ISSN: 0973-7510
E-ISSN: 2581-690X
Microorganisms inhabit extreme environments are novel in terms of phylogeny and also valuable source of extracellular hydrolytic enzymes. The present study focused on isolation and characterization of moderately thermophilic heterotrophic bacteria from Thar Desert using culture dependent method and assessment of their potential for production of industrially important enzymes. Screening of bacteria from sand dune samples collected from arid region of India led to isolation of total eight moderately thermophilic bacterial strains. All strains were characterized morphologically, biochemically and 16S rRNA gene sequencing analysis. The phylogenetic analysis of strains revealed that these strains affiliated to genera Bacillus (5 strains), Geobacillus (2 strains) and Aeribacillus (1 strain), which were able to grow between 25-60 °C. The present study documented the occurrence of species Geobacillus thermodenitificans, Bacillus paralicheniformis and Aeribacillus pallidus in sand dunes of Thar Desert, India. These moderately thermophilic strains showed an ability to produce one or more extracellular enzymes like amylase, protease, lipase and asparaginase. Thus, the strains isolated from Thar Desert could be potential candidates for industrial applications. The present study showed that bacterial strains exist in the same ecological niche might have ability to produce distinct enzymes.
16S rRNA gene, Bacillus, Phylogenetic analysis, Thar Desert, Moderately thermophiles
Temperature is one of the most important factor for classification of the bacteria. The optimal temperature for growth of mesophilic bacteria is around 37 °C, whereas, for moderately thermophilic bacteria, it is about 45-55 °C for moderately thermophilic bacteria, 60-80 °C for thermophilic bacteria and above 80 °C for hyperthermophilic bacteria.1–3. The enzymes produced by mesophilic bacteria have limited stability at high temperature. In the recent years, thermophilic bacteria are paid more attention because they are the potential source of thermostable extracellular hydrolytic enzymes. Thermostable protease enzyme Alcalase from Bacillus licheniformis is commercially used in processing of soy meal, protein-fortifed beverages, etc.4. Likewise, thermostable cellulases are able to avoid neutralization step in bioprocess conversation to improve lignocellulose conversion to biofuels5. Moreover, thermostable enzymes reduce the risk of contamination at higher temperatures and work against many solvents, detergent, acidic and alkaline pH in different industrial bio-processes 6,7. Thermophilic bacteria thrive in harsh environmental conditions like as geothermal vents, hot springs and desert soil8. The Thar Desert is large arid and semi-arid region in north-western part of India. It occupies about 2.34 million km2 area encompassing Rajasthan, Haryana Gujarat and South-Western Punjab9. Sandy soil of desert is a key factor that makes the mobile and fixed sand dunes by aeolian processes. Generally sand dunes represent the extreme drought condition that is hostile for the existence of life10. Thus, only extremphilic microorganisms can survive in such kind of enviornments. A few studies have been carried out to find out microbial diversity in Thar Desert11–13. Efforts have been made to explore unculturable bacterial diversity in Thar Desert soils by denaturing gradient gel electrophoresis14. The earlier studies from different arid regions of Thar Desert indicate the presence of potential thermophilic bacteria which are good source of lipase and amylase enzymes13,15. The study conducted on screening of extracellular enzymes from Yuhushiella sp. TD 032 isolated from rocky soil of Thar Desert but later it was found to be mesophilic bacterium16. The Thar Desert region of India is unique habitat with high temperature in summer and low temperature in winter and poor in availability of water because of sandy soil that make the desert soil an extreme ecosystem. Considering presence of these harsh conditions and paucity of work on culturable thermophilic bacteria and their ability for production of extracellular enzymes, the present study was undertaken to isolate and characterise strains of eubacteria using conventional methods and 16S rRNA gene sequencing. The work was extended for investigation of industrially important enzymes from isolated moderately thermophilic strains to evaluate their potential applications.
Site description, sample collection and physico-chemical analysis
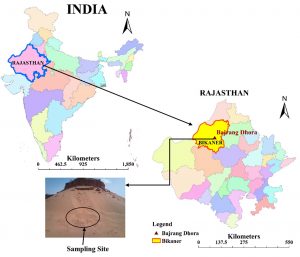
Thar Desert also known as Great Indian Desert covers about 2.34 million km2 area within India and mainly Bikaner, Barmer, Jaisalmer and Jodhpur districts of Rajasthan encompassing 61% boundary of Thar Desert. It is a large arid region dominated by fixed and mobile sand dunes. The sand dune samples were collected from Bajrang Dhora (28°03’18.41″ N 73°15’58″ E) which is located almost 11 km away from Bikaner (Rajasthan), India (Fig. 1). Samplings were carried out in the month June, 2014 and February 2015. Sand dune samples were collected from 10 cm depth of dune surface with the help of sterile spatula and transferred in to sterilized zip lock polythene bags and transported aseptically to laboratory. Physico-chemical conditions (temperature, pH, conductivity, total dissolve solids and salinity) of the soil during sampling were determined at the sampling site by multi-parameter PCSTestrTM35 (Eutech instrument, Oakton, Singapore).
Fig. 1. Location of sampling sites at Bajrang Dhora, Thar Desert, Bikaner, Rajasthan, India. (Outline map source: Google map)
Isolation and purification of bacterial isolates
Serial dilution method was used for bacterial isolation. Sandy soil sample (1 g) was suspended in 100 mL of sterilized distilled water, slightly homogenized and spreaded onto nutrient agar plates. The plates were incubated at 55 ºC for 24 h13. The colonies grown on the plates were purified by successive streaking on nutrient agar plates. Isolated bacterial strains were then subsequently stored at 4 ºC in refrigerator for further study.
