ISSN: 0973-7510
E-ISSN: 2581-690X
Antimicrobial activity of certain species of Lactobacillus bacteria were known to produce a certain type of bacteriocin called Plantaricin (Pln). In this study, the PlnF (plnF) gene (666 bp) encoding plantaricin was synthetically constructed based on a sequence of predefined and valuable amino acid compositions and subcloned into pET-28 (+) expression vector. Thereafter transformation of recombinant vector into BL21 (DE3) competent E. coli was achieved and to verify the result of cloning, the PCR colony technique was conducted. The expression of the inserted gene was analyzed by the SDS-polyacrylamide gel electrophoresis method, which detected a band of approximately 30 KD molecular weight for interested protein as a natural bioactive bacteriocin.
Lactobacillus, Cloning, Synthetic Gene, Plantaricin F. Expression, Bacteriocin
A bacteriocin is a group that is synthesized in the ribosome and has potent antimicrobial activity against some foodborne pathogens. The bacteriocin peptides that are produced by L. plantarum are recognized as plantaricin and are commonly reported as a class II bacteriocin, a very wide-ranging class with a variety of bactericidal and or bacteriostatic mechanisms.1 The bacteriocin produced by L. plantarum for example plantaricin (A, E, F, J, and K) all these types belong to class (II) bacteriocin.2 Alternatively, the plantaricin EF is a two-peptide formed by the lactic acid bacteria L. plantarum C11, and the two-fold peptides that form this bacteriocin are termed as Plantaricin E (PlnE) and Plantaricin F (PlnF). Plantaricin F (PlnF) produced by L. plantarum is mostly utilized in the food industry to prevent microbial contamination.3 Plantaricin produced by L. plantarum was classified into diverse types according to the presence or absence of several Pln genes. Moreover, the gene locus that encodes for plantaricin is genetically clustering in either simple or complex operon.4 In 1925, the first bacteriocin (colicin) was produced,5 and since that time several study attempts occurred to create and describe diverse types of bacteriocins. The furthermost important requirement to these studies is the large-scale manufacture of bioactive bacteriocin. Although the purification of these native peptides can be done from their regular strains, the process itself is extremely laborious and time-consuming.6 The development of biotechnology makes the production of native or recombinant bacteriocins more efficient by using synthetic genes and a good expression system.7
Gene synthesis is a method for synthesizing a concerned gene in vitro without an original DNA template. Furthermore, commercial synthesis of the desired gene is considering a more rapid step, it is time saving and is developed to be a cost effective alternate for classic cloning techniques and other molecular biology processes.8 Thus, our study has undertaken the cloning of synthetically constructed Pln F genes and expressed them in E. coli BL21(DE3), to produce a natural antimicrobial agent (plantaricin F).
Bacterial strain and plasmid
pUC57 (Thermo Fisher Scientific) was utilized as a cloning vector and E. coli DH5a (Invitrogen) competent cells were utilized for transformation. pET28a (+) was purchased (Novagen) and applied as an expression vector. E.coli BL21(DE3) was purchased (Novagen) and applied for recombinant protein transformation and expression.
Recombinant Vector Construction
The nucleotide sequences with approximately 666 bp of desired protein were synthesized (ShineGene, China) according to the amino acids sequences (Figure 1) of plantaricin F gene which was previously described.9
Figure 1. The mature peptide of (pln F) gene is in red color, while the black color is the amino acid sequences of the (His tag, Thrombin site, T7 tag, Trx tag, S tag, and Enterokinase site).
The synthetic gene fragment was already cloned into the plasmid vector pUC57 with EcoRI at 5’ end and XhoI at 3’ end. Cloning (pUC57) and expression vector (pET28a (+) was transformed first into E. coli DH5a cells using heat shock method.10,11 The transformed bacterial cells were then picked for plasmid extraction step by using the high pure plasmid isolation kit (Roche gel extraction kit Germany), according to the manufacturer’s protocols. The extracted plasmid vectors were electrophoresed in 1% agarose gel.12
After verifying the process of plasmid extraction, both vectors (pUC57 and pET28a(+)) were digested with EcoRI and XhoI. Once the pUC57 vector was digested, the digestion reaction was electrophoresed on 1% agarose gel and the desired fragment was extracted from the gel using Thermo Fisher Scientific for subcloning in the expression vector pET28a(+), which was also digested with EcoRI at 5’ and XhoI at 3’. The inserted fragment was ligated into digested pET28a(+) vector by utilizing the T4 DNA ligase enzyme. A ligation reaction was conducted in 20 μl reactions, wherein 3 μl of insert clone was mixed with 12 μl of the digested plasmid. Then 1 μl T4 DNA ligase was incorporated and the final volume of 20 μl was adjusted by adding distilled water. The reaction was incubated at room temperature for 2h and then refrigerated for 18h.
