ISSN: 0973-7510
E-ISSN: 2581-690X
The rise of pre-extensively drug-resistant (pre-XDR) and extensively drug-resistant (XDR) tuberculosis (TB) represents a serious threat to global health. This research investigates resistance patterns of Mycobacterium tuberculosis (MTB) clinical isolates to fluoroquinolones (FQs) and second-line injectable drugs (SLIDs). The samples were collected as part of the Revised National Tuberculosis Control Programme (RNTCP) at Damien Urban TB and Leprosy Center, Nellore, Andhra Pradesh, India. The study further explores genetic mutations linked to drug resistance. A sum of 954 clinical isolates were examined over a year using GenoType MTBDRsl VER 2.0. Among these, 759 isolates were sensitive to both FQs and SLIDs, while 103 exhibited resistance to one or more SLIDs. A total of 86 isolates exhibited resistance to FQs, 11 to SLIDs, and 6 to both. Genetic analysis of the gyrA gene revealed frequent mutations at codons 90, 91, and 94, with the highest occurrence at position 94. Within the 16S rRNA rrs gene, the G1484T mutation was dominant, followed by A1401G. Additionally, alterations in the eis promoter region, especially the C-14T substitution, were observed. Fifteen isolates displayed hetero-resistance, meaning both drug-resistant and drug-susceptible bacterial populations coexisted within the same sample. Furthermore, 31 isolates carried unidentified mutations, emphasizing the genetic complexity of MTB resistance mechanisms. These findings highlight the necessity for continuous surveillance and genetic analysis to better understand and manage drug-resistant TB.
Pre-XDR-TB, XDR-TB, Fluoroquinolone Resistance, Second-line Injectable Drugs, Genetic Alterations, Mycobacterium tuberculosis, Antimicrobial Resistance
Tuberculosis (TB), is a contagious illness caused by the bacterium Mycobacterium tuberculosis (MTB) and remains a major challenge to public health worldwide. According to the epidemiological data by the World Health Organization (WHO) for the year 2018, an estimated incidence of 10 million new cases of tuberculosis occurred worldwide. Mortality attributed to TB reached approximately 1.5 million deaths, with a notable proportion (approximately 251,000) occurring among people living with HIV. Furthermore, it is predicted that nearly one-quarter of the global population carries a latent tuberculosis infection, characterized by the presence of viable M. tuberculosis bacilli without causing active disease symptoms. Approximately 5%-15% of people carrying latent MTB infection have a life-time risk of progressing to active tuberculosis disease due to immune-compromised condition, HIV infection, diabetes, malnutrition, consumption of alcohol, or smoking. 87% of tuberculosis cases reported in 2018 were from 30 high-burden countries among which two-third of the newly diagnosed cases occurred in eight countries, namely, South Africa, Nigeria, Pakistan, Philippines, Indonesia, China, Bangladesh and India.1
Timely, early and accurate diagnosis with subsequent treatment with appropriate anti-tubercular drugs is the best way to manage tuberculosis. Treatment for tuberculosis (TB) involves a regimen of multiple antibiotics, typically lasting six months to a year. Isoniazid (INH) along with rifampicin (RIF), ethambutol, and pyrazinamide are used as the first line of drugs (FLDs) for treating tuberculosis. However, the rise and spread of drug-resistant MTB strains pose a substantial obstacle to effective global tuberculosis control efforts. Specifically, the evolution of multidrug-resistant tuberculosis (MDR-TB), defined by resistance to both rifampicin (RIF) and isoniazid (INH), two key first-line antitubercular agents, presents a critical public health crisis. Treatment of MDR-TB necessitates complex multidrug regimens, encompassing first-line drugs (FLDs) like pyrazinamide, and ethambutol, alongside second-line drugs (SLDs) including fluoroquinolones (FQs) such as moxifloxacin (MOX), ofloxacin (OFX), and levofloxacin (LEV), and second-line injectable drugs (SLIDs) like amikacin.2,3 Mycobacterium tuberculosis isolates exhibiting MDR or rifampicin resistance, coupled with resistance to at least one FQ, are classified as pre-extensively drug-resistant tuberculosis (pre-XDR TB).