Determination of optimum temperature and pH for growth
The physiological growth studies were carried out in the nutrient broth medium as described previously 17. The temperature range for optimum growth was determined by incubating the isolates from 25 to 60 ºC. The effect of pH on growth of isolates was tested with the pH range from 4.0 to 10.0. All growth tests were done at 50 ºC for 24 h and the growth was measured at OD600 nm by using spectrophotometer (HALO DB-30 UV-Visible double beam, Dynamica, Singapore).
Morphological studies of colony and cell
The appearance of colony such as shape, colour, opacity, margin and elevation was observed by magnifying lens (10X) after the growth of isolated strains on nutrient agar plates. The cell size was measured using ocular micrometer. Gram staining was performed using standard18. The various morphological features of bacterial strains were observed at 1000X using Axio Lab.A1 compound microscope (Carl Zeiss, Germany).
Biochemical characterization
A series of biochemical tests were performed at 50 ºC for 24 h at pH 7.0 using standard protocols19. Briefly, catalase activity was determined by adding a 3 % (w/v) H2O2 solution on colonies of isolates. Oxidase test was done by placing a loopful bacterial isolate on oxidase disc (Himedia, India) and the change in color was observed. Indole test was performed in tryptone broth after adding 0.5 ml Kovac’s reagent and waited for color change. Methyl Red (MR) and Voges-Proskauer (VP) tests were performed for acid production ability of strains. Utilization of citrate by isolates was observed on Simmons citrate agar (Himedia, India) medium slants. Urease enzyme production from urea was determined with suspension of bacterial strains inoculated in Christensen’s urea broth (CUB). Reduction of nitrate was determined in nitrate broth medium. Fermentation of carbohydrates was performed using triple sugar iron agar medium.
Screening of isolates for extracellular hydrolytic activities
Extracellular protease production
The qualitative screening of protease enzyme production by isolates was achieved using skim milk in nutrient agar medium with following components (g l-1): skim milk, 10; peptic digest of animal tissue, 5; sodium chloride, 5; beef extract, 1.5; yeast extract, 1.5; and agar, 15 and pH adjusted to 7.0 before autoclaving. Isolates were streaked on medium plates. A zone of hydrolysis around the colonies was considered as an evidence of proteolytic activity 20.
Extracellular amylase production
The amylase production was determined by growing the isolates on soluble starch containing nutrient agar medium with the following constituents (g l-1): soluble starch, 10; peptic digest of animal tissue, 5; sodium chloride, 5; beef extract, 1.5; yeast extract, 1.5; and agar, 15 (pH adjusted to 7.0 before autoclaving). Isolates were streaked in straight lines on medium plates. After incubation, individual plates were flooded with Gram’s iodine solution (3% (w/v), KI and 0.3% (w/v) iodine). A clear zone around the growth indicated hydrolysis of starch21.
Extracellular lipase production
Lipase activity of the isolates was detected by screening for turbid halo or zones of precipitation around colonies growing on nutrient agar plates containing 1% Tween-8022.
Extracellular asparaginase production
The ability of isolates to produce asparaginase was determined using nutrient agar medium with L-asparagine. The composition of the test medium was as follows (g l-1): L-asparagine, 20; glucose, 2; potassium chloride, 0.5; magnesium sulfate heptahydrate, 0.5; monopotassium phosphate, 1.52; agar, 20 and phenol red, 0.09 and pH 7.0. Clear pink zone around the colonies after the incubation of petri plates indicated the asparaginase production23.
Extracellular inulinase production
The production of inulinase enzyme by isolates was screened using inulin agar medium plates containing following constituents (g l-1): ammonium sulfate, 0.5; magnesium sulfate heptahydrate, 0.2; inulin, 2; monopotassium phosphate, 3; agar, 20 and pH 7.0. After incubation, plates were flooded with Lugol’s iodine solution for 3 to 5 minutes and washed with distilled water. The colorless zone around the colonies indicated inulinase enzyme production24.
Extracellular cellulase production
Cellulolytic activity of isolates was detected on carboxymethylcellulose (CMC) agar plates. Isolates were streaked on CMC agar medium plates and incubated at 50 °C for 24 h to allow the secretion of cellulase. After the incubation, plates were flooded with congo red (1% w/v) for 15 min. Subsequently, congo red solution was discarded and 1M NaCl solution added on plates for 15 min. The hydrolysis zone around the colonies indicated the cellulolytic activity 6.
Genomic DNA extraction
Genomic DNA of isolates was extracted using modified freeze/thaw method25. Briefly, bacterial culture was pelleted out at 9,000 g for 5 min and lysis solution (500 µl) added (mixed gently by inverting 3-6 times) and incubated at -20 °C for 15 minutes. Later, tubes were incubated at 70 °C for 15 min in water bath and this step was repeated. Cells were suspended in 600 µl of Chloroform and gently emulsified by inversing tubes for 3-5 times and centrifuged at 9,000 g for 3 min. Upper aqueous phase containing DNA was transferred to a new tube and equal amount of Propan-2-OL (iso-propyl alcohol) was added and mixed gently by inversion at room temperature and centrifuged at 9,000 g for 3 min. Supernatant was removed completely and DNA pellet was dissolved in 100 µl of 1.2 M NaCl solution. 300 µl of absolute cold ethanol added and the DNA was precipitated in 10 min at -20 °C. Later, the tubes were centrifuged at 9,000 g for 5 min. Ethanol was removed and pellet washed once with 70% cold ethanol and tubes were centrifuged at 9,000 g for 5 min. Lastly, the pellet was dissolved in 50 µl deionized water (pH 7.0) by gentle vortexing and kept at -20 °C for further use.
