ISSN: 0973-7510
E-ISSN: 2581-690X
Rhizospheric soils from cultivated olive (Olea europaea) trees of Chemlali, Chetoui, Quaissi, and Djalat cultivars were assessed for their bacterial abundance and diversity and were further screened for production of exopolysaccharides and exoenzymes (cellulase, chitinase, amylase, protease, lipase, and peroxidase). The results of the present study indicate that Chetoui cultivar revealed higher diversity, followed by Chemlali > Quaissi > Djalat, wherein, bacilli, enteric bacteria, and pseudomonads were abundantly present as specific bacterial groups associated with the Chetoui rhizosphere. Moreover, the exopolysaccharide (EPS)-producing bacteria of Chetoui cultivar (68.4%) presented the highest efficiency, followed by Djalat (23.5%) > Chemlali (7 %) > Quaissi (1%). These results revealed that the Chetoui cultivar presented highest enzyme activities, followed by Chemlali > Djalat > Quaissi, with a distinct abundance of peroxidase- and chitinase-producing bacteria, which may play a pivotal role in adapting olives to the environmental stresses. From this preliminary study, we confirmed that olive rhizosphere microbial diversity is essentially driven by the geographical origin and genotype of olive cultivars. Furthermore, we recommended the Chetoui olive cultivar rhizosphere as a potential reservoir for exoenzyme- and EPS-producing bacteria useful for future biotechnological applications.
Chetoui cultivar, Diversity, Exoenzymes, Exopolysaccharide-producing bacteria, Olea europaea, Rhizosphere
Olea europaea L. (olive tree), is one of the oldest domesticated oil trees in the Mediterranean area since several millennia and has acquired undisputable ecological and socioeconomic importance, over its longevity and genetic diversity. Its rhizosphere is considered an unexplored source for novel bioactive compounds produced by various bacteria1.
Limited research is available on olive rhizosphere associated microbiota due to the lack of robust assessments methods; however, several studies were conducted to explore the olive rhizosphere microbial diversity, structure, and complexity2-4.
Various techniques have been used to investigate both endophytic and rhizospheric olive microbiota, including media culture, fatty acid methyl ester (FAME) or phospholipid fatty acid analysis (PLFA)5, denaturing gradient gel electrophoresis (DGGE)6, fluorescent terminal restriction fragment length polymorphism (FT-RFLP)2, and high-throughput sequencing7.
Microbial communities inhabiting the interior of olive root (endophytic) are less diverse than those living in the vicinity of their roots (rhizospheric) and are mainly shaped by the olive cultivar genotype; moreover, the environmental, pedological, and agricultural practices such as tillage, irrigation, fertilization, and pesticide application also influence the microbial presence. Thus, information regarding the microbial communities associated with organic rhizospheres will help farmers in selecting efficient and sustainable agropractices and management8.
Olive was initially cultivated in 1980, in the Al-Jouf region, north Saudi Arabia, by the biggest Saudi agricultural companies. The seedlings were mostly imported from Syria, Italy, and Spain. The dominant cultivars are Arbequina, Arbosana, Cornicabra, Manzanillo, Nabbali, Picual, and Sorani9. Olive cultivation in the Kingdom of Saudi Arabia has increased rapidly; however, research on the olive rhizosphere microbial communities is limited, as reported in a previous study10.
In some previous studies, the phenotypic bacterial diversity of certain North African plant rhizospheres was studied and screened for amylase production11. The present study is the first to assess the rhizospheric bacterial diversity of some olive tree cultivars in Al-Jouf state and screen for exoenzymes and exopolysaccharide production for future biotechnological applications.
Olive Trees Cultivars and Soil Sampling
Olea europaea var. europaea trees of Chemlali, Chetoui, Quaissi, and Djalat cultivars used in the present study were supplied from the experimental field situated in the Olive Research Unit, Camel and Pasture Research Center (ORU/CPRC) (Skaka, Al Jouf area, Northern Saudi Arabia). Rhizospheric soil samples were collected from the four olive tree cultivars at 20-30 cm depth from the circumference of the tree canopy2. The samples were stored at a cool temperature for future lab processing.