Recombinant Vector Transformation
The ligated plasmid was then transformed into competent E. coli DH5a to produce high amounts of plasmid vector. Post-transformation, the positive colonies were screened and verified by colony PCR utilizing T7 universal primers. T7 Primer Forward: 5’ TAATACGACTCACTATAGGG 3’, T7 Primer Reverse: 5’GCTAGTTATTGCTCAGCGG3’. PCR program was included: initial denaturation (94°C, 4 min), denaturation (94°C, 30 s) for 30 cycles, annealing step (55°C, 30 s), extension step at 72°C for 1 min, and final extension (72°C, 12 min), the reaction was held at 15°C for 5 min. The PCR results were verified using 1% gel electrophoresis. The positively transformed colonies were selected for verifying the presence of the recombinant pET28a(+) and desired gene fragment, through digestion of extracted plasmids with EcoRI and XhoI. The recombinant pET28a(+) was transformed into competent cells E.coli BL21(DE3) bacterial cells for advancing into the protein expression process.
Expression of Recombinant Plantaricin
The expression of recombinant plantaricin was conducted according to the modified procedure of.13 Both pET28a(+) expression vector-subcloned synthetic plnF genes were singly expressed in E. coli BL21 (DE3). For selection of recombinant clones, kanamycin 50 μg/ml was incorporated to the medium. E. coli BL21 (DE3) was transformed with pET28a (+) single to produce E. coli BL21 (DE3); pET28a no insert was used as a negative control. Culture of transformed E. coli BL21 (DE3) with (pET28a-plnF) overnight was inoculated into 100 ml LB broth that contained kanamycin 50 μg/ml and was incubated at 37°C until the OD 600 value of 0.4 was attained. Expression was conducted and induced by incorporating 1 mm of Iso pro-pylβ-D-thiogalactopyranoside (IPTG) to the culture and incubated for 5h at 22°C and 37°C in broth medium at 150 pm. The culture were centrifuged at 8,000 g at 4°C for 10 min for harvested cells and then cell pellets resuspended in the lysis buffer (pH 8) comprising (20 mM imidazole, 20 mM Tris-HCl, and 500 mM NaCl) after that disrupted by sonication (0.5 cycles of output30, pulse on 30 s and pulse off 30 s) for 5 min. The lysate was centrifuged for 30 min at 18000 g and 4°C. The pellet and supernatant were then resuspended in the same lysis buffer. SDS polyacrylamide gel electrophoresis 12% (SDS PAGE) was used for separating the fractions of pellet and supernatant.
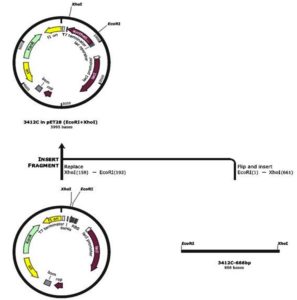
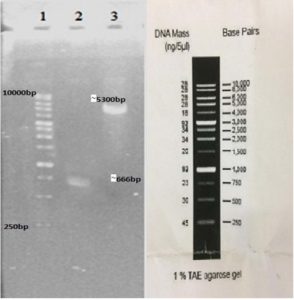
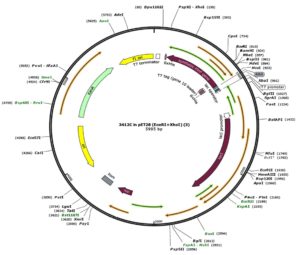
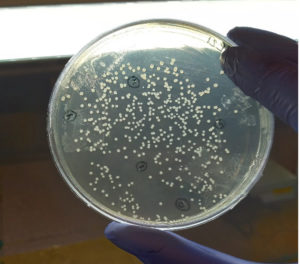
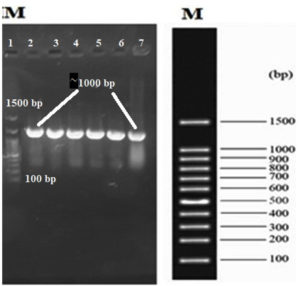
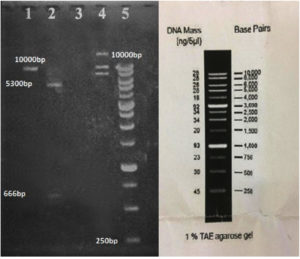
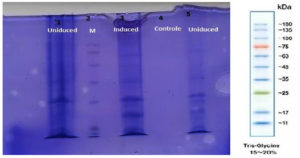
In our study, the synthetic plantaricin F gene was inserted into the multi-cloning site of the pET-28a-c(+) plasmid vector (Figure 2). In this figure, all steps for constructing recombinant plasmid vectors map and complete design strategy were accomplished by utilizing SnapGene software (5.2.4). Alternatively, Figure 3 reveals the agarose gel electrophoresis of approximately 666 bp band conforming to the fragment of synthetic plantaricin F gene which was digested successfully with XhoI and EcoRI restriction enzymes and cloned into pET-28a(+) vector. The recombinant plasmid vector pET-28a(+)-PlnF as depicted in Figure 4 was transformed into competent E. coli BL21(DE3). Kanamycin (50 mg mL-1) was used for the initial selection of transformants bacterial cells to produce transformants cells harboring recombinant plasmid vector (Figure 5) and named as pET-28a-c(+)-plnF-E.coliBL21 (DE3). Figure 6 depicts the results of positive PCR colony; in this figure the target band approximately 1000 was successfully amplified by using T7 universal primer. Additionally, Figure 7 depicts the results of recombinant pET28a(+) digestion to verify the cloning process of the desired gene. In this figure, the recombinant vector was successively linearized post EcoRI digestion. The results of SDS polyacrylamide gel electrophoresis were demonstrated in Figure 8. In this figure, the array of proteins present in the pellets and supernatant fraction were detected in the 12% (SDS PAGE), Lane 3 represented the total cell protein with approximately 27–30 KDa, a stained band that corresponding to the expected size.