16S rRNA gene Sequencing and phylogenetic analysis
The 16S rRNA gene of genomic DNA was amplified using PCR master mix (GeneiTM Company) with the universal primers 27F and 1495R26. The PCR thermal cycling conditions were as follows: Hot start at 98 °C for 5 min, followed by 35 cycles denaturation at 95 °C for 30 s, primer annealing at 56 °C for 45 s, elongation at 72 °C for 1.45 min with final extension at 72 °C for 5 min. Amplified genomic DNA was purified with GeneJET PCR purification kit (Thermo Fisher) as per the manufacturer’s protocol. Purified products were sequenced commercially (Eurofins Genomics India, Bangalore) to get partial sequences 16S rRNA gene.
The similarity search of 16S rRNA gene sequences of isolated strains was done through the Eztaxon database27 and parallel search also performed using NCBI database using BLASTN (www.ncbi.nlm.nih.gov/blastn) to retrieve closely related sequences for phylogenetic analysis. Initially, a large group of sequences (>1100 bp) was selected and later a smaller subset was employed for analysis based on the relatedness of the sequences and excluding taxa of uncertain affiliation. The multiple sequences alignment was performed using programme MUSCLE28 inbuilt in the phylogenetic analysis software MEGA 729. The aligned sequences were further refined manually to obtain unambiguous data. Finally, the sequences of type strains and other bacterial strains mostly allied to sequences of the present study were employed for a synoptic view of the phylogenetic tree. The phylogenetic tree using 16S rRNA gene fragment of 1117 nucleotides was constructed by the maximum likelihood (ML) method using the program MEGA 729 applying a T92+G model of nucleotide substitution, which gave the best fit to this dataset according to the Bayesian Information Criterion (BIC). Bootstrapping of 1000 replications was calculated to find out confidence values for the edges of the maximum likelihood tree. Planococcus citreus (X62172) was chosen as the outgroup.
Nucleotide sequence accession numbers
The nucleotide sequences determined in this study have been deposited in the GenBank (NCBI database) under accession numbers MH560567-MH560574.
Physico-chemical properties of sand dunes samples of Thar Desert
In the present study, the physico-chemical properties of sand dune samples named Sp1S, Sp2S, Sp3S and Sp4S (June 2014), Sp2W, Sp3W and Sp4W (February 2015) from Thar Desert were determined, as shown in Table 1. The Thar Desert sand dunes in both seasons represent the variation in temperature between 55 to 25 °C. The sand samples showed properties from neutrophilic to slightly alkaliphilic (pH 7.6 to 8.4). The electrical conductivity (µs) and total dissolved solids (ppm) in the sand samples were found to be more in winter season in comparison to summer season (Table 1).
Table (1):
Physicochemical analysis of Sand Dune Samples of Thar Desert.
Sample |
pH |
Salinity (ppm) |
Conductivity (µs) |
TDS (ppm) |
|---|---|---|---|---|
Sp1S |
7.6 |
ND |
0.05 |
0.02 |
Sp2S |
7.5 |
ND |
0.11 |
0.06 |
Sp3S |
7.7 |
ND |
0.05 |
0.02 |
Sp4S |
7.8 |
ND |
0.05 |
0.03 |
Sp2W |
8.2 |
40 |
0.07 |
50.5 |
Sp3W |
8.3 |
52.4 |
0.1 |
71.5 |
Sp4W |
8.4 |
77.8 |
0.155 |
110 |
ppm=part per million; µs=microsiemens; ND=not detected
Colony and cell morphology
The present study revealed that all isolated strains (1S, 2AS, 2BS, 3S, 2AW, 3W and 4W) were motile, gram positive rods, spore forming and arranged in chain form which indicated that isolates belonged to Bacillus species. All isolated strains showed growth on nutrient agar plate within 24 h of incubation at 50 °C. Morphologically, all isolates showed some variation in shape, size, color, light transmission, margin, opacity, elevation of the colonies (Table 2 & 3). Isolates were found basically in two different shapes; circular and irregular. The colonies of isolates were white and creamy- white in color. The cell size varied in the range of width 0.1-0.5 µm and length 0.3-1.6 µm. Colonies of strains 1S, 2AS, 2BS, 3S and 4S were observed to be opaque and strains 2AW, 3W and 4W formed translucent colonies on nutrient agar plates.
Table (2):
Morphological features of isolates.
Isolate name |
Shape |
Colour |
Opacity |
Elevation |
Margin |
Light transmission |
|---|---|---|---|---|---|---|
1S |
Irregular |
Creamy- white |
Rough |
Raised |
Undulate |
Opaque |
2AS |
Circular- large |
Creamy-white |
Rough |
Flat |
Entire |
Opaque |
2BS |
Circular-small |
Creamy-white |
Smooth |
Raised |
Entire |
Opaque |
3S |
Irregular |
White |
Rough |
Raised |
Undulate |
Opaque |
4S |
Circular-moderate |
Creamy-white |
Smooth |
Raised |
Entire |
Opaque |
2AW |
Irregular |
White |
Rough |
Flat |
Curled |
Translucent |
3W |
Circular-large |
Creamy-white |
Smooth |
Raised |
Entire |
Translucent |
4W |
Circular-moderate |
White |
Smooth |
Raised |
Entire |
Translucent |
Physiological and biochemical characterization
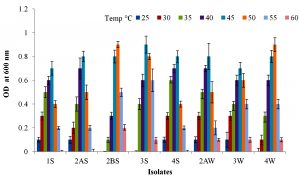
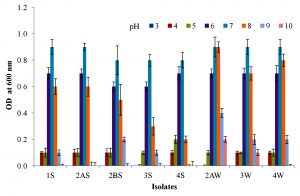
Physiological parameters such as temperature and pH were optimized for the growth of all investigated strains (Fig. 2 & 3). The optimum temperature range for growth of all the strains was determined by incubating at 25 to 60 °C for 24 h. It was noted that strains 1S, 2AS, 4S and 3W were able to grow between 25-55 °C with optimum temperature of 45 °C. Strains 2BS, 3S, 2AW and 4W were able to grow between 30-60 °C with optimum temperature of 50, 45, 45 and 50 °C respectively. The optimum pH range for growth of all strains was observed 7.0-8.0. All strains showed maximum growth at slightly alkaline condition.