Total Rhizospheric Bacteria and Fungi Isolation
Total viable counts (TVC) of bacteria and fungi were assessed with the dilution plate method by suspending the rhizospheric soil samples in a sterile saline solution; the suspensions were serially diluted, and triplicates of 100 µl samples were plated on nutrient agar (NA) and potato dextrose agar (PDA). The plates were incubated at 28°C for 48 h for bacteria and 1 week for fungi.
Specific Bacterial Group Isolation
Olive rhizosphere bacterial diversity was evaluated via specific bacterial group isolation on selective media as follows: bacilli on nutrient agar after boiling the soil samples for 10 min at 80°C; enteric bacteria on MacConkey agar medium; pseudomonads on Kings B agar medium containing gram per liter (20 peptone, 1.5 K2HPO4, 20 agar, 1.5 g MgSO4 × 7 H2O, and 15 mL glycerin). Actinomycetes on modified glycerol yeast extract agar medium (MGYEA) containing gram per liter (25 peptone, 2 yeast extract, 15 agar, and 5 ml glycerol) at 28°C for 1 week.
Screening for Exopolysaccharide-Producing Bacteria
Exopolysaccharide (EPS)-producing bacteria were screened by spreading 100 µl rhizospheric samples, prepared as aforementioned, on nutrient agar sucrose 1% medium (NAS), and incubated for 24-72 h at 28°C, for developing mucoid colonies with steaky texture that indicated EPS production. To confirm the presence of gram-negative EPS-producing bacteria, the colonies were subcultured on MacConkey agar medium, and the positive results were characterized by the appearance of mucoid colonies.
Screening for Exoenzyme-Producing Bacteria
Rhizospheric samples collected from the four olive cultivars as described in the sampling section were serially diluted and screened for several enzymatic activities using semiquantitative plate assay. Amylase, cellulase, and chitinase were assessed separately on 0.5% starch, cellulose, and colloidal chitin agar plates, respectively; whereas, protease, lipase, and peroxidase were assessed on 0.5% skim milk, tributyrin, and Congo red agars, respectively. The plates were incubated for 24-48 h at 28°C for amylolytic, lipolytic, and proteolytic activities, and for 5 days for cellulase, chitinase, and peroxidase activities, expressed by clearing zone appearance surrounding the colonies.
Olives Rhizosphere Bacterial Abundance and Diversity
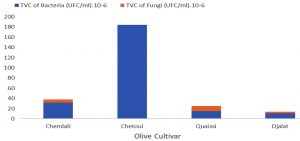
The olive rhizosphere microbiological study illustrated in Fig. 1 revealed variable results between the investigated cultivars. Considering the TVC of bacteria, we observed that the bacteria were present in the following order: Chetoui > Chemlali > Quaissi > Djalat; moreover, fungi in all cultivars were scarce and were completely absent in the Chetoui cultivar rhizosphere. This was presumably due to the presence of endophytic arbuscular mycorrhiza or other antagonistic microorganisms as reported by several researchers2,8,13.
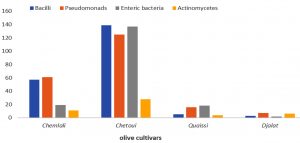
Comparable results were obtained with olive rhizosphere bacterial diversity illustrated in Fig. 2, where the Chetoui cultivar revealed highest diversity, followed by Chemlali > Quaissi > Djalat, with a large abundance of bacilli, enteric bacteria, and pseudomonads as specific bacterial groups associated with the Chetoui rhizosphere. These findings were in accordance with studies that confirmed the occurrence of Proteobacteria presented by pseudomonads and enteric bacteria as well as Firmicutes presented by bacilli as natural inhabitants of olive rhizosphere, which act as effective antagonists against phytopathogens14,15. Furthermore, Fernandez-Gonzalez, et al. (2019)8 reported the dominance of Actinobacteria and Proteobacteria as root endophytes by average relative abundances of 64% and 26%, respectively, thus highlighting that cultivars Chemlal de Kabylie (Algeria), Llumeta (Spain), and Mavreya (Greece), may play pivotal role in adapting olive cultivars to difficult environmental stresses. Moreover, Mridha et al.)10 emphasized the role of both arbuscular mycorrhizal fungi and plant growth-promoting rhizobacteria in enhancing olive growth and health when used as microbial inoculants.