Isolates resistant to rifampicin or exhibiting MDR phenotype, along with resistance to at least one FQ and one Group A drug such as bedaquiline, or linezolid, are designated as XDR TB.4 In this article, MDR-TB refers to resistance to at least RIF and INH, pre-XDR TB is defined as MDR-TB with additional resistance to either FQs or SLIDs, and XDR-TB referred to resistance to FQs along with at least one SLID, including amikacin, kanamycin, or capreomycin, consistent with earlier WHO classifications. Among the SLDs, FQs constitute an important component of anti-tubercular drug regimen, most importantly against MDR TB. FQs are broad-spectrum antibiotics that inhibit the growth of M. tuberculosis by binding to DNA gyrase, an ATP-dependent type II topoisomerase that helps in unwinding of double-stranded DNA during DNA replication, RNA synthesis and recombination by introducing negative supercoils. DNA gyrase is a four-part enzyme made up of two different types of subunits, known as A and B. The genes responsible for producing these subunits are gyrA and gyrB. In recent years, antibiotics like moxifloxacin, gatifloxacin, and levofloxacin have been extensively utilized for managing MDR-TB. However, extensive use of FQs for treating bacterial infections, especially for respiratory and as an anti-tubercular drug, has led to the development of FQ resistance. Recent studies from India have reported high rates of fluoroquinolone resistance in Mycobacterium tuberculosis, predominantly associated with gyrA/gyrB-mediated resistance.5,6 Phenotypic resistance to FQs is connected with mutations within the quinolone resistance-determining region (QRDR) of the gyrA and gyrB genes, which encode the DNA gyrase enzyme. Mutations in gyrA confer high-level FQ resistance, whereas gyrB mutations are associated with lower-level resistance. Notably, FQ resistance is commonly linked to mutations in the QRDR, particularly at codons 88-94 of the gyrA gene and codons 500 and 538 of the gyrB gene.2 Recent Indian study further identify transmission risks associated with these gyrA hotspots in high-burden regions.6
Additional SLDs employed in the management of MDR-TB include injectable agents such as aminoglycosides (AGs), specifically kanamycin (KAN), or amikacin (AMK) and the cyclic polypeptide capreomycin (CAP). A significant degree of cross-resistance has been noticed among AGs such as AMK and KAN. Hence, structurally unrelated CAP is used as an alternative for MDR-TB when AG resistance occurs. MTB clinical isolates have demonstrated instances of shared-resistance between AMK/KAN and CAP. Furthermore, CAP resistance is linked to higher treatment failure and mortality rates. AMK/KAN and CAP exert anti-tubercular activity through the inhibition of protein translation. Resistance to SLIDs is frequently attributed to single nucleotide polymorphisms (SNPs) within the rrs gene, which encodes the 16S ribosomal RNA. Mutations in the 1400-1500 region of the rrs gene, especially at positions 1401, 1402, and 1484, are common and linked to specific drug resistance. In the rrs region, G1484T mutation caused high resistance to all three SLIDs In particular, the A1401G point mutation in the rrs gene confers relatively lower resistance to capreomycin and augmented resistance to amikacin/kanamycin. Conversely, the C1402 substitution within the rrs gene yields lower resistance to kanamycin (KAN), increased resistance to capreomycin, and retains susceptibility to Earlier studies have indicated that mutations in the -10 to -14 region and at the -37 promoter region of the eis gene, which encodes aminoglycoside acetyltransferase, result in low-level resistance to KAN. The C-14T SNP within the eis promoter region is the most frequently observed variant, correlating with low-level resistance to kanamycin (KAN).2,7 Additionally, mutations within the tlyA gene, which encodes a 2′-O-methyltransferase responsible for modifications to helix 44 of the 16S rRNA at nucleotide C1409 and helix 69 of the 23S rRNA at nucleotide C1920, contribute to capreomycin resistance.8,9