Table (3):
Biochemical characterization and phylogenetic relatedness of strains isolated from Thar Desert.
Characteristics |
1S |
2AS |
2BS |
3S |
4S |
2AW |
3W |
4W |
|---|---|---|---|---|---|---|---|---|
Gram staining |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
Cell size (width, length µm) |
0.1-0.3×1-2 |
0.2-0.3×1-3 |
0.5-0.8×1-4 |
0.5-0.8×1-8 |
0.1-0.4×1-3 |
0.2-0.3×1-5 |
0.3-0.6×1-3 |
0.4-0.8×1-6 |
Motility |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
Endospore |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
Temperature range (°C) |
25-55 |
30-55 |
30-60 |
30-60 |
25-55 |
25-60 |
30-60 |
30-60 |
pH range |
6-8 |
6-8 |
6-8 |
6-8 |
6-8 |
6-11 |
6-10 |
6-8 |
Indole |
– |
– |
– |
– |
– |
– |
– |
– |
Triple sugar Iron |
+ |
+ |
– |
– |
+ |
+ w |
+ |
+ |
MR |
– |
– |
– |
– |
+ |
– |
+ |
– |
VP |
+ |
+ |
– |
– |
+ |
+ |
+ |
+ |
Nitrate |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
H2S |
– |
– |
– |
– |
– |
– |
– |
– |
Citrate |
+ |
+ |
– |
– |
+ |
+ |
+ |
+ |
Catalase |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
Oxidase |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
Phylogenetic similarity |
Bacillus subtilis subsp. inaquosorum |
Bacillus subtilis subsp. subtilis |
Aeribacillus pallidus |
Geobacillus thermodenitrificans subsp. calidus |
Bacillus subtilis subsp. inaquosorum |
Bacillus paralicheniformis |
Bacillus paralicheniformis |
Geobacillus thermodenitrificans subsp. calidus |
+ indicates positive; – indicate negative; +w indicates weakly positive
The physiological and biochemical characteristics of strains observed by classical methods (Table 3) showed similarity with phylogenetically related strains. Strain 1S and 4S exhibited similar characteristics to phylogenetically close Bacillus subtilis subsp. inaquosorum as it was gram positive, rod shaped catalase positive, nitrate reduction positive, citrate positive, motile and optimum growth 45 °C. The strain 2BS showed alike characteristics to Aeribacillus pallidus as it was small rod shaped, gram positive, motile, catalase positive, oxidase negative, unable to degrade starch and casein and optimum growth at 50 °C. Likewise, the strain 2AS was phylogenetically related to Bacillus subtilis hswx88 which showed features such as gram positive, motile, indole negative, nitrate reduction positive, starch hydrolysis positive, urease production negative. The strains 3S and 4W showed similar characteristics as Geobacillus thermodenitrificans subsp. calidus, as it was gram positive, rod shaped, motile, catalase positive, oxidase positive, nitrate reduction positive, amylase production positive and optimum growth was observed at 45 °C and 50 °C respectively. The 2AW and 3W strains exhibited similarity with Bacillus paralicheniformis, as they were gram positive, rod shaped, motile, catalase positive, nitrate reduction positive, able to hydrolysis of starch, casein and optimum growth at 45 °C.
Production of extracellular hydrolytic enzymes
The isolated strains were tested for their ability to produce six industrially important hydrolytic enzymes (Table 4). Interestingly, it was noted that except 2BS and 3S all other strains showed multi hydrolytic enzyme production ability. The strains 1S, 2AS, 2BS, 3S, 4S, 2AW, 3W and 4W were able to produce amylase enzyme. The enzymes such as protease, lipase and cellulase were produced by 1S, 2AS, 4S, 2AW, 3W and 4W. Nevertheless, inulinase enzyme production was only shown by strains 2AS, 2AW and 3W. It is worth to mention that strain 2AW and 3W were able to produce asparaginase enzyme along with other enzymes.
Table (4):
Screening of industrially putative enzymes from isolated strains.
Strain no. |
Amylase |
Protease |
Cellulase |
Asparaginase |
Inulinase |
Lipase |
|---|---|---|---|---|---|---|
1S |
+ |
+ |
+ |
– |
– |
+ |
2AS |
+ |
+ |
+ |
– |
+ |
+ |
2BS |
+w |
– |
– |
– |
– |
– |
3S |
+ |
– |
– |
– |
– |
– |
4S |
+ |
+ |
+ |
– |
– |
+ |
2AW |
+ |
+ |
+ |
+ |
+ |
+ |
3W |
+ |
+ |
+ |
+ |
+ |
+ |
4W |
+ |
+ |
+ |
– |
– |
+ |
+ indicates positive; – indicates negative; +w indicates weakly positive
Phylogenetic analysis
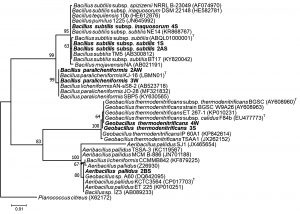
16S rRNA gene nucleotide sequencing data of representative strains were analyzed for similarity with BLAST search (http://www.ncbi.nlm.nih.gov). The sequencing results revealed that the genus Bacillus was predominant rather than Geobacillus and Aeribacillus in sand dunes of Thar Desert. The five strains (1S, 2AS, 4S, 3W and 2AW) were found to belong to genus Bacillus. Other two strains (3S, 3W and 4W) were related to genus Geobacillus and one of strains (2BS) was affiliated to genus Aeribacillus (Fig. 4).
