ISSN: 0973-7510
E-ISSN: 2581-690X
Pseudomonas aeruginosa is a major cause of urinary tract infections. This organism has extended resistance to antimicrobials along with multiple virulence factors, making it difficult to treat. In this study, 49 isolates from urine samples were identified as P. aeruginosa and serotyped by the slide agglutination method. The sensitivity of isolates against 10 antipseudomonal drugs was determined. Phenotypically, lipase, protease, hemolysin, and biofilm production were detected. Genes for the type III secretion system, elastase B, and exotoxin A were detected by PCR. Serotype O11 was the most predominant serotype among test isolates. High levels of resistance were observed against ceftazidime, cefepime, piperacillin, and piperacillin/tazobactam while 10.2% of isolates were resistant to amikacin. MDR was detected in 20.4% of the isolates and was significantly associated with strong biofilm producers. About 95.9% and 63.3% of P. aeruginosa isolates had proteolytic and lipolytic activity, respectively. Among the genes detected, the exoY gene was the most prevalent gene (79.6%), while the exoU gene was the least frequent one (10.2%). toxA and lasB genes were amplified in 63.27% and 75.5% of the isolates, respectively. In addition, the exoU gene was significantly associated with MDR isolates. The high incidence of exoS, exoT, exoY, lasB, and toxA genes in uropathogenic P. aeruginosa implies that these genes can be considered markers for virulent isolates. Furthermore, the coexistence of exoU and exoS genes, even in 6% of isolates, poses a significant treatment challenge because those isolates possess both the invasive and cytotoxic properties of both effector proteins.
Serotypes, Type III secretion system, antimicrobial resistance, P. aeruginosa, Urinary tract infections
Urinary tract infections (UTIs) represent a great health problem that affects people of all ages.1 These infections may be hospital-acquired. Catheterization of the urinary tract is an important predisposing factor to this type of infection by introducing opportunistic pathogens into the urinary tract.2 P. aeruginosa is commonly isolated from complicated UTIs. This bacterium is widespread in healthcare facilities and can get accessed through tap water, food, disinfectant, medical supplies, and healthcare staff resulting in severe nosocomial infections, especially among immunocompromised patients.1,3
Treatment of infections caused by P. aeruginosa is a great challenge facing physicians all over the world. The bacterium has both intrinsic and acquired resistance to many varieties of antimicrobials, which limits the therapeutic options available. Besides, multiple virulence factors, either cell-associated (e.g., lipopolysaccharides, flagellum, and adhesins) or cell-free (e.g., protease, elastase, lipase, hemolysins, exotoxin A) have been linked to P. aeruginosa pathogenicity, allowing it to persist in a wide range of settings. Some of these virulence factors are needed for colonization, while others help the invasion process.4,5
The most potent virulence toxin is exotoxin A (encoded by the toxA gene). The majority of clinical isolates of P. aeruginosa produce this toxin. It has destructive effects on host cells including suppression of protein synthesis, apoptosis of host cells, and impairment of the host cellular immune response.6,7
The second important virulence factor is the type III secretion system (TTSS) which plays a significant role in serious infections caused by P. aeruginosa. This system transports the effector proteins, ExoS, ExoT, ExoU, and ExoY, to the cytosol of host cells to start their harmful effects. Both ExoS and ExoT can disrupt actin cytoskeleton and cause host cell death in addition to inhibition of phagocytosis.8 ExoU is an exoenzyme with phospholipase activity. It has rapid and potent cytotoxic activity that damages the host cells including macrophages. Besides, exoU increases the expression of inflammation genes. ExoU-mediated cell death is characterized by a fast loss of plasma membrane integrity, which is associated with necrosis.9,10 ExoY possesses adenylate cyclase activity, which disrupts the actin cytoskeleton and promotes the synthesis of the second messengers cGMP and cUMP in host cells.11 Another virulence factor secreted by P. aeruginosa isolates is LasB elastase, which is encoded by the lasB gene. This enzyme targets host proteins such as collagen and elastin causing degradation of host tissues which then facilitates the invasion process.12
The capacity of P. aeruginosa to induce an infection has been linked to its potential to form biofilms. Biofilms can escape the host defense mechanisms as well as resist antimicrobial therapy.13 P. aeruginosa has a high tendency for sticking to surfaces forming biofilms. Thus, long-term bladder catheterization predisposes to adhesion and biofilm formation by P. aeruginosa leading to recurrent UTIs.14
Based on the O-antigen portion of their lipopolysaccharide molecule, P. aeruginosa isolates were divided into 20 serotypes by the international antigenic system (IATS). Several investigations have found a link between various virulence factors of P. aeruginosa isolates and their serotypes.15-17
Characterization of uropathogenic P. aeruginosa will help in the selection of the proper therapeutic strategy and improve the treatment outcome. Therefore, the present study aimed to investigate the antimicrobial resistance, serotypes, biofilm formation, and TTSS genotypes of P. aeruginosa isolated from urinary tract infections in Egypt. The correlations between the studied virulence factors, antimicrobial resistance, and recorded serotypes were also investigated.
Isolation and identification of P. aeruginosa
The research ethics committee of Faculty of Pharmacy, Mansoura University authorized the experiments carried out in this work (Code number: 2021-236). Forty-nine isolates of P. aeruginosa were isolated from urine samples from urinary tract infected patients admitted to the Urology and Nephrology center, Mansoura University, Mansoura, Egypt. The identification of P. aeruginosa isolates was based on their gram reaction, growth on cetrimide agar, oxidase production, and pyocyanin pigment production.
Serotyping of P. aeruginosa isolates
Isolates of P. aeruginosa were serotyped by the slide agglutination method using 4 polyvalent and 16 monovalent antisera (Bio-Rad®, France), and the test was done following the manufacturer’s instructions. The serotype groups were determined as described in the International Antigen Typing Scheme (IATS).18 The isolates that did not agglutinate with any antisera were described as non-typeable.
Antimicrobial susceptibility testing
The antimicrobial activity of ten antipseudomonal drugs was determined using the Kirby- Bauer disc diffusion method according to Clinical Laboratory Standard Institute guidelines.19 The used antimicrobial discs were ceftazidime (CAZ, 30 μg), cefepime (FEP, 30 µg), piperacillin (PRL, 100μg), imipenem (IPM, 10μg), meropenem (MEM, 10 μg), levofloxacin (LEV, 5μg), ciprofloxacin (CIP, 5μg), amikacin (AK, 30μg), gentamicin (CN, 10μg), and piperacillin/tazobactam (TPZ, 100 µg /10 µg) (Bioanalyse, Turkey). Resistance to 3 or more classes of antimicrobials was considered multidrug resistance (MDR).