Chetoui and Chemlali are Tunisian cultivars, whereas Quaissi and Djalat were of Syrian origin. Considering the aforementioned results, we noticed that the Tunisian cultivars were superior to the Syrian cultivars in bacterial counts and diversity, which proves the role of the olive genotype in shaping the rhizospheric and endophytic microbial communities, as reported by Muller et al.3.
EPS-Producing Bacteria
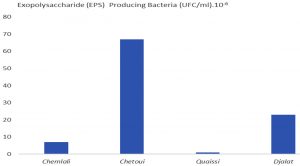
The results illustrated in Fig. 3 revealed variable counts of EPS-producing bacteria between the investigated cultivars, based on the following descending order: Chetoui (68.4 %) > Djalat (23.5%) > Chemlali (7 %) > Quaissi (1%), whereas Fig. 4 clearly illustrates the mucoid colonies with steaky texture on MacConkey agar medium indicated (EPS) production and confirmed the negativity of their Gram, and may have failed, Erwinia, Enterobacter, Klebsiella genera or Pseudomonas aeruginosa, Ralstonia, and Azotobacter vinelandii. As reported in a previous study16, the genus Paenibacillus presented maximum rhizospheric and endophytic EPS-producing bacteria17. In contrast, Fhoula et al. (2013)18 isolated good EPS-LAB producers from olive rhizosphere, mainly belonging to the species W. confusa, W. paramesenteroides, and Ln. mesenteroides.
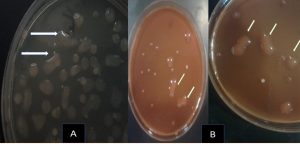
Fig. 4. Rhizospheric exopolysaccharide producing bacteria grown on NAS medium (A) MacConkey agar medium (B) mucoid colonies with steaky texture indicated (EPS) production.
EPS-producing bacteria improve the soil texture and aggregation, and also attach bacteria to plant roots19, thus protecting bacteria and plants from drought and oligotrophy by augmenting the absorption of water and nutrients from the soil. Moreover, this may help in relieving the saline stress by reducing the sodium content available for plant absorption20. Furthermore, EPSs are typically correlated with bacterial and plant resistance and protection against biotic and abiotic stress factors such as pathogens, salinity, desiccation, drought, heavy metals, and UV radiations21,22.
Exoenzyme-producing bacteria
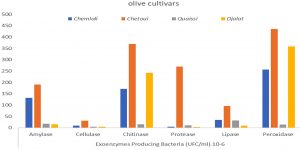
As illustrated in Fig. 5, the rhizospheric exoenzyme-producing bacteria isolated from olive cultivars revealed the superiority of the Chetoui cultivar in all tested enzyme activities, followed by Chemlali > Djalat > Quaissi. Distinct differences were found in microbial enzyme activities of the investigated rhizospheres in the following order: peroxidase > chitinase > amylase > protease > lipase > cellulase. Considering that the chitinase and peroxidase-producing bacteria (stress enzymes) were higher in Chetoui and Djalat cultivars, they may presumably play a pivotal role in adapting the olives to difficult environmental stresses. From the enzymatic profiles, it can be concluded that hydrolytic enzymes (cellulase, amylase, lipase, chitinase, and protease) linked to carbon and nitrogen cycles represent a useful index for soil organic matter evolution, as reported by Geissler et al.23. In contrast, Sofo et al. (2014)24 assessed the soil enzyme activities of β-glucosidase, dehydrogenase, FDA hydrolase, and protease sampled from olive orchard soil subjected to classical and sustainable practices. The results revealed that proteases and glucosidases activities were higher in the groves managed by sustainable agricultural treatments.