The spread of XDR-TB has severely hindered global TB control, especially in HIV-prevalent regions. In 2017, an estimated 558,000 MDR/RR-TB cases and 230,000 related deaths were reported globally, with the majority in India and China. Approximately 8.5% of MDR-TB cases were identified as XDR-TB.10 According to data from the WHO, roughly 20.8% of MDR-TB cases exhibited fluoroquinolone resistance in 2018, while 6.2% of these cases were classified as XDR-TB in 2018.1,8,11 India recorded approximately 2.69 million new TB cases in 2018, with 9,700 reported deaths.12,13 Between 2018 and 2020, fluoroquinolone resistance in tuberculosis cases in India ranged from 27.4% to 29.6%, while resistance to SLIDs was observed in 1.3% to 1.5%. Additionally, 5.3% to 6.3% of cases showed resistance to both drug types. The persistence of these resistance patterns is evident in the latest Indian research data.14 Resistance to SLD hinders tuberculosis control strategies, which arises from several contributing factors, including primary infection with MDR-TB, suboptimal or inconsistent treatment regimens, inadequate patient adherence to prescribed protocols, prior exposure to SLDs, irregular or erratic drug utilization, and the administration of substandard SLD medications.15
Timely and early detection of resistance of MTB to SLDs is essential for effective management of MDR-TB and to prevent the development of XDR-TB. While culture-based drug susceptibility testing (DST) remains the most trusted approach, it is characterized by labor-intensive procedures, prolonged turnaround times, and technical complexity. However, the advent of rapid nucleic acid-based molecular tests, including Xpert MTB/RIF and line probe assays (LPAs), has substantially enhanced the management of MDR-TB through significant reductions in testing turnaround times (TATs), specifically 90 minutes for Xpert MTB/RIF, 48 hours for LPAs, and approximately 42 days for culture-based DST.16,17
The Bactec MGIT-960 remains the gold-standard test to detect SLD resistance. However, in 2016, WHO introduced gyrA and rrs gene-based GenoType MTBDRsl V1.0 for detecting mutations conferring SLD resistance in FQ and SLID target genes from smear positive clinical specimens and cultured isolates. To enhance the sensitivity of the GenoType MTBDRsl V1.0, a modified version known as GenoType MTBDRsl V2.0, which includes mutations/SNPs in gyrB and eis promoter region, was introduced recently. GenoType MTBDRsl V2.0 developed by Hain Life Science, built on reverse hybridization technology, detects SLD resistant mutations associated with resistant MTB. Recently, the GenoType MTBDRsl version 2.0 (VER 2.0) LPA has been used for fast, accurate diagnosis of MTB and identification of resistance mutations in FQ resistance determining regions of gyrA (codons 90, 91, and 94) and gyrB genes (codons 536-541) and SLID resistance conferring regions of the rrs gene (1401, 1402, and 1484 nucleotide positions) and promoter region of eis genes (10-14). Furthermore, mutations in the tlyA gene, which encodes a putative 2′-O-methyltransferase (TlyA), confer resistance to capreomycin (CAP). Diagnostic performance studies conducted with GenoType MTBDRsl version 2.0 indicated good sensitivity (FQ 83.6%-100%; AG/CP drugs 94.4%-100%) and specificity (FQ 89.2%-100%; AG/CP drugs 91.4%-98.5%).16,18 High detection rates of fluoroquinolone resistance have also been observed in Northern Indian cohorts using complementary molecular assays such as Xpert MTB/XDR, supporting its utility in high-burden settings.19
Several studies suggest geographic variations in mutations within SLD target genes.20 Comprehending these geographical disparities in SLD resistance mutation frequencies is critical for mitigating the rapid dissemination of MDR-TB. While limited reports exist concerning MDR-TB prevalence and associated drug resistance mutations in this specific region, a dearth of information persists regarding the actual occurrence of pre-XDR TB and XDR-TB.21,22 Real-world WGS data from Indian patients underscore this gap, revealing elevated pre-XDR proportions driven by FQ mutations.23 Also, associated mutations in drug target genes are unknown. Furthermore, the corresponding mutations in drug resistance associated genes remain uncharacterized. Consequently, present study sought to determine the resistance patterns of Mycobacterium tuberculosis clinical isolates to fluoroquinolones (FQs) and second-line injectable drugs (SLIDs) and elucidate the type and frequency of mutations within the gyrA, gyrB, rrs genes, and the eis gene promoter region, utilizing the GenoType MTBDRsl version 2.0 assay.