The extreme environments which exist on Earth are the rich source of variety of microorganisms with special characteristics. Low water availability and high temperature during summer and diurnal fluctuations make the Thar Desert an extreme envrionment for microbial life11. In the present study, total eight moderately thermophilic bacterial strains were screened out from different sand dune samples of Thar Desert, Bikaner, Rajasthan using conventional studies such as morphological, physiological, biochemical characterization and molecular identification by 16S rRNA gene sequencing. The results obtained from effect of different temperature range (25-60 °C) revealed that all studied strains were belonged to moderately thermophilic. The 16S rRNA gene sequence similarity (1419 bp) exhibited that isolated strains were related to different species of bacilli such as 1S and 4S to Bacillus subtilis subsp. inaquosorum30 ; 3S and 4W to Geobacillus thermodenitrificans subsp. calidus31 ; 2BS to Aeribacillus pallidus 32 ; 3W and 2AW to Bacillus paralicheniformis33 and 2AS to Bacillus subtilis hswx8834.
The results obtained in the present study for biochemical characterization such as oxidase, catalase, MR-VP, citrate utilization, indole production, triple sugar iron utilization, nitrate reduction were consistent with earlier thermophilic bacteria isolated from various geothermal areas. Moreover, biochemical characters of moderately thermophilic bacteria vary from strain to strain even if they are isolated from same ecological niche as revealed from our study and previous findings13,17,35,36 .
The hydrolytic properties of thermostable enzymes have long driven in many industries such as food, cosmetic, agriculture, pharmaceutical, leather because they increase the rate of reaction, reduces the risk of contamination, increases reactant solubility, increases the stability of enzyme37. Among of thermophilic bacteria of genus Bacillus and Geobacillus are dominant for the production potential thermostable enzymes15,30,38. Nevertheless, many thermophilic bacteria are still unexplored which possess ability to produce variants of thermostable enzymes. It is obvious from the findings of the present study that strains isolated from sand dunes of Thar Desert are able to produce at least one and more than two hydrolytic enzymes. Worldwide, thermophilic bacteria having hydrolytic activities were mostly found in hot spring17,35. Moreover, greater hydrolytic activity was observed in gram positive thermophilic bacteria rather than gram negative from different hot springs6,35,36. The extracellular thermostable enzymes of thermophilic bacteria have great interest for both fundamental research and industrial application39. The number of thermophilic bacteria affiliated to genus Bacillus sp. and Geobacillus sp. like Bacillus licheniformis, Anoxybacillus sp. NBY46, Bacillus mycoides NBM19, Bacillus pumilus NBM31, Bacillus sp. NBM43, Bacillus subtilus NBM48, Geobacillus sp. TP-2, Aeribacillus sp. O3, Bacillus sp. O2, O7, O8, O14, O17 and O19 have been isolated from different geothermal springs and reported as producer of various thermostable hydrolytic extracellular enzymes such as amylase, protease, cellulase, gelatinase, xylanase, lecithinase and lipase6,17,40.
The enzyme asparaginase has been reported as a source of high therapeutic agent for acute lymphocytic leukemia and Hodgkin’s lymphomas and also able to reduce acrylamide level commercial fried foods22,41. Recently, the asparaginase enzyme of Bacillus licheniformis was used to treat acute lymphoblastic leukemia (ALL) and in vitro evaluation of anti-cancerous properties42,43. In the previous studies, asparaginase production has been reported from thermophilic bacteria belonging to Bacillus sp. HB844, Bacillus subtilis hswx88 33, Geobacillus thermoleovorans N7 originated mostly from hot springs. In our investigations, only two strains (2AW and 3W) belonging to Bacillus paralicheniformis showed ability to produce asparaginase. To the best of our knowledge, this is the first report to show asparaginase production by Bacillus strains originated from sand dunes of Thar Desert.
Likewise, inulinase enzyme producing thermophilic bacteria have great industrial interest because this enzyme is used to hydrolyse inulin for production of fuel ethanol45. The strains LT-2, LT-4, LT-5, LT-6, LT-7, LT-8 (Bacillus licheniformis) and LMa-9 (Aneuribacillus migulanus) isolated from sediment and water samples of geothermal areas in Patagonia, Argentina46 and the strain Bacillus sp. SG7 isolated from thermal water samples from the region of Velingrad, Bulgaria was able to produce extracellular inulinase47. Interestingly, the strains (2AW and 3W) those were able to produce asparaginase also showed potential to produce extracellular inulinase. Besides these strains, 2AS also exhibited positive test for this enzyme. So far, none of study described the inulinase producing bacteria isolated from Thar Desert, India. The ability of production of amylase, protease, lipase, asparaginase, inulinase and cellulase enzymes by strains isolated from Thar Desert indicates their potential use in different industries like textile, food, pharmaceutical, detergent, leather, agrochemicals, etc.