Olive rhizosphere microbial communities are remarkably complex and diverse, and are mainly shaped by the olive cultivar genotype. Culture-dependent techniques are less efficient for understanding this high complex diversity and interactivity, whereas culture-independent approaches such as high-throughput sequencing technologies and rhizosphere microbiomics may provide the full microbial composition at the species level. Moreover, they offer wealth information for comprehending the complex interactions implicated in olive tree rhizosphere microbial ecology.
Furthermore, we conclude that the Chetoui olive cultivar rhizosphere was the most diverse and the best potential reservoir for exoenzymes and EPS-producing bacteria useful for future biotechnological applications; however, future work is necessary to screen other plant growth-promoting traits or to test their biological control activities against major olive pathogens.
ACKNOWLEDGMENTS
This project has been supported by Jouf University under research project No. (627/39). We thank the deanship of scientific research for their financial support, and also gratefully acknowledges the contribution of Olive Research Unit – Camel and Pasture Research Center – Sakaka city, for providing olive cultivars rhizospheric soil samples
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct, and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
ETHICS STATEMENT
Not applicable.
AVAILABILITY OF DATA
All datasets analyzed in the study are included in the manuscript and presented as Figs.
- Shenlong Z, Erli N, Ainong S, Beiquan M. Genetic Diversity Analysis of Olive Germplasm (Olea europaea L.) With Genotyping-by-Sequencing Technology. Front Genet. 2019:10:1-11.
Crossref - Aranda S, Montes-Borrego M, Jimenez-Diaz RM, Landa BB. Microbial communities associated with the root system of wild olives (Olea europaea L. subsp. europaea var. sylvestris) are good reservoirs of bacteria with antagonistic potential against Verticillium dahliae. Plant Soil. 2011;343:329-345.
Crossref - Muller H, Berg C, Landa B, Auerbach A, Moissl-Eichinger C, Berg G. Plant genotype-specifc archaeal and bacterial endophytes but similar Bacillus antagonists colonize Mediterranean olive trees. Front Microbiol. 2015;6:1-9.
Crossref - Martins F, Pereira JA, Bota P, Bento A, Baptista P. Fungal endophyte communities in above- and belowground olive tree organs and the efect of season and geographic location on their structures. Fungal Ecol. 2016;20:193-201.
Crossref - Drenovsky RE, Elliott GN, Graham KJ, Scow KM. Comparison of phospholipid fatty acid (PLFA) and total soil fatty acid methyl esters (TSFAME) for characterizing soil microbial communities. Soil Biology and Biochemistry. 2004;36(11):1793-1800.
Crossref - Pascazio S, Crecchio C, Ricciuti P, Assunta MP, Xiloyannis C, Sofo A. Phyllosphere and carposphere bacterial communities in olive plants subjected to different cultural practices. Int J Plant Biol. 2015;6(1):15-19.
Crossref - Takahashi S, Tomita J, Nishioka K, Hisada T, Nishijima M. Development of a Prokaryotic universal primer for simultaneous analysis of Bacteria and Archaea using next-generation sequencing. PLoS One. 2014;9:1-9.
Crossref - Fernandez-Gonzalez AJ, Villadas PJ, Cabanas Cabanas CG-L, et al. Defining the root endosphere and rhizosphere microbiomes from the World Olive Germplasm Collection. Sci Rep. 2019;9:20423.
Crossref - Faiez A, Alhassen A, Khaled M, Hechmi M. The current status of the Saudi Arabian Olive sector and its future development. International Conference on Agricultural Technologies in Arid Lands. KACST, Saudi Arabia. Abstract No. OP21. 2013.