This investigation took place at the Damien Urban Tuberculosis and Leprosy Center in Nellore, Andhra Pradesh, India. where the tuberculosis diagnosis adhered to the protocols of the Revised National Tuberculosis Control Program (RNTCP), India. Damien TB research centre is a Central TB Division, India, certified TB diagnostic laboratory for testing drug susceptibility of MTB at phenotypic and genotypic levels. The study analyzed samples collected between January and December 2019. Clinical drug-resistant MTB isolates such as MDR TB or RIF mono-resistant isolates confirmed by first-line line probe assay (LPA) (GenoType MTBDRplus Ver 2.0, Hain Lifescience,) or the GeneXpert MTB/RIF assay were selected to study resistance to FQ and SLIDs using second-line LPA (GenoType MTBDRsl Ver 2.0, Hain Lifescience). Clinical sputum specimens underwent digestion and decontamination employing the N-acetyl-L-cysteine/sodium hydroxide (NALC/NaOH) technique, in accordance with the protocol outlined by Kent and Kubica.24 All sputum samples exhibiting acid-fast bacilli (AFB) positivity upon microscopy, as well as smear-negative or scanty AFB samples demonstrating growth on MTB-specific Lowenstein-Jensen (L-J) medium, were included for drug-resistance profiling using LPA analysis. All microbiological procedures were executed within a Class II biosafety cabinet. The Institutional Ethics Committee of SVS Medical College, Mahabubnagar, Telangana, India, approved this study (IRB No. SVSMC/IEC 07/2020).
Line probe assay/GenoType MTBDRsl V2.0 assay
The GenoType MTBDRsl V2.0 (Hain Lifesciences, Nehren, Germany), a qualitative laboratory test conducted as per manufacturer’s guidelines (http://www.hainlifescience.de). The GenoType MTBDRsl V2.0 assay operates based on polymerase chain reaction (PCR) combined with DNA strip technology. The GenoLyse® kit extracted DNA from processed sputum samples and LJ-grown isolates, which was then used for PCR amplification in LPA. Amplification mixtures A (AM-A) and B (AM-B) provided with the kit were thawed and a master mix was prepared (10 microliter of AM-A and 35 microliter of AM-B) in a separate tube. The master mix was gradually added to purified DNA (5 µl), making a total volume of 50 µl. A separate tube with 5 µl nuclease-free water and 45 µl master mix served as a negative control. DNA amplification was performed using a thermal cycler and hybridization of PCR products was done at 45 °C in Twin Cubator/GT-Blot. Strips were developed as per the standard procedures mentioned in the instruction manual. The results were analyzed and compared with the evaluation sheet provided with the kit. Strip contained twenty-seven reaction zones, including conjugate control (CC), amplification control (AC), MTB complex (TUB), gyrA, gyrB, rrs, eis locus control and wild type and corresponding mutant bands of target gene. Within the targeted genetic region, the lack of the native, unmodified sequence, coupled with the identification of a corresponding altered sequence, signifies resistance of the bacterial strain to the corresponding antimicrobial agent. The presence of the native sequence without mutations indicates drug susceptibility. Isolates showing both WT and MUT bands were classified as hetero-resistant.25
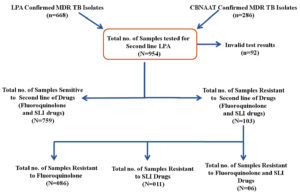
In this study involving 954 specimens, the susceptibility to SLDs was assessed. SLD resistance was identified through WT and MUT probe hybridization in gyrA/gyrB (fluoroquinolone resistance) and rrs/eis (injectable drug resistance) using the GenoType MTBDRsl v2.0 assay. The identification of all WT probe signals for the relevant drug target genes, indicating the absence of mutations, was interpreted as evidence of susceptibility to the respective SLD. The absence of any WT probe band for the drug target gene, with or without the presence of a mutation (MUT) band (i.e., detectable mutations), was interpreted as resistance (Figure 1). Out of 954 isolates, 668 isolates were LPA confirmed and 286 isolates were CB-NAAT confirmed isolates. 759 (79.55%) primary drug-resistant MTB isolates were susceptible to FQ and SLDs, 103 (10.79%) isolates were SLD-resistant and 92 (9.64%) isolates showed results as invalid. Among the 103 second line drug-resistant MTB isolates, 86 (86/103-83.49%; 86/954-9.01%) isolates were mono-resistant to FQ, 11 (11/103-10.67%; 11/954-1.15%) isolates were mono-resistant to SLIDs and 6 (06/103-05.82%; 06/954-0.63%) isolates were resistant to both FQ and SLIDs (Figure 2).