On the basis of phylogenetic analysis, the strains 1S and 4S were closely related to type strain B. subtilis subsp. inaquosorum (CP013984) with 99.92 and 100% respectively 29,46. The sequencing results revealed that the genus Bacillus was predominant rather than Geobacillus and Aeribacillus in sand dunes of Thar Desert. The four strains (1S, 2AS, 4S and 2AW) were found to be belonged to genus Bacillus. The strains 1S and 4S were closely related to type strain B. subtilis subsp. inaquosorum (CP013984) with 99.92 and 100% respectively. The phylogenetic tree consists of two main clades and each clade is divided into two sub-clades. Two distinct sub-clades of Bacillus genus were observed in the phylogenetic tree (Fig. 4). One sub-clade virtually intricate group of closely related species including Bacillus subtilis subsp. subtilis. Most of the sequence from isolates originated from India are yet unpublished and needs further validation (data not shown). The present study described the morphological, biochemical and production of hydrolytic enzymes of strain related to Bacillus subtilis subsp. subtilis. Another sub-clade consisted sequences related to Bacillus licheniformis with the type strain of B. paralicheniformis32. It is clearly indicated from our results that earlier reported species belong to paralicheniformis instead of licheniformis.
The Thar Desert soil is an evidence of ecological niches of Aeribacillus pallidus as the strain 2BS showed the closeness to type strain KCTC3564 of Aeribacillus pallidus. Most of the strains of genus Aeribacillus isolated from hot springs, geothermal oil field and desert soils48,49. The strain Aeribacillus pallidus SAT4 was isolated from Thar Desert, Pakistan used to produce antibacterial peptide 50. The sequence of 16S rRNA gene was not included in tree because of a short sequence (432 bp) available in GenBank. The alkalistable carbonic anhydrase producing Aeribacillus pallidus TSHB1 has been isolated from Choti Anhoni hot spring of Pipariya, Madhya Pradesh, India51. These findings reveal that Aeribacillus is widely distributed in different habitat.
The strains 3S and 4W were closely related to type strain F84B of Geobacillus thermodenitrificans subsp. calidus isolated from well-pipeline sediment sample in Kizilcahamam, Turkey 30. Recently, most of the strains of this genus were isolated from compost, deep surface oil reservoirs, hot springs and soils36,52,53. Geobacillus thermodenitrificans strains have also been isolated from Tattapani hot spring sediment, North West Himalayas and from Damodar river, India54. In the earlier studies, a few of other strains were reported from intestine of a mackerel (fish) and rhizosphere of plant55,56. The present study exhibited the occurrence of Geobacillus thermodenitrificans in sandy soil of Thar Desert of India.
The present study revealed that Thar Desert is a treasure for moderately thermophilic bacterial diversity. The presence of strains related to species such as Geobacillus thermodenitrificans, Bacillus paralicheniformis, Bacillus subtilis and Aeribacillus pallidus in Thar Desert of India can tolerate two extreme conditions (high temperature and aridity). The investigated strains were able to produce thermostable amylase, cellulase, protease, inulinase, asparaginase and lipase enzymes. Thus, microorganisms thriving in such harsh environments should be explored extensively for phylogenetic studies and also for their biotechnological applications.
Acknowledgements
The authors would like to thank Department of Microbiology, Central University of Rajasthan, for providing the basic infrastructural facilities required for the present work. DKN is thankful to University Grant Commission, New Delhi and AKT to Central University of Rajasthan for providing financial support in the form of fellowships.
Conflict of Interest
The authors declare that they have no conflicts of interest.
- Zeikus J G. Thermophilic bacteria: ecology, physiology and technology. Enzyme Microb Technol, 1979; 1 (4): 243–252.
- Stetter K O, Fiala G, Huber G, Huber R, Segerer A. Hyperthermophilic microorganisms. FEMS Microbiol Lett, 1990; 75 (2): 117–124.
- Rawlings D E. Mesophilic, autotrophic bioleaching bacteria: description, physiology and role. In Biomining; Springer, 1997; pp. 229–245.
- Barzkar N, Homaei A, Hemmati R, Patel S. Thermostable marine microbial proteases for industrial applications: scopes and risks. Extremophiles, 2018; 22(3): 335-346.
- Bhalla A, Bansal N, Kumar S, Bischoff K M, Sani R K. Improved lignocellulose conversion to biofuels with thermophilic bacteria and thermostable enzymes. Bioresour Technol, 2013; 128: 751–759.
- Mohammad B T, Al Daghistani H I, Jaouani A, Abdel-Latif S, Kennes C. Isolation and Characterization of Thermophilic Bacteria from Jordanian Hot Springs: Bacillus licheniformis and Thermomonas hydrothermalis Isolates as Potential Producers of Thermostable Enzymes. Int J Microbiol, 2017; pp. 1-12.
- Gomes J, Steiner W. The biocatalytic potential of extremophiles and extremozymes. Food Technol Biotechnol, 2004; 42 (4): 223–235.
- Antranikian G, Vorgias C E, Bertoldo C. Extreme environments as a resource for microorganisms and novel biocatalysts. In Marine biotechnology I; Springer, 2005; pp. 219–262.
- Sinha R K, Bhatia S, Vishnoi R. Desertification control and rangeland management in the Thar desert of India. In Rala Report no. 200; 1996; pp. 115–123.
- Tsoar H. Sand dunes mobility and stability in relation to climate. Phys A Stat Mech its Appl, 2005; 357 (1): 50–56.
- Bhatnagar A, Bhatnagar M. Microbial diversity in desert ecosystems. Curr Sci, 2005; pp. 91–100.