Phenotypic detection of some virulence factors of P. aeruginosa isolates
Qualitative detection of Lipase and protease enzymes
Lipase enzyme production was detected on nutrient agar plates supplemented with Tween 80. Following incubation, lipase-producing colonies showed a zone of precipitation surrounding them.20 Skimmed milk/ brain heart infusion agar was used to detect the proteolytic activity of the tested isolates. After 24-48 h of incubation, protease producers showed a zone of clearance surrounding growth.21
Quantitative detection of Hemolysin
Test isolates with optical density 0.257± 0.002 at λ600 nm were inoculated in nutrient broth and incubated for 48 h at 28°C with shaking at 120 rpm. The hemolytic activity was assessed using human erythrocytes suspension in hemolysin assay buffer (2% v/v). For each isolate, the supernatant was collected by centrifugation, mixed with an equal volume of erythrocyte suspension, and incubated at 37°C for 2 h. The mixture was then centrifuged, and the optical density of the supernatant at λ540 nm was determined. Control experiments for spontaneous lysis (negative control) and complete lysis (positive control, 0.2% sodium dodecyl sulfate) were carried out. The percentage (%) of lysed erythrocytes was calculated.22
% of lysed erythrocytes = [(X-B)/ (T-B)] × 100
B is the absorbance of the negative control, T is the absorbance of the positive control, and X is the absorbance of the test sample.
Quantitative detection of biofilm
Colonies of overnight cultures were suspended in tryptic soy broth to yield 1 McFarland turbidity. 100 μl of each bacterial suspension was inoculated in triplicate into the wells of a microtiter plate and incubated for 24 h at 37°C. The content of wells was aspirated. The wells were then washed three times with phosphate-buffered saline. Absolute methanol was added for 15 min to fix adherent cells, then the plates were emptied and left to dry. Crystal violet (1%) was added for 20 min and excess stain was washed away. After air drying of the plates, glacial acetic acid [33% (V/V)] was added. Finally, a microtiter plate reader was used to measure the optical density of each well at λ492nm. Negative control wells were included. Consequently, P. aeruginosa isolates were classified as strong/moderate/weak slime producers or as non-adherent.20
Molecular detection of the type III secretion system, exotoxin A and elastase B
Extraction of the tested isolates’ genomic DNA
A single colony from overnight grown culture was picked up and suspended in 0.1 ml of DNase / RNase-free water. The bacterial cell suspensions were held in a thermocycler at 95°C for 10 min, cooled on ice, and centrifuged. The supernatants were kept at -20°C in aliquots of 5 μl untill their use as template DNA in PCR.23
Polymerase chain reaction for the detection of virulence genes
The virulence genes (exoS, exoT, exoU, exoY, toxA, and lasB) were amplified by PCR as described previously.24 Positive control and negative control reactions were performed simultaneously. Primer sequences, annealing temperature, and expected amplicon sizes are presented in Table 1.
Table (1):
Distribution of antimicrobial resistance patterns among P. aeruginosa isolates.
| Target gene |
Primer sequence (5´ – 3´) | Amplicon size (bp) | Annealing Temp. | Ref. |
|---|---|---|---|---|
| lasB | F TCATCACCGTCGACATGAAC | 490 | 60 °C | [22] |
| R TGCCCTTCTTGATGTCGTAG | ||||
| exoT | F CAATCATCTCAGCAGAACCC | 1159 | [25] | |
| R TGTCGTAGAGGATCTCCTG | ||||
| exoY | F TGCCATAGAATCCGTCCTC | 145 | [26] | |
| R GATGACCGCCGATTATGAC | ||||
| exoS | F AGGCATTGCCCATGACCTTG | 372 | ||
| R ATACTCTGCTGACCTCGCTC | ||||
| exoU | F CTAGAAGAGAAAGGCATGCTCG | 274 | [22] | |
| R CTATGCGTGGGAGTACATTGAG | ||||
| toxA | F GACAACGCCCTCAGCATCAACAGC | 390 | 65 ° C | |
| R CGCTGGCCCATTCGCTCCAGC |
For the 49 isolates of P. aeruginosa, all data obtained in the previous tests were scored using a binary code system. Then, analysis was done by the Past 4.03 application through the unweighted pair group method with arithmetic mean (UPGMA) and Dice coefficient. A dendrogram was then constructed based on the analyzed data.
Statistical analysis of data
The Spearman rank correlation coefficient test was used to determine whether there is a significant association between the virulence factors, biotypes, and serotypes studied using SPSS software (version 13; SPSS Inc.). At a P-value of ≤0.05, the results were statistically considered significant.
Serotyping of P. aeruginosa isolates
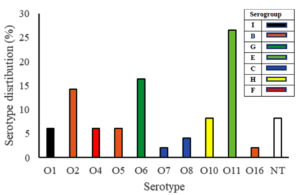
Ten serotypes (O1, O2, O4 – O8, O10, O11, and O16) with 7 serogroups (B, C, E, F, G, H, and I) were detected in P. aeruginosa isolates. Serotype O11 was the most prevalent (26%) followed by serotype O6 (16.3%) and serotype O2 (14%). The least frequent serotypes were O7 and O16 (one isolate each, 2%). Four isolates of P. aeruginosa (8%) were non-typeable, Fig. 1.
Antimicrobial susceptibility testing
Resistance to ceftazidime and cefepime was demonstrated by all tested P. aeruginosa isolates. A high resistance rate was also recorded for piperacillin (95.9%) and piperacillin/tazobactam (91.8%). The resistance rate to meropenem, imipenem, and gentamicin was relatively low (16.3%, 20.4%, and 24.5%, respectively). About 34.7% of isolates were resistant to both ciprofloxacin and levofloxacin. Most P. aeruginosa isolates (89.8%) were susceptible to amikacin. The antibiogram of P. aeruginosa isolates is shown in Table 2. Twelve antimicrobial resistance patterns were detected (A1: A12). Resistance pattern A3 was the most frequent pattern among the tested isolates (46.9%). Multidrug resistance was detected in 10 isolates (20.4%, patterns A5, A9, A10, A11, and A12) and three of them were resistant to all tested antimicrobials.
Table (2):
Distribution of antimicrobial resistance patterns among P. aeruginosa isolates.