Figure 2. Diagrammatic representative the construction of recombinant plasmid vector pET-28a-c(+)-PlnF. The restriction enzymes XhoI and EcoRI was used to digest both the target gene and the plasmid vector
Figure 3. Gel electrophoresis of the cloning process. Lane 1: molecular marker ladder (Yekta Tajhiz Azma, Iran). Lane 2: insert fragment digested with EcoRI and XhoI (~666 bp). Lane 3: the desired portion of pET2a(+) after digestion with EcoRI and XhoI (~5300 bp)
Figure 4. The recombinant plasmid vector pET-28a-c(+)-PlnF: The constructed design of pET-28a-c(+) carrying the synthetic fragment of PlnF introduced into the MCS and controlled by strong bacteriophage T7 expression region
Figure 5. Transformed E. coli BL21 (DE3) competent cells on Kanamycin (50mg mL-1) LB selectable medium
Figure 6. Gel electrophoresis of the colony PCR process to verify the presence of the recombinant pET28a(+) after bacterial transformation. Lane 1: molecular marker ladder (Yekta Tajhiz Azma, Iran). Lane 2 to 6: positive colonies. Lane numbers start from the left
Figure 7. Digestion of the recombinant pET28a(+) to verify the cloning process of our desired gene. Lane 1: linearization of the digested vector after digestion with EcoRI. Lane 2: digestion of the recombinant vector with EcoRI and XhoI which also shows our desired gene fragment out of the vector. Lane 3: empty. Lane 4: the recombinant vector after plasmid extraction which shows three bands for linear, supercoiled, and circular plasmids lane 5:Marker ladder ( Yekta Tajhiz Azma, Iran )
The first major challenge in our study is the attempt to separately isolate the Pln F gene from Pln E gene and subsequently, the PlnF gene is expressed (pre-mature peptide) separately from PlnE gene. To achieve this goal, the gene encoding PlnF (plnF) has been synthetically constructed depending on the plantaricin amino acids sequences and cloned into the pUC57 vectors. The synthesis of the desired gene according to their amino acid sequences is considered modern and efficient than the conventional cloning procedure and has several advantages. The major advantage is that the gene synthesis allows for codon to be optimized and subsequently to increase the efficiency of protein expression.8
Since we had decided to use pET28a(+) as an expression vector, the synthetic DNA fragment was designed with the restriction site for EcoRI and XhoI (at 5’ and 3’, respectively). The synthetic gene fragment was already cloned into the plasmid vector pUC57 with EcoRI and XhoI (at 5’ and 3’, respectively). The introduction of these two artificial restriction sites in the synthetic DNA fragment was a significant step to facilitate ligation with the target vector. Moreover, both enzymes have demonstrated good results and are 100% functional in the “H buffer”. Before proceeding to the rest of the sub-cloning steps, each cloning (pUC57) and expression vector (pET28a (+)) were transformed primarily into E. coli DH5a cells using the heat shock method, and this step is crucial to increase the concentration of vectors used in our study.14
The second step was to select a proper expression vector. In our study pET-28a-c(+) was used as a suitable vector. By using this system, the desired gene fragment was expressed easily and made it easier to be isolated and purified and the vectors pET-28a-(+) hold an N-terminal His -Tag®/thrombin/T7-Tag® configuration plus an elective C-terminal His-Tag sequence, this also facilities protein purification step from transformed bacteria.