7.40% (02/27) of the LPA confirmed RIF mono-resistant isolates, 12.90% (08/62) of the LPA confirmed MDR TB isolates, 6.04% (35/579) of the LPA confirmed INH mono-resistant isolates and 14.33% (41/286) of the CB-NAAT confirmed RIF-resistant isolates have shown FQ mono resistance. 01.20% (07/579) of the LPA confirmed INH mono-resistant isolates and 01.39% of the (04/286) CB-NAAT confirmed RIF-resistant isolates were SLID mono-resistant. 04.83% (03/62) of the LPA confirmed MDR TB isolates and 01.04% (03/286) of the CB-NAAT confirmed RIF-resistant isolates were extremely drug-resistant, exhibiting resistance to both FQs and SLIDs. Of the 27 LPA confirmed RIF mono-resistant MTB isolates, except two isolates, the rest of the isolates were susceptible to FQs and SLIDs (Table 1).
Table (1):
Second-line anti-TB drug resistance patterns among MTB clinical isolates categorized by first-line drug resistance status
| No. | Isolates information | Total no. of isolates | 2nd line LPA invalid | Susceptible to 2nd line of drugs | Isolates resistant to second-line of MTB drugs | ||
|---|---|---|---|---|---|---|---|
| FQ mono resistant | SLID-mono resistant | XDR (resistant to FQ and SLIDs) | |||||
| 1 | LPA RIF mono-resistant | 027 | 7 (25.92%) | 18 (66.66%) | 02 (07.40%) | 00 (0%) | 00 (0%) |
| 2 | LPA MDR TB | 062 | 6 (09.67%) | 45 (72.58%) | 08 (12.90) | 00 (0%) | 03 (04.83) |
| 3 | LPA INH mono-resistant | 579 | 49 (08.46%) | 488 (84.28%) | 35 (06.04%) | 07 (01.20%) | 00 (0%) |
| 4 | CB-NAAT RIF resistant | 286 | 30 (10.48%) | 208 (72.72) | 41 (14.33) | 04 (01.39%) | 03 (01.04%) |
| Total | 954 | 092 | 759 | 86 | 11 | 06 | |
Demographic profile analysis of the samples included in the study indicated that 5.03% (48/954), 33.22% (317/954) and 61.74% (589/954) of the first line drug-resistant MTB isolates were from age groups less than 20, between 20 and 40 and more than 40, respectively. The majority of the samples studied were from males (80.29%; 766/954) compared to females (19.70%; 188/954). When the treatment history was analyzed, 66.14% (631/954) of the samples were from new TB cases and 33.85% (323/954) from chronic infection cases. Of the 86 FQ mono-resistant MTB isolates, 4 were from individuals under 20, 28 from the 20-40 age group, and 54 from those over 40. Among SLID mono-resistant cases, 5 were in patients 20-40 age group, and 6 were in the over 40 age group. Similarly, four XDR cases were from patients under 20, and two were from the 20-40 age group. 10 (01.30%) isolates from males and 1 (000.53%) isolate from a female were SLID mono resistant. 00.39% of MTB isolates from males and 01.59% of the MTB isolates from females had the XDR phenotype. FQ mono-resistant, SLID mono-resistant and XDR phenotype were observed in 06.33% (40/631), 01.13% (07/631) and 00.47% (03/631) of the new cases, respectively. 14.24% (46/323), 01.23% (04/323) and 00.93% (03/323) of the MTB isolates from chronic infections were FQ mono-resistant, SLID mono-resistant and XDR phenotype, respectively (Table 2).