Code |
Resistance pattern |
No. of resistant isolates (%) |
|---|---|---|
A1 |
FEP/CAZ |
2 (4.08) |
A2 |
FEP/PRL/CAZ |
1 (2.04) |
A3 |
FEP/PRL/TPZ/CAZ |
23 (46.9) |
A4 |
IPM/FEP/PRL/TPZ/CAZ |
1 (2.04) |
A5 |
CIP/LEV/ CN/FEP/PRL/CAZ |
1 (2.04) |
A6 |
CIP/LEV/FEP/PRL/TPZ/CAZ |
7 (14.29) |
A7 |
IPM/MEM/FEP/PRL/TPZ/CAZ |
3 (6.12) |
A8 |
IPM/MEM/CN/FEP/PRL/TPZ/CAZ |
2 (4.08) |
A9 |
CIP/LEV/ CN/FEP/PRL/TPZ/CAZ |
4 (8.16) |
A10 |
CIP/LEV/ CN/AK/FEP/PRL/TPZ/CAZ |
1 (2.04) |
A11 |
IPM/CIP/LEV/ CN/AK/FEP/PRL/TPZ/CAZ |
1 (2.04) |
A12 |
IPM/MEM/CIP/LEV/ CN/AK/FEP/PRL/TPZ/CAZ |
3 (6.12) |
Correlation between serotypes of P. aeruginosa isolates and their antibiogram
Among the most frequent serotypes (O2, O6, O10, and O11), the isolates of serotype O11 were the most sensitive to all tested antimicrobials. A positive significant correlation between quinolone resistance and serotypes O6 (P=0.008) and O10 (P=0.004) was observed. In contrast, quinolone resistance was negatively associated with serotype O11 (P=0.017). In addition, 37.5% and 50% of serotypes O6 and O10, respectively showed multidrug resistance, Table 3.
Table (3):
Correlation between antimicrobial resistance and the commonly detected serotypes.
| Serotype | Antimicrobials, No. of resistant isolates (%) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| CIP | LEV | AK | CN | IPM | MEM | PRL | TPZ | MDR | |
| O2 | 1(14.3) | 1(14.3) | 1(14.3) | 2(28.6) | 1(14.3) | 1(14.3) | 7 (100) | 7 (100) | 1(14.3) |
| Correlation Coefficient | -0.175 | -0.175 | 0.055 | 0.039 | -0.062 | -0.023 | 0.084 | 0.122 | -0.062 |
| Significance | 0.229 | 0.229 | 0.707 | 0.792 | 0.672 | 0.878 | 0.565 | 0.405 | 0.672 |
| O6 | 6 (75) | 6 (75) | 2 (25) | 3(37.5) | 3(37.5) | 1(12.5) | 8 (100) | 8 (100) | 3(37.5) |
| Correlation Coefficient | 0.374 | 0.374 | 0.216 | 0.134 | 0.187 | -0.046 | 0.091 | 0.132 | 0.187 |
| Significance | 0.008** | 0.008** | 0.136 | 0.360 | 0.197 | 0.755 | 0.533 | 0.367 | 0.197 |
| O10 | 4(100) | 4(100) | 1 (25) | 2 (50) | 1(25) | 1(25) | 4 (100) | 3 (75) | 2(50) |
| Correlation Coefficient | 0.409 | 0.409 | 0.146 | 0.177 | 0.034 | 0.070 | 0.062 | -0.183 | 0.219 |
| Significance | 0.004** | 0.004** | 0.318 | 0.224 | 0.817 | 0.633 | 0.675 | 0.207 | 0.131 |
| O11 | 1 (7.7) | 1 (7.7) | 0 | 1 (7.7) | 2(15.4) | 2(15.4) | 11(84.6) | 10(76.9) | 1(7.7) |
| Correlation Coefficient | -0.341 | -0.341 | -0.203 | -0.235 | -.075 | -.015 | -0.343 | -0.327 | -0.190 |
| Significance | 0.017* | 0.017* | 0.163 | 0.104 | 0.609 | 0.917 | 0.016* | 0.022* | 0.192 |
** Significant at 0.01 level; * Significant at 0.05 level.
Phenotypic detection of protease, lipase, hemolysin, and biofilm
Most P. aeruginosa isolates (95.9%) were proteolytic on skimmed milk-BHI agar (Isolates 1 and 4 were negative), while 63.3% of isolates were positive lipase producers. For hemolysin production, 28 isolates (57.14%) were non-hemolytic while 16 isolates (32.7%) caused lysis of less than 10% of RBCs in the suspension. The highest % of lysed RBCs were observed with isolates No. 20 and 33 (75.49 % and 79.83 %, respectively).
Biofilm production was assessed using a microtiter plate assay. P. aeruginosa isolates were classified into 3 categories: strongly adherent (10 isolates, 20.4%), moderately adherent (30 isolates, 61.2%), and weakly adherent (9 isolates, 18.4%).
Correlation between the studied virulence factors, antimicrobial susceptibility, and serotypes of P. aeruginosa isolates
The association between lipase and hemolysin production was studied. Fig. 2 shows that hemolysin production had a significant negative relationship with lipase production, P= 0.01.
The correlations between strong and weak biofilm producers and antimicrobial resistance, lipase, and hemolysin production were also investigated, Table 4. There were significant correlations between each of the following pairs: strong biofilm production and gentamicin resistance (P=0.036), strong biofilm production and MDR (P=0.009), weak biofilm production, and quinolone resistance (P=0.026). In addition, all weak biofilm producers were lipase producers (P=0.011) while one isolate only caused lysis of RBCs (P=0.034).
Table (4):
Correlation between biofilm production and antimicrobial resistance, hemolysin, and lipase production.
| Biofilm category | Antimicrobial agents Resistant isolates (%) | Lipase | Hemolysin | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CIP | LEV | AK | CN | IPM | MEM | PRL | TPZ | MDR | |||
| Strong | 5(50) | 5(50) | 2(20) | 5(50) | 3(30) | 2(20) | 10(100) | 10(100) | 5(50) | 4(40) | 6(60) |
| Correlation Coefficient | 0.163 | 0.163 | 0.164 | 0.300 | 0.121 | 0.050 | 0.104 | 0.151 | 0.372 | -0.244 | 0.175 |
| Significance | 0.264 | 0.264 | 0.261 | 0.036* | 0.409 | 0.731 | 0.475 | 0.300 | 0.009** | 0.091 | 0.228 |
| Weak | 7(77.8) | 7(77.8) | 1(11.1) | 3(33.3) | 0 | 0 | 9(100) | 8(88.9) | 3(33.3) | 9(100) | 1(11.1) |
| Correlation Coefficient | 0.319 | 0.319 | 0.014 | 0.098 | -0.240 | -0.210 | 0.098 | -0.051 | 0.152 | 0.361 | -0.304 |
| Significance | 0.026* | 0.026* | 0.923 | 0.505 | 0.096 | 0.148 | 0.504 | 0.727 | 0.297 | 0.011* | 0.034* |
** Significant at 0.01 level.
* Significant at 0.05 level.