The use of pET plasmid vectors as expression vectors have another advantage since the most effective system of this vector was designed for cloning and regulating the high expression of recombinant proteins in transformed E. coli.15 As pET plasmids vector have a strong promoter derived from bacteriophage T7 transcription, this promoter makes the production of recombinant protein (Synthetic plnF gene) so active, specific, and possessive to the amount of production of target RNA which can be comparable to the amount of ribosomal RNA in a cell.16
Alternatively, the insertion of the target gene under the control of a strong promoter as T7 expression systems may also cause a problem. The tricky thing in using such an inducible vector as T7 systems is that the RNA polymerase enzyme based on T7 promoter is highly active, and a minor basal level may lead to significant expression of recombinant peptides even in the absence of a certain inducer.17 Consequently, if the target recombinant protein is effectively toxic to the bacterial host cell, the establishment of such a target vector in the expression host cell could be difficult or even hopeless, or subsequently, make the expression system unsettled leading to the occurrence of mutations.16 To overcome this problem, the BL21(DE3)-pET-28a (+) system was used in this study, as these vectors have a lac operator gene that offers enough lac repressor to suppress both the T7 and lac promoter in this system and the chromosomal gene for T7 RNA polymerase in cell genomes. In this manner, the crucial level of target recombinant protein at uninduced cells is significantly reduced, but this rate of induction leads to the standard excessive levels of expression.18,19 To proceed with the cloning steps, the PCR colony was succeeded and the expected bands were amplified successfully which agree with,20 which approved the significance of the PCR colony step as a high throughput screening step for verifying and selecting genetically transformed cells and this creates a promising, easier, reliable, and rapid routine colony PCR (cPCR) technique.
The last step in the cloning project was to investigate the in vitro expression of the recombinant DNA in BL21(DE3) -pET-28a (+). To induce expression, Isopropyl-β-D-thiogalactopyranoside (IPTG) was incorporated as a final concentration of 1 mM into the broth medium and incubated in the shaker incubator for 5 h at 22°C and 37°C at 150 rpm, and the results were verified on SDS-polyacrylamide gel.
Approximately 27–30 KD of suspected recombinant protein bands were detected on 12% polyacrylamide gel, the size of band was quite similar to the value resulting from the calculated amino acids sequence. The high molecular weight of the current protein can be attributed to the additional part which is added to the original sequences. As it is a synthetic protein it has normally extra sequences like (thioredoxin -TrxA) which are added to the fusion partner which increase the acidity and molecular weight of particular protein. Successful (thioredoxin-TrxA) fusion partners were normally translated, and it also has determinants which allow for suitable purifications, which can be easily and completely cleaved from the desired peptides products, ideally, reducing the occurrence of inclusion body. Additionally, thioredoxin may also act to avoid the accumulation and precipitation of fused proteins, which enabled the desired protein to adopt their exact tertiary structural folds.9,21
By genetic engineering, the synthetic plantaricin F gene was successfully constructed and subcloned into a proper expression system for improved production of recombinant bioactive proteins against pathogenic bacteria.
ACKNOWLEDGMENTS
The authors would like to thank Professor Fatemeh Rahbarizadeh and all technicians at the Department of Medical Biotechnology, Faculty of Medical Sciences, Tarbiat Modares University, Tehran, I.R. Iran for their help and cooperation. The authors would also like to thank Department of Veterinary Microbiology, College of Veterinary Medicine, University of Basrah, Basrah, Iraq for their support.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
This article does not contain any studies with human participants or animals performed by any of the authors.
- Ahmad V, Khan MS, Jamal MS,Alzohairy MA, Al Karaawi MA, Siddiqui MU. Antimicrobial potential of bacteriocins: in therapy, agriculture and food preservation. Int J Antimicrob Agents. 2017;49(1):1-11.