- Songara D, Kaur S. DNA Based Identification and Characterization of Thermophilic Streptomyces sp. From Desert Soil of Rajasthan. Int J Curr Microbiol App Sci, 2013; 2 (10): 418–427.
- Gaur D, Jain P K, Bajpai V. Production of extracellular a-amylase by thermophilic Bacillus sp. isolated from arid and semi-arid region of Rajasthan, India. J Microbiol Biotechnol Res, 2017; 2 (5): 675–684.
- Kumar Gothwal R, Kumar Nigam V, Mohan M K, Sasmal D, Ghosh P. Extraction of bulk DNA from Thar Desert soils for optimization of PCR-DGGE based microbial community analysis. Electron J Biotechnol, 2007; 10 (3): 400–408.
- Bhosale H, Shaheen U, Kadam T. Characterization of a hyperthermostable alkaline lipase from Bacillus sonorensis 4R. Enzyme Res, 2016; pp. 1-11.
- Ibeyaima A, Rana J, Dwivedi A, Gupta S, Sharma S K, Saini N, Sarethy I P. Characterization of Yuhushiella sp. TD-032 from the Thar Desert and its antimicrobial activity. J Adv Pharm Technol Res, 2016; 7(2): 32.
- Baltaci M O, Genc B, Arslan S, Adiguzel G, Adiguzel A. Isolation and Characterization of Thermophilic Bacteria from Geothermal Areas in Turkey and Preliminary Research on Biotechnologically Important Enzyme Production. Geomicrobiol J, 2017; 34 (1): 53–62.
- Gram C. The differential staining of Schizomycetes in tissue sections and in dried preparations. Fortschitte der Med, 1884; 2: 185–189.
- Sharma A, Pandey A, Shouche Y S, Kumar B, Kulkarni G. Characterization and identification of Geobacillus spp. isolated from Soldhar hot spring site of Garhwal Himalaya, India. J Basic Microbiol, 2009; 49 (2): 187–194.
- Banerjee U C, Sani R K, Azmi W, Soni R. Thermostable alkaline protease from Bacillus brevis and its characterization as a laundry detergent additive. Process Biochem, 1999; 35 (1–2): 213–219.
- Sharma A, Satyanarayana T. High maltose-forming, Ca2+-independent and acid stable a-amylase from a novel acidophilic bacterium, Bacillus acidicola. Biotechnol Lett, 2010; 32 (10): 1503–1507.
- Gutiérrez C, González C. Method for simultaneous detection of proteinase and esterase activities in extremely halophilic bacteria. Appl Microbiol, 1972; 24 (3): 516.
- Bhagat J, Kaur A, Chadha B S. Single step purification of asparaginase from endophytic bacteria Pseudomonas oryzihabitans exhibiting high potential to reduce acrylamide in processed potato chips. Food Bioprod Process, 2016; 99: 222–230.
- Li A-X, Guo L-Z, Fu Q, Lu W-D. A simple and rapid plate assay for screening of inulindegrading microorganisms using Lugol’s iodine solution. African J Biotechnol, 2011; 10 (46): 9518–9521.
- Mirmohammadsadeghi H, Abedi D, Mohmoudpour H R, Akbari V. Comparison of five methods for extraction of genomic DNA from a marine Archaea, Pyrococcus furiosus. Pak J Med Sci, 2013; 29(1)Suppl: 390-394..
- Chen X, Du X, Wang W, Zhang J, Sun Z, Liu W, Li L I, Sun T, Zhang H. Isolation and identification of cultivable lactic acid bacteria in traditional fermented milk of Tibet in China. Int J dairy Technol, 2010; 63 (3): 437–444.
- Jimat D N, Mohamed I B F, Azmi A S, Jamal P. Purification And Partial Characterization Of L-Asparaginase Enzyme Produced By Newly Isolate Bacillus Sp. IIUM Eng J, 2017; 18 (2): 1–10.
- Edgar R C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res, 2004; 32 (5): 1792–1797.
- Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol, 2016; 33 (7): 1870–1874.
- Rooney A P, Price N P J, Ehrhardt C, Swezey J L, Bannan J D. Phylogeny and molecular taxonomy of the Bacillus subtilis species complex and description of Bacillus subtilis subsp. inaquosorum subsp. nov. Int J Syst Evol Microbiol, 2009; 59 (10): 2429–2436.
- Cihan A C, Ozcan B, Tekin N, Cokmus C. Geobacillus thermodenitrificans subsp. calidus, subsp. nov., a thermophilic and a-glucosidase producing bacterium isolated from Kizilcahamam, Turkey. J Gen Appl Microbiol, 2011; 57 (2): 83–92.
- Scholz T, Demharter W, Hensel R, Kandler O. Bacillus pallidus sp. nov., a new thermophilic species from sewage. Syst Appl Microbiol, 1987; 9 (2): 91–96.
- Dunlap C A, Kwon S-W, Rooney A P, Kim S-J. Bacillus paralicheniformis sp. nov., isolated from fermented soybean paste. Int J Syst Evol Microbiol, 2015; 65 (10): 3487–3492.
- Pradhan B, Dash S K, Sahoo S. Screening and characterization of extracelluar L-asparaginase producing Bacillus subtilis strain hswx88, isolated from Taptapani hotspring of Odisha, India. Asian Pac J Trop Biomed, 2013; 3 (12): 936–941.
- Zahoor S, Javed M M, Babarl M E. Characterization of a novel hydrolytic enzyme producing Thermophilic bacterium isolated from the hot spring of Azad Kashmir-Pakistan. Brazilian Arch Biol Technol, 2016; 59: 1-13.