Moreover, the correlation between the tested virulence factors and the most frequently encountered serotypes was analyzed. Hemolysin production was more prevalent in serotype O2 isolates (71.4%) while lipase production was detected more in serotype O10 isolates. In addition, a high percentage of isolates of serotypes O2, O6, and O11 isolates (71.4%, 75%, and 92.3%, respectively) were strong to moderate biofilm producers. Statistically, none of these observations proved significant.
PCR-based detection of genes encoding TTSS, exotoxin A, and elastase B
For TTSS genes, exoY was the most prevalent gene (39 isolates, 79.6%) followed by exoS (33 isolates, 67.3%) and exoT (31 isolates, 63.3%) while the exoU gene was the least frequent (5 isolates, 10.2%). The toxA and lasB genes were amplified in 31 isolates (63.27%) and 37 isolates (75.5%), respectively. Three isolates (6.12%) did not amplify any of the tested genes. Table 5 illustrates the coexistence profiles of the tested genes. Twenty-one profiles were recorded and 11 of them were presented by 2 or more isolates with the highest predominance for profile P20 (8 isolates, 16.33%).
Table (5):
Virulence genotypes of P. aeruginosa isolates.
Code |
Gene profile |
No of isolates |
|---|---|---|
P1 |
No genes |
3 |
P2 |
toxA |
1 |
P3 |
toxA, exoS |
1 |
P4 |
lasB, exoY |
1 |
P5 |
toxA, lasB, exoT |
1 |
P6 |
toxA, lasB, exoY |
2 |
P7 |
lasB, exoU, exoY |
1 |
P8 |
lasB, exoT, exoY |
2 |
P9 |
exoS, exoT, exoY |
3 |
P10 |
lasB, exoS, exoY |
1 |
P11 |
toxA, exoS, exoT |
1 |
P12 |
toxA, exoU, exoY |
1 |
P13 |
lasB, exoS, exoT |
1 |
P14 |
toxA, lasB, exoS, exoY |
6 |
P15 |
toxA, lasB, exoS, exoT |
2 |
P16 |
lasB, exoS, exoT, exoY |
5 |
P17 |
toxA, exoS, exoT, exoY |
2 |
P18 |
lasB, exoS, exoU, exoY |
1 |
P19 |
toxA, lasB, exoT, exoY |
4 |
P20 |
toxA, lasB, exoS, exoT, exoY |
8 |
P21 |
toxA, lasB, exoS, exoT, exoU, exoY |
2 |
Correlation studies detected a positive significant correlation between the lasB and exoY genes (P=0.003) and the exoS and exoT genes (P=0.05). In addition, the lasB gene was significantly detected in isolates sensitive to quinolones (P=0.049). exoU gene was significantly associated with MDR isolates (P=0.000) in contrast to the exoT gene that had a negative association with MDR isolates (P=0.014).
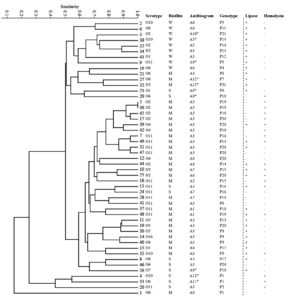
Cluster analysis of the constructed dendrogram divided thirty-eight isolates of P. aeruginosa into 10 clusters with a similarity of 70%. Eleven isolates with a lower similarity percentage (< 70%) were not included in these clusters. The largest cluster had 12 isolates that were moderate biofilm producers of A3 and A8 biotypes. The second large cluster had 5 isolates that were also moderate biofilm producers of the A3 biotype. Only two isolates (Nos. 5 and 36) had 100 % similarity with respect to the analyzed data, Fig. 3.
P. aeruginosa is well-known as a major cause of UTIs especially those associated with catheterization. In addition, this bacterium contributes significantly to the morbidity and death rates associated with nosocomial infections. In the current study, 49 uropathogenic isolates were identified as P. aeruginosa. Although the study population was different from those involved in previous studies, the high prevalence of O11 and O6 serotypes was also recorded.15,17,27-29 Lu et al.17 reported the failure of the standard technique to serotype about 8% of their isolates which is similar to this study where four isolates (8%) were non-typeable. Other studies recorded non-typeable isolates in higher proportions.30-32
The antibiogram of the infecting isolates is very helpful for physicians to select the appropriate antimicrobial to shorten the treatment period and improve the outcome. In UTIs, improper antimicrobial will delay the treatment process and increase the risk for urosepsis.33 In this study, the antimicrobial susceptibility testing demonstrated high levels of resistance (91.8%-100%) for ceftazidime, cefepime, piperacillin, and piperacillin/tazobactam. Previous studies conducted in Egypt also revealed a significant rate of resistance to these antimicrobials.34-40 However, other investigations in Egypt reported a lower rate of resistance (29%-43%).41,42 The same discrepancies in resistance rate were observed in different countries, where some studies recorded a high resistance rate,43-45 while others recorded a relatively low resistance rate with respect to the used antimicrobials.46-48 Isolates of the current study showed low to moderate resistance (20.4%-34.7%) to gentamicin, ciprofloxacin, levofloxacin, imipenem, and meropenem. Although some reports coincide with current data,34,37,49 several research groups reported high resistance levels to quinolones, gentamicin, and/or carbapenems.36,40,50 In agreement with previously published data,34,41, 51-53 amikacin showed the highest efficacy against tested isolates.
Multidrug resistance of P. aeruginosa is alarming everywhere, but the problem is escalating in Egypt and other developing countries because there are no regulations that limit the prescription of antibiotics even the broad-spectrum ones. Antibiotics are also sold without a doctor’s prescription as over-the-counter medications. In the current study, 20.4% of the isolates were MDR compared to 48.8% -100% of the MDR demonstrated by many research groups in Egypt.34,35,37-39,42 In addition, 37.5% and 50% of isolates belonging to serotypes O6 and O10 isolates were MDR and a significant association was found between these two serotypes and quinolone resistance. These results agree partially with Abdel-Rhman and Rizk who found a significant association between MDR and O6 and O10 serotypes.39 In contrast to many earlier studies,54,55 P. aeruginosa isolates of serotype O11 were the most sensitive to all tested antimicrobials, and a negative significant correlation was detected between these isolates and quinolone resistance.