Crossref - Kjos M, Nes IF, Diep DB. Class II one-peptide bacteriocins target a phylogenetically defined subgroup of mannose phosphotransferase systems on sensitive cells. Microbiology. 2009; 155(9):2949-2961.
Crossref - Mustopa AZ, Murtiyaningsih H, Fatimah F, Suharsono S. Cloning and Heterologous Expression of Extracellular Plantaricin F Produced by Lactobacillus plantarum S34 Isolated from “Bekasam” in Lactococcus lactis. Microbiol Indones. 2016;10(3):3.
Crossref - Diep DB, Straume D, Kjos M, Torres C, Nes IF. An overview of the mosaic bacteriocin pln loci from Lactobacillus plantarum. Peptides. 2009;30(8):1562- 1574.
Crossref - Mesa-Pereira B, Rea C, Cotter PD, Hill C, Ross RP. Heterologous Expression of Biopreservative Bacteriocins With a View to Low Cost Production. Front Microbiol. 2018;9:1654:1.
Crossref - Rodriguez J, Martinez MI, Horn N, Dodd HM. Heterologous production of bacteriocins by lactic acid bacteria. Int J Food Microbiol. 2003;80(2):101-116.
Crossref - Cintas LM, Herranz C, Hernandez PE. “Natural and heterologous production of bacteriocins,” in Prokaryotic Antimicrobial Peptides: From Genes to Applications, eds D. Drider and S. Rebuffat (New York, NY: Springer),2011; 115-143.
Crossref - Schwartz M, Lo H, Zhou, Yang JP. Gene Synthesis: A Cost-Effective Alternative to Traditional Molecular Cloning. J Biomol Tech. 2011;22(Suppl): S31.
- Kusdianawati K, Mustopa AZ, Suharsonoa S, Budiarto BR, Fatimah F, Danuri H. Construction, expression, and purification of recombinant pre-mature peptide of plantaricin F from F Lactobacillus plantarum S34 in Escherichia coli. Indones J Agric Sci. 2015;1(16):31-38.
Crossref - Green MR, Sambrook J. Cloning and Transformation with Plasmid Vectors. In: Molecular Cloning: A Laboratory. Manual (4th ed). Cold Spring Harbor: Cold Spring Harbor Laboratory Press. 2012:157-260.
- Hanahan D.Studies on transformation of Escherichia coli with plasmids. J Mol Biol.1983; 166(4):557-580.
Crossref - Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. 2nd ed. New York: Cold Spring Harbor Laboratory Press.1989:412.
Crossref - Tham HY, Song A, Yusoff K, Ta, GH. Effect of different cloning strategies in pET-28a on solubility and functionality of a staphylococcal phage endolysin. Biotechniques. 2020;69(3):161-170.
Crossref - Tu Z, He G, Li KX, et al. An improved system for competent cell preparation and high efficiency plasmid transformation using different Escherichia coli strains. Electron J Biotechnol. 2005;8(1):114-120.
Crossref - Al-Kanany FN, Othman RM. Cloning and Expression of Pseudomonas aeruginosa AlkB Gene in E. coli. J Pure Appl Microbiol. 2020;14(1):389-396.
Crossref - Aguero JA ,Sanchez O, Barrera M, Toledo yJR. Molecular cloning and expression of a fragment of the gene codifying for the protein Erns of classical swine fever virus. Rev Salud Anim. 2008;30(2):85-92.
- Tabor S. Expression using the T7 RNA polymerase/promoter system. Curr Protoc Mol Biol. 2001; Chapter 16:Unit16.2.
Crossref - Studier FW. Protein production by autoinduction in high-density shaking cultures. Protein Expr Purif. 2005;41:207-234.
Crossref - Dubendorf JW , Studier FW. Controlling basal expression in an inducible T7 expression system by blocking the target T7 promoter with lac repressor. J Mol Biol. 1991;5;219(1):45-59.
Crossref - Nouemssi SB, Ghribi M, Beauchemin R, Meddeb-Mouelhi F, Germain H, Desgagne-Penix I. Rapid and Efficient Colony-PCR for High Throughput Screening of Genetically Transformed Chlamydomonas reinhardtii. 2020;10(9):186.
Crossref - LaVallie ER, Lu Z, Diblasio-Smith EA, Collins-Racie LA, McCoy JM. Thioredoxin as a Fusion Partner for Production of Soluble Recombinant Proteins in Escherichia coli. Methods Enzymol. 2000;326:322-340.
Crossref
© The Author(s) 2023. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.