Extended-spectrumβ-lactamases among Enterobacteriaceae isolated from urinary tract infections in Gaza Strip, Palestine. BioMed Research International. 2019;4041801.
Crossref - Avendano-Ortiz J, Ponce-Alonso M, Llanos-Gonzalez E, et al. The impact of colistin resistance on the activation of innate immunity by lipopolysaccharide modification. Infect Immun. 2023;91(2):e0001223.
Crossref - Fatma M, Moustafa N, Gaber SA, Elsaid RG, Mohamev RA. Molecular mechanisms of colistin resistance among multi-drug resistant (MDR) Klebsiella pneumoniae and Escherichia coli isolated from ICU patients and their susceptibility towards Eravacycline. Microbes and Infectious Diseases. 2023;4(1):127-137.
Crossref - Meletis G, Oustas E, Botziori C, Kakasi E, Koteli A. Containment of carbapenem resistance rates of Klebsiella pneumoniae and Acinetobacter baumannii in a Greek hospital with a concomitant increase in colistin, gentamicin and tigecycline resistance. New Microbiol. 2015;38(3):417-421.
- Monaco M, Giani T, Raffone M, et al. Colistin resistance superimposed to endemic carbapenem-resistant Klebsiella pneumoniae: A rapidly evolving problem in Italy, November 2013 to April 2014. Eurosurveillance. 2014;19(42).
Crossref - Ejaz H, Younas S, Qamar MU, et al. Molecular epidemiology of extensively drug-resistant MCR encoded colistin-resistant bacterial strains co-expressing multifarious β-lactamases. Antibiotics. 2021;10(4):467.
Crossref - Aghapour Z, Gholizadeh P, Ganbarov K, et al. Molecular mechanisms related to colistin resistance in Enterobacteriaceae. Infect Drug Resist. 2019;12:965-975.
Crossref - Wise MG, Estabrook MA, Sahm DF, Stone GG, Kazmierczak KM. Prevalence of MCR-type genes among colistin-resistant Enterobacteriaceae collected in 2014-2016 as part of the Inform Global Surveillance Program. PLOS ONE. 2018;13(4):e0195281.
Crossref - Gharaibeh MH, Alyafawi DA, Elnasser ZA, Lafi SQ, Obeidat HM. Emergence of MCR-1 gene and carbapenemase-encoding genes among colistin-resistant Klebsiella pneumoniae clinical isolates in Jordan. J Infect Public Health. 2022;15(8):922-929.
Crossref - El Sayed Zaki M, Abou ElKheir N, Mofreh M. Molecular study of Colistin resistant clinical isolates of Enterobacteriaceae species. J Clin Mol Med. 2018;1(1).
Crossref - Ibrahim E, Ahmed Y, Mohamed A, Ibrahim W. Detection of colistin resistant gram negative bacilli in intensive care unit patients admitted to Ain Shams University Hospitals. Microbes and Infectious Diseases. 2020;2(1):92-99.
Crossref - Wang R, van Dorp L, Shaw LP, et al. The global distribution and spread of the mobilized colistin resistance gene MCR-1. Nat Commun. 2018;9(1):1179.
Crossref - Elbediwi M, Li Y, Paudyal N, et al. Global burden of Colistin-resistant bacteria: Mobilized Colistin Resistance Genes Study (1980-2018). Microorganisms. 2019;7(10):461.
Crossref - Wang X, Wang Y, Zhou Y, et al. Emergence of colistin resistance gene MCR-8 and its variant in Raoultella Ornithinolytica. Front Microbiol. 2019;10:228.
Crossref - Sundaramoorthy NS, Suresh P, Selva Ganesan S, GaneshPrasad A, Nagarajan S. Restoring colistin sensitivity in colistin-resistant E. coli: Combinatorial use of marr inhibitor with efflux pump inhibitor. Sci Rep. 2019;9(1):19845.
Crossref - Ni W, Li Y, Guan J, et al. Effects of efflux pump inhibitors on colistin resistance in multidrug-resistant gram-negative bacteria. Antimicrob Agents Chemother. 2016;60(5):3215-3218.
Crossref - Park YK, Ko KS. Effect of carbonyl cyanide 3-chlorophenylhydrazone (CCCP) on killing Acinetobacter baumannii by Colistin. J Microbiol. 2015;53(1):53-59.
Crossref - Ngbede EO, Adekanmbi F, Poudel A, et al. Concurrent resistance to carbapenem and colistin among Enterobacteriaceae recovered from human and animal sources in Nigeria is associated with multiple genetic mechanisms. Front Microbiol. 2021;12:740348.
Crossref - Panigrahi K, Pathi BK, Poddar N, et al. Colistin resistance among multi-drug resistant gram-negative bacterial isolates from different clinical samples of ICU patients: Prevalence and clinical outcomes. Cureus. 2022;14(8):e28317.
Crossref - Sorour A, Ibrahim K, Hegab A. Prevalence of acquired colistin resistance among gram negative bacilli isolated from patients admitted at Cairo University Hospitals. Egypt J Med Microbiol. 2022;31(1):97-104.
Crossref - Shrief R, El-Ashry AH, Mahmoud R, El-Mahdy R. Effect of Colistin, fosfomycin and Meropenem/Vaborbactam on carbapenem-resistant Enterobacterales in Egypt: A cross-sectional study. Infect Drug Resist. 2022;15:6203-6214.
Crossref - De La Cadena E, Mahecha M, Velandia AM, et al. Identification of MCR-1 genes and characterization of resistance mechanisms to colistin in Escherichia coli isolates from Colombian hospitals. Antibiotics. 2023;12(3):488.
Crossref - Garcia-Bustos V, Cabanero-Navalon MD, Salavert Lleti M. Resistance to beta-lactams in gram-negative bacilli: Relevance and potential therapeutic alternatives. Rev Esp Quimioter. 2022;35(Suppl2):1-15.
Crossref
© The Author(s) 2023. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.