The pathogenesis of P. aeruginosa in different illnesses has been linked to its ability to express multiple virulence factors. Previous studies had shown that among the multiple virulence factors produced by P. aeruginosa, individual virulence factors may control and contribute to the outcome of various infections. Like other infections, the establishment of UTIs requires colonization of host tissues, which is facilitated by many virulence factors of the bacterium. In this study, most isolates were proteolytic on skimmed milk-BHI agar while 63.3% of isolates were positive lipase producers. Proteases, according to Gupta et al., play an important role in the pathogenesis of P. aeruginosa in UTIs because they enhance bacterial colonization of renal tissues by degrading a variety of substrates that aid tissue invasion and dissemination.56 Lipases also destroy host tissues.57 The case was different for hemolysin production, where 21 isolates (42.86%) were hemolytic and 16 of them caused lysis of less than 10% of the RBCs and there was a significant negative association between lipase and hemolysin production. Gupta et al. observed that mutant strain producing hemolysin but not producing proteases and elastase was incapable of colonizing the renal tissues of the used mice model. This may suggest that hemolysin has a minimal role in UTIs compared to proteases and elastases.56
The ability of P. aeruginosa to cause infections has been linked to its tendency to form biofilms. Its capacity to adhere to catheter surfaces makes catheterized patients more likely to get UTIs.58,59 In addition, urea, the major solute in urine may encourage the biofilm development of P. aeruginosa through alterations in the cell membrane or production of additional extracellular matrix.60 In this study, all isolates had biofilm-forming capacity, where 20.4%, 61.2%, and 18.4% of them were strong, moderate, and weak biofilm producers, respectively. The same finding was recorded in two previous studies conducted in Egyptian hospitals where 100% of the isolates were biofilm producers.38,61 Also, a lower percentage of biofilm production by P. aeruginosa was reported by other studies.37,62-65
In contrast to studies that found no relationship between biofilm and MDR,38,62,63 this research concurred with other reports that found a significant link between biofilm production and MDR.37,65,66 In addition, weak biofilm production was associated with quinolone resistance which agrees with previous investigations.63,67,68 Most weak biofilm producers were nonhemolytic but all of them had lipolytic activity. This also may reflect the importance of lipase production in the pathogenesis of P. aeruginosa. Besides, the current work found no significant correlation between the serotypes predominant in the isolates and any of the investigated virulence factors, contradicting the findings of Mittal et al.69 and Visca et al.70
Some virulence factors are recognized as major in acute infections caused by P. aeruginosa, among which is TTSS. The distribution of TTSS genotypes among P. aeruginosa clinical isolates can help in understanding the epidemiology of such infections. The distribution frequency of TTSS genes varied between studies. In this study, the exoS gene was prevalent in 67.3% of isolates, which was consistent with previous reports.38,71-73 Also, the exoU gene was detected in only 10.2% of isolates; however, higher prevalence rates were reported in Egypt and other countries.38,72,74,75 While the exoT gene was amplified in 63.3% of isolates and it was significantly associated with exoS gene, a recent study in Egypt recorded exoT gene in only 6.7% of isolates.38 Other research studies showed high prevalence rates of the exoT gene.10,74 The exoY gene was the most predominant gene (79.6%), which is consistent with earlier studies.38,74 Statistical analysis revealed a significant association between MDR and exoU gene, which coincides with previous reports.72,73 Few isolates (6%) harbored both exoS and exoU genes which agrees with prior studies.72,73,76,77 These isolates will possess the invasive and cytotoxic properties of both toxins. Although the exoU gene was found in only a few isolates, earlier studies correlated its existence with high mortality rates.77-79 In the current investigation, the elastase encoding gene lasB was detected in 75.5% of isolates and it was statistically associated with exoY gene. Similar results were reported in Egypt38, 80; however, higher prevalence was observed in studies conducted by Benie et al. (89.2%),81 and Babour et al. (100%).82 About 63.3% of the P. aeruginosa isolates amplified the toxA gene which is regarded as an important virulence factor in catheter-associated UTIs.83 A higher frequency of this gene was reported in Iran (95.7%)9 but recently in Egypt, a lower frequency (45.6%) was recorded.38
Based on biotypes, serotypes, virulence factor production, and PCR results, the dendrogram grouped 38 isolates into 10 clusters (70% similarity), while 11 isolates had similarities lower than 70%. The 17 isolates that represented the two largest clusters were moderate biofilm producers and of definite biotypes but were of different serotypes. This confirms that serotyping is still a more useful tool to discriminate between P. aeruginosa isolates.84 Two isolates showed 100% similarity, which may indicate cross-infection among hospitalized patients.39
In this study, isolates of serotype O11 were the most sensitive to all antimicrobials tested compared to isolates of other commonly encountered serotypes. Amikacin was the most effective antimicrobial against test isolates. Although MDR was detected in 20.4% of isolates, it was significantly associated with strong biofilm production, making treatment more difficult. Most of the isolates had proteolytic and lipolytic but nonhemolytic activity, which may reflect the importance of lipase and protease enzymes in the pathogenesis of P. aeruginosa in UTIs. The current work found no significant correlation between the serotypes predominant in the test isolates and any of the investigated virulence factors. In addition, the high prevalence of exoS, exoT, exoY, lasB, and toxA genes in uropathogenic P. aeruginosa isolates suggests identifying these genes as a maker of virulent strains. Although exoU gene was detected in only 10.2% of isolates, it is commonly associated with a high mortality rate. Furthermore, the coexistence of exoU and exoS genes even in a small % of isolates constitutes a great challenge in treatment as those isolates will have both the invasive and cytotoxic properties of the two effector proteins, especially that exoU gene was found significantly in MRD isolates. Isolates of different serotypes were distributed in all clusters in the dendrogram, emphasizing that serotyping is still a more useful tool to discriminate between P. aeruginosa isolates.
ACKNOWLEDGMENTS
The author would like to thank Urology and Nephrology Center at Mansoura University, Egypt, for providing the Clinical Isolates used in this study.
FUNDING
None.
ETHICS STATEMENT
This study was approved by the Institutional Ethics Committee, Faculty of Pharmacy, Mansoura University, Egypt with reference number:2021-236.
AVAILABILITY OF DATA
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
- Wolska K, Kot B, Jakubczak A. Phenotypic and genotypic diversity of Pseudomonas aeruginosa strains isolated from hospitals in Siedlce (Poland). Braz J Microbiol. 2012;43(1):274-82.
Crossref - Mittal R, Aggarwal S, Sharma S, Chhibber S, Harjai K. Urinary tract infections caused by Pseudomonas aeruginosa: a minireview. J Infect Public Health. 2009;2(3):101-111.
Crossref - Newman JW, Floyd RV, Fothergill JL. The contribution of Pseudomonas aeruginosa virulence factors and host factors in the establishment of urinary tract infections. FEMS Microbiol Lett. 2017;364(15):fnx124.
Crossref - Breidenstein EB, de la Fuente-Nunez C, Hancock RE. Pseudomonas aeruginosa: all roads lead to resistance. Trends Microbiol. 2011;19(8):419-426.