Table (2):
Demographic profile of the drug-resistant clinical isolates of the study
| Category | Total Isolates (%) | 2nd Line LPA Invalid (%) | Susceptible to 2nd Line (%) | FQ Mono-resistant (%) | SLID Mono-resistant (%) | XDR (%) |
|---|---|---|---|---|---|---|
| Age | ||||||
| <20 | 48 (5.03%) | 7 (14.58%) | 37 (77.08%) | 4 (8.33%) | 0 (0%) | 0 (0%) |
| 20-40 | 317 (33.22%) | 26 (8.20%) | 254 (80.12%) | 28 (8.83%) | 5 (1.57%) | 4 (1.26%) |
| >40 | 589 (61.74%) | 59 (10.01%) | 468 (79.45%) | 54 (9.16%) | 6 (1.06%) | 2 (0.34%) |
| Sex | ||||||
| Male | 766 (80.29%) | 75 (9.80%) | 608 (79.37%) | 70 (9.13%) | 10 (1.30%) | 3 (0.39%) |
| Female | 188 (19.70%) | 17 (9.04%) | 151 (80.31%) | 16 (8.51%) | 1 (0.53%) | 3 (1.59%) |
| History of TB Treatment | ||||||
| New Case | 631 (66.14%) | 59 (9.37%) | 522 (82.72%) | 40 (6.33%) | 7 (1.13%) | 3 (0.47%) |
| Chronic Case | 323 (33.85%) | 33 (10.21%) | 237 (73.37%) | 46 (14.24%) | 4 (1.23%) | 3 (0.93%) |
Out of 86 FQ mono-resistant isolates, highest number of isolates, 54 (62.79%) were in >40 years age group followed by 32.55% (28/86) in 20-40 years age group and 4.65% (04/86) in <20 years age group. 81.39% (70/86) of the FQ mono-resistant isolates were males and 18.60% (16/86) were females. Furthermore, 46.51% (40/86) of the FQ mono-resistant isolates were new cases and 53.48% (46/86) of cases were from chronic infections. Among the SLID mono-resistant, 54.54% (06/11) of isolates were in >40 years age group and 45.45% (05/11) were in 20-40 years age group. 90.90% (10/11) of SLID mono-resistant isolates were from males and 09.09% (01/11) of the isolates from females. Furthermore, highest numbers (07/11; 63.63%) of SLID mono-resistant isolates were from new TB cases followed by chronic infections 36.36% (4/11). 66.66% (04/06) of the XDR isolates were in 20-40 years age group and 33.33% (02/06) isolates from >40 years age group. The number of XDR isolates were same from males (3/6; 50%) and females (3/6; 50%) and new (3/6; 50%) and chronic infections (3/6; 50%) (Table 2).
The type and frequency of mutations conferring resistance to SLD target genes such as gyrA, gyrB, rrs and eis were analyzed in first line drug-resistant isolates. Among the 86 FQ mono-resistant isolates, mutations in gyrA were observed in 77.90% (67/86) isolates, and those in gyrB were observed in 22.09% (19/86) isolates. Analysis of the gyrA gene in 86 FQ mono-resistant MTB isolates identified the D94G mutation (codons 92-96) as the most common, found in 27.90% (n = 24) of cases. The A90V mutation in the gyrA gene (codons 89-93) was the second most common, detected in 18.60% (n = 16) of isolates. Notably, the gyrA gene exhibited an unknown/uncharcheterised profile in 10.46% of the tested isolates (n = 9). Additional mutations identified within the gyrA gene included the following: S91P, observed in 3.48% of isolates (n = 3); D94A, also present in 3.48% of isolates (n = 3); and D94N or D94Y, detected in 3.48% of isolates (n = 3). A heteroresistant phenotype was observed in 9 isolates (10.46%) with D94G mutation being the most prominent and was observed in 7 isolates (8.13%). Eleven isolates exhibited resistance solely to second-line injectable drugs, characterized by notable mutations at the A1401G locus (two isolates) and the G1484T locus (Seven isolates). Among the 11 isolates, 5 were heteroresistant and 2 with unknown mutations in the 1402 region of the rrs gene.
Codon 94 exhibited the highest frequency of mutations, including multiple substitutions (D94G, D94A, D94N/Y) (n = 38) followed by codon 90 (n = 17) and then codon 91 (n = 03) of the gyrA gene in FQ mono-resistant isolates. Aspartic acid (D) was replaced with glycine (G) in 31 isolates, aspartic acid (D) to asparagine (N) in 4 isolates and aspartic acid (D) to alanine (A) in 3 isolates. Mutation in codon 90 resulted in the conversion of amino acid alanine (A) to valine (V) in 16 isolates and mutation in codon 91 resulted in conversion of serine (S) to proline (P) in 3 isolates. As mentioned earlier, there were 9 unknown mutations in the gyrA gene, 19 in gyrB, further among the 11 SLID mono-resistant isolates highest mutations were observed at codon 1484 (n = 07), followed by codon 1401 (n = 02) and two unknown mutations. Mutations at codon position 1484 resulted in conversion of glycine (G) to threonine (T) and mutations at 1401 lead to alanine (A) to glycine (G) conversion. In XDR isolates, mutations in codon 94 (n = 03), 91 (n = 02) and 90 (n = 01) in the gyrA gene, codon 1484 (n = 02), 1402 (n = 01) and 1401 (02) were mutated in the rrs gene. Serine (S) to proline (P) substitution was observed in two isolates, alanine (A) to valine (V) substitution in one isolate. Aspartic acid (D) to alanine (A) and glycine (G) conversion was observed in one isolate each. Alanine (A) to glycine (G) substitution at 1401 codon was observed in two isolates. Glycine (G) to threonine (T) at codon 1484 in two isolates. A C-14t mutation was detected at the -2 promoter region of the eis gene in one XDR isolate (Table 3).