- Mridha MAU, Al-Qarawi AA, Al-Oud SS, Al-Barakah FN. Status and need of research on growth improvement of Olive (Olea europaea L.) with microbial inoculants in Saudi Arabia (Article) J Pure Appl Microbiol. 2013;7(3):1861-1868.
- Slimane C, Cheba B, Benourrad A. Screening and phenotypic diversity of amylase producing rhizospheric bacteria from some North African plants. Procedia Technology. 2016;22:1197-1204.
Crossref - Remel. Microbiology Products: Instructions for use of MacConkey Agar. 2005. http://www.remelinc.com. [28 Jun 2013]
- Cabanas CGL, Schiliro, E, Valverde-Corredor A, Mercado-Blanco J. The biocontrol endophytic bacterium Pseudomonas fluorescens PICF7 induces systemic defense responses in aerial tissues upon colonization of olive roots. Front Microbiol. 2014:5:427: 1-14.
Crossref - Cabanas CG-L, Ruano-Rosa D, Legarda G, Pizarro-Tobias P, Valverde-Corredor A, Carlos Trivino J, Roca A, Mercado-Blanco J. Bacillales Members from the olive rhizosphere are efective biological control agents against the defoliating pathotype of Verticillium dahliae. Agriculture. 2018; 8: 1-23.
Crossref - Maldonado-Gonzalez MM, Schiliro E, Prieto P, Mercado-Blanco J. Endophytic colonization and biocontrol performance of Pseudomonas fuorescens PICF7 in olive (Olea europaea L.) are determined neither by pyoverdine production nor swimming motility. Environ Microbiol. 2015;17:3139-3153.
Crossref - Iqbal A, Bhatti HN, Nosheen S, Jamil A, Malik MA. Histochemical and physicochemical Study of Bacterial Exopolysaccharides. Biotechnol. 2002;1(1):28-33.
Crossref - Rafigh SM, Yazdi AV, Vossoughi M, Safekordi AA, Ardjmand M. Optimization of culture medium and modeling of curdlan production from Paenibacillus polymyxa by RSM and ANN. Int J Biol Macromol. 2015;70:463-473.
Crossref - Fhoula I, Najjari A, Turki Y, Jaballah S, Boudabous A, Ouzari H. Diversity and Antimicrobial Properties of Lactic Acid Bacteria Isolated from Rhizosphere of Olive Trees and Desert Truffles of Tunisia. Bio Med Research International. 2013;2013:ID405708.
Crossref - Yegorenkova IV, Tregubova KV, Ignatov VV. Paenibacillus polymyxa rhizobacteria and their synthesized exoglycans in interaction with wheat roots: Colonization and root hair deformation. Curr Microbiol. 2013;66:481-486.
Crossref - Upadhyay SK, Singh JS, Singh DP. Exopolysaccharide-producing plant growth-promoting rhizobacteria under salinity condition. Pedosphere. 2011;21(2):214-222.
Crossref - Arunima B,Allison MT, Kaitlynn CS, et al. Soil microbial EPS resiliency is influenced by carbon source accessibility. Soil Biology and Biochemistry. 2020;151:108037.
Crossref - Timmusk S, Copolovici D, Copolovici L, Teder T, Nevo E, Behers L. Paenibacillus polymyxa biofilm polysaccharides antagonise Fusarium graminearum. Sci Rep. 2019;9(1):662.
Crossref - Geisseler D, Horwath WR. Relationship between carbon and nitrogen availability and extracellular enzyme activities in soil. Pedobiologia. 2009;53(1):87-98.
Crossref - Sofo A, Ciarfaglia A, Scopa I, et al. Soil microbial diversity and activity in a Mediterranean olive orchard using sustainable agricultural practices A. Soil Use and Management. 2014;30:160-167.
Crossref
© The Author(s) 2020. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.