Crossref - Kerr KG, Snelling AM. Pseudomonas aeruginosa: a formidable and ever-present adversary. J Hosp Infect. 2009;73(4):338-344.
Crossref - Tanom A, Farajnia S, Peerayeh SN, Majidi J. Cloning, expression and characterization of recombinant exotoxin A-flagellin fusion protein as a new vaccine candidate against Pseudomonas aeruginosa infections. Iran Biomed J. 2013;17(1):1-7.
Crossref - Wolf P, Elsasser-Beile U. Pseudomonas exotoxin A: from virulence factor to anti-cancer agent. Int J Med Microbiol. 2009;299(3):161-176.
Crossref - Khodayary R, Nikokar I, Mobayen MR, et al. High incidence of type III secretion system associated virulence factors (exoenzymes) in Pseudomonas aeruginosa isolated from Iranian burn patients. BMC Res Notes. 2019;12(1):1-6.
Crossref - Bahador N, Shoja S, Faridi F, et al. Molecular detection of virulence factors and biofilm formation in Pseudomonas aeruginosa obtained from different clinical specimens in Bandar Abbas. Iran J Microbiol. 2019;11(1):25.
Crossref - Berthelot P, Attree I, Plesiat P, et al. Genotypic and phenotypic analysis of type III secretion system in a cohort of Pseudomonas aeruginosa bacteremia isolates: evidence for a possible association between O serotypes and exo genes. J Infect Dis. 2003;188(4):512-518.
Crossref - Morrow KA, Frank DW, Balczon R, Stevens T. The Pseudomonas aeruginosa exoenzyme Y: a promiscuous nucleotidyl cyclase edema factor and virulence determinant. Handb Exp Pharmacol. 2017;238:67-85.
Crossref - Hamood AN, Griswold J, Colmer J. Characterization of elastase-deficient clinical isolates of Pseudomonas aeruginosa. Infect Immun. 1996;64(8):3154-3160.
Crossref - Azimi S, Kafil HS, Baghi HB, et al. Presence of exoY, exoS, exoU and exoT genes, antibiotic resistance and biofilm production among Pseudomonas aeruginosa isolates in Northwest Iran. GMS Hyg Infect Control 2016;11:Doc04.
Crossref - Drenkard E. Antimicrobial resistance of Pseudomonas aeruginosa biofilms. Microbes Infect. 2003;5(13):1213-1219.
Crossref - Faure K, Shimabukuro D, Ajayi T, Allmond LR, Sawa T, Wiener-Kronish JP. O-antigen serotypes and type III secretory toxins in clinical isolates of Pseudomonas aeruginosa. J Clin Microbiol. 2003;41(5):2158-2160.
Crossref - Hostacka A, Majtan V. Serotyping and virulence factors of Pseudomonas aeruginosa clinical isolates. Acta Microbiol Immunol Hung. 1997;44(2):141-146. PMID: 9330662
- Lu Q, Eggimann P, Luyt C-E, et al. Pseudomonas aeruginosa serotypes in nosocomial pneumonia: prevalence and clinical outcomes. Crit Care. 2014;18(1):R17.
Crossref - Legakis N, Aliferopoulou M, Papavassiliou J, Papapetropoulou M. Serotypes of Pseudomonas aeruginosa in clinical specimens in relation to antibiotic susceptibility. J Clin Microbiol. 1982;16(3):458-463.
Crossref - CLSI. Performance standards for antimicrobial susceptibility testing: approved 28th ed. CLSI Wayne, PA; 2018.
- Hassan R, El-Naggar W, Habib E, El-Bargisy R. Comparative studies on Staphylococcus aureus isolates associated with infections in diabetic and non-diabetic patients. Egypt J Med Microbiol. 2012;21(2).
Crossref - Sokol PA, Ohman DE, Iglewski BH. A more sensitive plate assay for detection of protease production by Pseudomonas aeruginosa. J Clin Microbiol. 1979;9(4):538-540.
Crossref - Eid D, EN W, Barwa R, El-Sokkary MA. Phenotypic and genotypic characterization of some virulence factors in Pseudomonas aeruginosa strains isolated from different clinical sources in Mansoura University Hospitals. New Egyptian Journal of Microbiology. 2012;32(2):151-67.
- Englen M, Kelley L. A rapid DNA isolation procedure for the identification of Campylobacter jejuni by the polymerase chain reaction. Lett Appl Microbiol. 2000;31(6):421-426.
Crossref - Williams JG, Kubelik AR, Livak KJ, Rafalski JA, Tingey SV. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Research. 1990;18(22):6531-6535.
Crossref - Finnan S, Morrissey JP, O’gara F, Boyd EF. Genome diversity of Pseudomonas aeruginosa isolates from cystic fibrosis patients and the hospital environment. J Clin Microbiol. 2004;42(12):5783-5792.
Crossref - El-Mowafy SA, Abd El Galil KH, El-Messery SM, Shaaban MI. Aspirin is an efficient inhibitor of quorum sensing, virulence and toxins in Pseudomonas aeruginosa. Microbial Pathogenesis. 2014;74:25-32.
Crossref - Estahbanati HK, Kashani PP, Ghanaatpisheh F. Frequency of Pseudomonas aeruginosa serotypes in burn wound infections and their resistance to antibiotics. Burns. 2002;28(4):340-348.
Crossref - Fonseca AP, Correia P, Sousa JC, Tenreiro R. Association patterns of Pseudomonas aeruginosa clinical isolates as revealed by virulence traits, antibiotic resistance, serotype and genotype. FEMS Immunol Med Microbiol. 2007;51(3):505-516.
Crossref - Vachee A, Scheftel J, Husson M, Izard D, Ross P, Monteil H. Tricentric study of the sensitivity of Pseudomonas aeruginosa serotyping to beta-lactams and aminoglycosides. Pathologie-biologie 1997;45(5):357-362.
- Rodriguez DS, Rizo AG, Monroy SP, Reyes T. Antimicrobial susceptibility and serotyping of Pseudomonas aeruginosa isolated from HIV/AIDS patients. Rev Cubana de Med Trop. 2002;54(2):142-146. PMID: 15849941
- Jamasbi RJ, Proudfoot EM. Phenotypic and genotypic characteristics of clinical isolates of pseudomonas aeruginosa: rate of occurrence and distribution of different serotypes, antimicrobial susceptibility profiles, and molecular typing. Laboratory Medicine. 2008;39(3):155-161.
Crossref - Stankovic-Nedeljkovic N, Kocic B, Tiodorovic B, Brankovic S, Mladenovic-Antic S. Serotyping and analysis of produced pigments kinds by Pseudomonas aeruginosa clinical isolates. Vojnosanit Pregled. 2011;68(11):923-929.