- Sen S K, Raut S, Satpathy S, Rout P R, Bandyopadhyay B, Mohapatra P K Das. Characterizing novel thermophilic amylase producing bacteria from Taptapani hot spring, Odisha, India. Jundishapur J Microbiol, 2014; 7 (12): e11800.
- Hasan F, Shah A A, Hameed A. Industrial applications of microbial lipases. Enzyme Microb Technol, 2006; 39 (2): 235–251.
- Balan A, Ibrahim D, Abdul Rahim R, Rashid A, Azzahra F. Purification and characterization of a thermostable lipase from Geobacillus thermodenitrificans IBRL-nra. Enzyme Res, 2012; pp. 1-7.
- Shrestha N, Chilkoor G, Vemuri B, Rathinam N, Sani R K, Gadhamshetty V. Extremophiles for microbial-electrochemistry applications: a critical review. Bioresour Technol, 2018; 255: 318-330.
- Sahay H, Mahfooz S, Singh A K, Singh S, Kaushik R, Saxena A K, Arora D K. Exploration and characterization of agriculturally and industrially important haloalkaliphilic bacteria from environmental samples of hypersaline Sambhar lake, India. World J Microbiol Biotechnol, 2012; 28 (11): 3207–3217.
- Schrappe M, Reiter A, Ludwig W-D, Harbott J, Zimmermann M, Hiddemann W, Niemeyer C, Henze G, Feldges A, Zintl F. Improved outcome in childhood acute lymphoblastic leukemia despite reduced use of anthracyclines and cranial radiotherapy: results of trial ALL-BFM 90. Blood, 2000; 95 (11): 3310–3322.
- Mahajan R V, Kumar V, Rajendran V, Saran S, Ghosh P C, Saxena R K. Purification and characterization of a novel and robust L-asparaginase having low-glutaminase activity from Bacillus licheniformis: in vitro evaluation of anti-cancerous properties. PLoS One, 2014; 9 (6): e99037.
- Sudhir A P, Agarwaal V V, Dave B R, Patel D H, Subramanian R B. Enhanced catalysis of l-asparaginase from Bacillus licheniformis by a rational redesign. Enzyme Microb Technol, 2016; 86: 1–6.
- Pritsa A A, Kyriakidis D A. L-asparaginase of Thermus thermophilus: Purification, properties and identificaation of essential amino acids for its catalytic activity. Mol Cell Biochem, 2001; 216 (2): 93–101.
- Gao W, Bao Y, Liu Y, Zhang X, Wang J, An L. Characterization of thermo-stable endoinulinase from a new strain Bacillus smithii T7. Appl Biochem Biotechnol, 2009; 157 (3): 498–506.
- Cavello I, Urbieta M S, Segretin A B, Giaveno A, Cavalitto S, Donati E R. Assessment of Keratinase and Other Hydrolytic Enzymes in Thermophilic Bacteria Isolated from Geothermal Areas in Patagonia Argentina. Geomicrobiol J, 2018; 35 (2): 156–165.
- Gavrailov S, Ivanova V. Isolation and characteristics of a thermophilic Bacillus strain, producer of inulinase. J Biosci Biotechnol, 2014; pp. 83-94
- Yasawong M, Areekit S, Pakpitchareon A, Santiwatanakul S, Chansiri K. Characterization of thermophilic halotolerant Aeribacillus pallidus TD1 from Tao dam hot spring, Thailand. Int J Mol Sci, 2011; 12 (8): 5294–5303.
- Yildirim V, Baltaci M O, Ozgencli I, Sisecioglu M, Adiguzel A, Adiguzel G. Purification and biochemical characterization of a novel thermostable serine alkaline protease from Aeribacillus pallidus C10: a potential additive for detergents. J Enzyme Inhib Med Chem, 2017; 32 (1): 468–477.
- Muhammad S A, Ahmed S. Production and characterization of a new antibacterial peptide obtained from Aeribacillus pallidus SAT4. Biotechnol Reports, 2015; 8: 72–80.
- Bose H, Satyanarayana T. Suitability of the alkalistable carbonic anhydrase from a polyextremophilic bacterium Aeribacillus pallidus TSHB1 in biomimetic carbon sequestration. Bioprocess Biosyst Eng, 2016; 39 (10): 1515–1525.
- Liu X, Dong Y, Zhang J, Zhang A, Wang L, Feng L. Two novel metal-independent long-chain alkyl alcohol dehydrogenases from Geobacillus thermodenitrificans NG80-2. Microbiology, 2009; 155 (6): 2078–2085.
- Gerasimova J, Kuisiene N. Characterization of the novel xylanase from the thermophilic Geobacillus thermodenitrificans JK1. Microbiology, 2012; 81 (4): 418–424.
- Priya I, Dhar M K, Bajaj B K, Koul S, Vakhlu J. Cellulolytic activity of thermophilic bacilli isolated from Tattapani hot spring sediment in North West Himalayas. Indian J Microbiol, 2016; 56 (2): 228–231.
- Collins F W J, O’Connor P M, O’Sullivan O, Rea M C, Hill C, Ross R P. Formicin–a novel broad-spectrum two-component lantibiotic produced by Bacillus paralicheniformis APC 1576. Microbiology, 2016; 162 (9): 1662–1671.
- Wang Y, Liu H, Liu K, Wang C, Ma H, Li Y, Hou Q, Liu F, Zhang T, Wang H. Complete Genome Sequence of Bacillus paralicheniformis MDJK30, a Plant Growth-Promoting Rhizobacterium with Antifungal Activity. Genome Announc, 2017; 5 (25): e00577-17.
© The Author(s) 2018. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.