Table (3):
Type and frequency of mutations in SLD target genes of MTB clinical Isolates
| Gene | Band missing | Gene region | Mutations present | Both FQ + SLID resistant | Mono ‘FQ’ Resistant | Mono ‘SLID’ Resistant |
|---|---|---|---|---|---|---|
| + | 89-93 | A90V | – | 1 | – | |
| + | 92-96 | D 94 G | – | 7 | – | |
| + | 92-96 | D 94 N | – | 1 | – | |
| gyr | gyrA WT1, WT2 | 85-90, 89-93 | UK | – | 1 | – |
| gyrA WT1, WT2 | 85-90, 89-93 | S91P | 1 | 0 | – | |
| gyrA WT1, WT2, WT3 | 85-90, 89-93, 92-96 | UK | – | 2 | – | |
| gyrA WT2 | 89-93 | A90V | 1 | 16 | – | |
| gyrA WT2 | 89-93 | S91P | 1 | 3 | – | |
| gyrA WT2 | 89-93 | UK | – | 1 | – | |
| gyrA WT3 | 92-96 | D94A | 1 | 3 | – | |
| gyrA WT3 | 92-96 | D94G | 2 | 24 | – | |
| gyrA WT3 | 92-96 | D94N or D94Y | – | 3 | – | |
| gyrA WT3 | 92-96 | UK | – | 4 | – | |
| gyrA WT3, gyrB WT | 92-96, 536-541 / 497-502 | UK | – | 1 | – | |
| gyrB WT | 536-541 or 497-502 | UK | – | 19 | – | |
| Total | 6 | 86 | ||||
| rrs | + | 1484 | G1484T | 1 | – | 5 |
| rrs WT1 | 1401 | A1401G | 2 | – | 2 | |
| rrs WT1 | 1402 | UK | 1 | – | 2 | |
| rrs WT2 | 1484 | G1484T | 1 | – | 2 | |
| eis | eis WT3 | -2 | C-14t | 1 | – | – |
| Total | 6 | – | 11 | |||
*UK: Unknown Mutations
A single isolate exhibited the D94A mutation in gyrA (codon 94) and the C-14T substitution in the eis promoter region within a RIF-resistant background. Mutational analysis of XDR MTB isolates revealed mutations in the gyrA and rrs genes, while no mutations were detected in gyrB. The mutational profile of XDR MTB isolates is shown in Table 4.
Table (4):
Mutational analysis of XDR-MTB isolates
| Type of Test performed for 1st Line of Drugs | Sample ID | rpoB | katG | inhA | rrs | gyrA | gyrB | eis |
|---|---|---|---|---|---|---|---|---|
| LPA | 1757 | + | + | – | G1484T | D94A | – | – |
| 5097 | + | + | – | 1402 UK | S91P | – | – | |
| 8220 | + | – | + | G1484T | A90V | – | – | |
| CB-NAAT | 2626 | + | – | – | A1401G | S91P | – | – |
| 8222 | + | – | – | A1401G | D94G | – | – | |
| 11900 | + | – | – | – | D94A | – | C-14T |
Multidrug-resistant tuberculosis and extensively drug-resistant tuberculosis represent substantial worldwide public health obstacles to tuberculosis containment. Data published in the 2019 WHO report indicate approximately 10 million incident tuberculosis cases and 1.5 million tuberculosis-related fatalities globally. Among the 1.5 million cases, the majority of deaths occurred in developing countries with 58% of them from Asia alone.11 India contributes to approximately 25% of the worldwide burden of MDR-TB cases. In countries like India, the widespread use of FQs and AGs, alone or combined, for non-mycobacterial infections may have contributed to MTB resistance to SLDs. Intriguingly, tuberculosis is an endemic disease in India and compounded by frequent or routine use of SLDs such as FQ, AM, KM and CPM, making it imperative to study the extent and magnitude of SLD resistance in MTB isolates in this country.11,26
The observed rise in resistance to second-line antituberculosis medications is a significant and escalating concern. Accurate epidemiological data on second line drug-resistance in Andhra Pradesh are currently scarce. Therefore, this investigation sought to find out the prevalence and characterize the mutation patterns associated with second-line drug-resistance in MTB cases within Andhra Pradesh, a state located in southern India, employing the GenoType MTBDRsl v2.0 assay. The study conducted on the first line of drug-resistant pulmonary tuberculosis isolates indicated prevalence of pre-XDR TB cases (n = 97-10.16%) such as FQ mono-resistant (86/954; 9.01%) and SLID mono-resistant (11/954; 1.15%) and XDR phenotypes (n = 06-0.63%) in this region.