Crossref - Wu C-T, Lee H-Y, Chen C-L, Tuan P-L, Chiu C-H. High prevalence and antimicrobial resistance of urinary tract infection isolates in febrile young children without localizing signs in Taiwan. J Microbiol Immunol Infect. 2016;49(2):243-248.
Crossref - Abbas HA, El-Ganiny AM, Kamel HA. Phenotypic and genotypic detection of antibiotic resistance of Pseudomonas aeruginosa isolated from urinary tract infections. Afr Health Sci. 2018;18(1):11-21.
Crossref - Mahmoud AB, Zahran WA, Hindawi GR, Labib AZ, Galal R. Prevalence of multidrug-resistant Pseudomonas aeruginosa in patients with nosocomial infections at a university hospital in Egypt, with special reference to typing methods. J Virol Microbiol. 2013;13:165-259.
Crossref - Basha AM, El-Sherbiny GM, Mabrouk MI. Phenotypic characterization of the Egyptian isolates “extensively drug-resistant Pseudomonas aeruginosa” and detection of their metallo-β-lactamases encoding genes. Bulletin of the National Research Centre. 2020;44(1):1-11.
Crossref - Mahmoud MF, Fathy FM, Gohar MK, Soliman MH. Biofilm Formation And Quorum Sensing lasRGene Of Pseudomonas Aeruginosa Isolated From Patients With Post-Operative Wound Infections. European Journal of Molecular & Clinical Medicine. 2021;8(2):2177-2189.
- Eladawy M, El-Mowafy M, El-Sokkary MMA, Barwa R. Antimicrobial resistance and virulence characteristics in ERIC-PCR typed biofilm forming isolates of P. aeruginosa. Microbial Pathog. 2021;158:105042.
Crossref - Abdel-Rhman SH, Rizk DE. Serotypes, antibiogram and genetic relatedness of Pseudomonas aeruginosa isolates from urinary tract infections at urology and Nephrology Center, Mansoura, Egypt. Adv Microbiol. 2018;8(08):625.
Crossref - Abaza AF, El Shazly SA, Selim HS, Aly GS. Metallo-beta-lactamase producing Pseudomonas aeruginosa in a healthcare setting in Alexandria, Egypt. Pol J Microbiol. 2017;66(3):297-308.
Crossref - Gad GF, El-Domany RA, Zaki S, Ashour HM. Characterization of Pseudomonas aeruginosa isolated from clinical and environmental samples in Minia, Egypt: prevalence, antibiogram and resistance mechanisms. J Antimicrob Chemother. 2007;60(5):1010-1017.
Crossref - Wassef M, Mahallawy H, Zafer M, Ghaith D, Hamid R. Lab based surveillance of multidrug resistant Pseudomonas aeruginosa in Cairo University Hospitals, Egypt. Egypt J Microbiol Exp. 2015;2(2):47-51.
Crossref - Yekani M, Memar MY, Alizadeh N, Safaei N, Ghotaslou R. Antibiotic resistance patterns of biofilm-forming Pseudomonas aeruginosa isolates from mechanically ventilated patients. Int J. 2017;5:85.
- Corehtash ZG, Ahmad Khorshidi FF, Akbari H, Aznaveh AM. Biofilm formation and virulence factors among Pseudomonas aeruginosa isolated from burn patients. Jundishapur J Microbiol. 2015;8(10):e22345.
Crossref - Ghorashi Z, Nezami N, Ghotaslou R, Ghorashi S. Pattern of Pseudomonas aeruginosa drug resistance in Tabriz Children Hospital. Pak J Biol Sci. 2010;13(8):400-404.
Crossref - Sader HS, Castanheira M, Duncan LR, Flamm RK. Antimicrobial susceptibility of Enterobacteriaceae and Pseudomonas aeruginosa isolates from United States medical centers stratified by infection type: results from the International Network for Optimal Resistance Monitoring (INFORM) surveillance program, 2015-2016. Diagn Microbiol Infect Dis. 2018;92(1):69-74.
Crossref - Khan MA, Faiz A. Antimicrobial resistance patterns of Pseudomonas aeruginosa in tertiary care hospitals of Makkah and Jeddah. Ann Saudi Med. 2016;36(1):23-28.
Crossref - Pathmanathan SG, Samat NA, Mohamed R. Antimicrobial susceptibility of clinical isolates of Pseudomonas aeruginosa from a Malaysian Hospital. Malays J Med Sci. 2009;16(2):27-32. PMCID: PMC3336164
- Elmaraghy N, Abbadi S, Elhadidi G, Hashem A, Yousef A. Virulence genes in Pseudomonas aeruginosa strains isolated at Suez Canal University Hospitals with respect to the site of infection and antimicrobial resistance. Int J Clin Microbiol Biochem Technol. 2019;2:8-19.
Crossref - Mohamed FA. Antibiotic susceptibility of Pseudomonas aeruginosa isolated from different clinical sources. Zagazig Journal of Pharmaceutical Sciences. 2019;28(2):10-17.
- Fazeli H, Sadighian H, Nasr-Esfahani B, Pourmand MR. Identification of class-1 integron and various β-lactamase classes among clinical isolates of Pseudomonas aeruginosa at children’s medical center hospital. J Med Bacteriol. 2012;1(3-4):25-36.
- Khan F, Khan A, Kazmi SU. Prevalence and susceptibility pattern of multi drug resistant clinical isolates of Pseudomonas aeruginosa in Karachi. Pak J Med Sci. 2014;30(5):951-954.
Crossref - Abdalla EB, Gehan ES, Ahmed M, Lotfy B. Phenotyping and genotyping of Pseudomonas aeruginosa urine isolates in Zagazig University Hospitals. 2008.
- Gailiene G, Pavilonis A, Kareiviene V. The peculiarities of Pseudomonas aeruginosa resistance to antibiotics and prevalence of serogroups. Medicina. 2007;43(1):36.
Crossref - Oliver A, Mulet X, Lopez-Causape C, Juan C. The increasing threat of Pseudomonas aeruginosa high-risk clones. Drug ResistUpdat. 2015;21:41-59.
Crossref - Gupta P, Gupta R, Harjai K. Multiple virulence factors regulated by quorum sensing may help in establishment and colonisation of urinary tract by Pseudomonas aeruginosa during experimental urinary tract infection. Indian J Med Microbiol. 2013;31(1):29-33.
Crossref - Gellatly SL, Hancock RE. Pseudomonas aeruginosa: new insights into pathogenesis and host defenses. Pathog Dis. 2013;67(3):159-173.
Crossref - Hall-Stoodley L, Stoodley P. Biofilm formation and dispersal and the transmission of human pathogens. Trends Microbiol. 2005;13(1):7-10.
Crossref - Trautner BW, Hull RA, Darouiche RO. Prevention of catheter-associated urinary tract infection. Curr Opin Infect Dis. 2005;18(1):37-41.
Crossref - Cole SJ, Records AR, Orr MW, Linden SB, Lee VT. Catheter-associated urinary tract infection by Pseudomonas aeruginosa is mediated by exopolysaccharide-independent biofilms. Infect Immun. 2014;82(5):2048-2058.
Crossref - Elhabibi T, Ramzy S. Biofilm production by multi drug resistant bacterial pathogens isolated from patients in intensive care units in Egyptian hospitals. J Microb Biochem Technol. 2017;9(4):151-158.
Crossref - Kamali E, Jamali A, Ardebili A, Ezadi F, Mohebbi A. Evaluation of antimicrobial resistance, biofilm forming potential, and the presence of biofilm-related genes among clinical isolates of Pseudomonas aeruginosa. BMC Research Notes. 2020;13(1):1-6.
Crossref - Cepas V, Lopez Y, Munoz E, et al. Relationship between biofilm formation and antimicrobial resistance in gram-negative bacteria. Microb Drug Resist. 2019;25(1):72-79.
Crossref - Banar M, Emaneini M, Satarzadeh M, et al. Evaluation of mannosidase and trypsin enzymes effects on biofilm production of Pseudomonas aeruginosa isolated from burn wound infections. PloS One 2016;11(10):e0164622.
Crossref - El-sayed H, Fahmy Y. Correlation between biofilm formation and multidrug resistance in clinical isolates of Pseudomonas aeruginosa. Microbes and Infectious Diseases. 2021;2(3):541-549.
- Abdulhaq N, Nawaz Z, Zahoor MA, Siddique AB. Association of biofilm formation with multi drug resistance in clinical isolates of Pseudomonas aeruginosa. EXCLI Journal. 2020;19:201-208.
Crossref - Soto S, Smithson A, Martinez J, Horcajada J, Mensa J, Vila J. Biofilm formation in uropathogenic Escherichia coli strains: relationship with prostatitis, urovirulence factors and antimicrobial resistance. J Urol. 2007;177(1):365-368.
Crossref - Fabrega A, Soto SM, Balleste-Delpierre C, Fernandez-Orth D, Jimenez de Anta MT, Vila J. Impact of quinolone-resistance acquisition on biofilm production and fitness in Salmonella enterica. J Antimicrob Chemother. 2014;69(7):1815-1824.
Crossref - Mittal R, Sharma S, Chhibber S, Aggarwal S, Gupta V, Harjai K. Correlation between serogroup, in vitro biofilm formation and elaboration of virulence factors by uropathogenic Pseudomonas aeruginosa. FEMS Immunol Med Microbiol. 2010;58(2):237-243.
Crossref - Visca A, Chiarini F, Mansi A, Vetriani C, Serino L, Orsi N. Virulence determinants in Pseudomonas aeruginosa strains from urinary tract infections. Epidemiol Infect. 1992;108(2):323-336.
Crossref - Shaver CM, Hauser AR. Relative contributions of Pseudomonas aeruginosa ExoU, ExoS, and ExoT to virulence in the lung. Infect Immun. 2004;72(12):6969-6977.
Crossref - Hassuna NA. Molecular detection of the virulent ExoU genotype of Pseudomonas aeruginosa isolated from infected surgical incisions. Surg Infect. 2016;17(5):610-614.
Crossref - Mitov I, Strateva T, Markova B. Prevalence of virulence genes among bulgarian nosocomial and cystic fibrosis isolates of Pseudomonas aeruginosa. Braz J Microbiol. 2010;41(3):588-595.
Crossref - Feltman H, Schulert G, Khan S, Jain M, Peterson L, Hauser AR. Prevalence of type III secretion genes in clinical and environmental isolates of Pseudomonas aeruginosa. Microbiology. 2001;147(10):2659-2669.
Crossref - Hassuna NA, Mandour SA, Mohamed ES. Virulence constitution of multi-drug-resistant Pseudomonas aeruginosa in upper Egypt. Infect Drug Resist. 2020;13:587.
Crossref - Veesenmeyer JL, Hauser AR, Lisboa T, Rello J. Pseudomonas aeruginosa virulence and therapy: evolving translational strategies. Crit Care Med. 2009;37(5):1777-1786.
Crossref - Georgescu M, Gheorghe I, Curutiu C, Lazar V, Bleotu C, Chifiriuc M-C. Virulence and resistance features of Pseudomonas aeruginosa strains isolated from chronic leg ulcers. BMC Infect Dis. 2016;16(Suppl 1):92.
Crossref - El Solh AA, Akinnusi ME, Wiener-Kronish JP, Lynch SV, Pineda LA, Szarpa K. Persistent infection with Pseudomonas aeruginosa in ventilator-associated pneumonia. Am J Respir Crit Care Med. 2008;178(5):513-519.
Crossref - Sullivan E, Bensman J, Lou M, Agnello M, Shriner K, Wong-Beringer A. Risk of developing pneumonia is enhanced by the combined traits of fluoroquinolone resistance and type III secretion virulence in respiratory isolates of Pseudomonas aeruginosa. Crit Care Med. 2014;42(1):48-56.
Crossref - Sonbol FI, Khalil MAEF, Mohamed AB, Ali SS. Correlation between antibiotic resistance and virulence of Pseudomonas aeruginosa clinical isolates. Turk J Med Sci. 2015;45(3):568-577.
Crossref - Benie CKD, Dadie A, Guessennd N, et al. Characterization of virulence potential of Pseudomonas aeruginosa isolated from bovine meat, fresh fish, and smoked fish. Eur J Microbiol Immunol. 2017;7(1):55-64.
Crossref - Babour IA, Mohamed MB, Shehabi AA. Molecular characterization of Pseudomonas aeruginosa isolates from various clinical specimens in Khartoum/Sudan: Antimicrobial resistance and virulence genes. The International Arabic Journal of Antimicrobial Agents. 2020;10(1).
Crossref - Goldsworthy MJH. Gene expression of Pseudomonas aeruginosa and MRSA within a catheter-associated urinary tract infection biofilm model. Bioscience Horizons. 2008;1(1):28-37.
Crossref - del Barrio-Tofino E, Sanchez-Diener I, Zamorano L, et al. Association between Pseudomonas aeruginosa O-antigen serotypes, resistance profiles and high-risk clones: results from a Spanish nationwide survey. J Antimicrob Chemother. 2019;74(11):3217-3220.
Crossref
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.