This study found a high FQ resistance rate (9.01%) among first-line drug-resistant isolates, including INH mono-resistant and MDR cases. Various studies conducted in India indicated the prevalence percentage of FQ resistance among MDR isolates as low as 3% to as high as 35%. The findings of this study align with earlier reports from India. A particularly concerning observation is the identification of FQ mono resistance drug-resistant Mycobacterium tuberculosis isolates. FQs are critical components of drug-resistant tuberculosis treatment regimens for multidrug-resistant TB cases.27 However, in TB-endemic nations such as India, FQs have also been used largely for the treatment of drug-sensitive MTB and against various community acquired bacterial infections owing to their broad spectrum activity and easy access to these drugs over-the-counter availability. The swift development of fluoroquinolone resistance in MDR-TB cases severely compromises tuberculosis treatment efficacy and presents a major obstacle to contemporary tuberculosis control strategies.26 The WHO End TB Strategy, a component of the Sustainable Development Goals, seeks to eliminate the global tuberculosis epidemic by the year 2035. Achieving this objective necessitates prompt diagnosis and treatment of individuals infected with both drug-resistant and susceptible MTB strains. The WHO End TB Strategy sets specific goals, including a 95% decline in tuberculosis-related deaths, a 90% decrease in the number of new TB cases, and the complete elimination of catastrophic expenses incurred by families affected by the disease.28 In 2017, 8.5% of the XDR MTB cases were reported among MDR TB cases in 127 WHO member countries.10 The majority of these cases-approximately 88% were concentrated in the South-East Asia and Europe regions. In 2017, a total of 10,800 cases of extensively drug-resistant tuberculosis (XDR-TB) were recorded in 77 countries. India reported 2,650 XDR-TB cases, making it the second-highest after the Russian Federation, which reported 3,661 cases, among the top five nations with the highest number of TB cases (Source: https://tbfacts.org/XDR-TB/).
The GenoType MTBDRsl VER 2.0 is a laboratory-based qualitative diagnostic test designed to detect the MTB complex and early, rapidly identify resistance to SLDs, including FQs and AGs/cyclic peptides (CPs) on smear-positive or smear-negative isolates. Additionally, the test rapidly identifies specific mutations linked to SLD resistance. The GenoType MTBDRsl VER 2.0 approved by WHO in 2016 can be used directly on patient samples or MTB culture isolate, and thus, eliminates additional delays associated with culturing the isolates. Version 2.0 includes two additional genes beyond those in Version 1.0: gyrB (linked to FQ resistance) and the eis promoter region (connected with low-level resistance to kanamycin or capreomycin).17 The inclusion of these additional targets has enhanced the assay’s ability to detect phenotypic resistance to fluoroquinolones and kanamycin. The diagnostic efficacy of GenoType MTBDRsl VER 2.0 and mutational analysis of second-line drug target genes under Indian settings was evaluated in various previous studies.17,29 The GenoType MTBDRsl version 2.0 (VER 2.0) offers a fast diagnostic test for MTB detection and the characterization of resistance-associated mutations. Diagnostic performance studies conducted with GenoType MTBDRslversion 2.0 indicated good sensitivity (FQ 83.6-100%; AG/CP drugs 94.4-100%) and specificity (FQ 89.2-100%; AG/CP drugs 91.4-98.5%).16 The phenomenon of heteroresistance, characterized by the occurrence of both drug sensitive (wild-type) and drug-resistant (mutant) MTB within a single sample or isolate, was detected at varying frequencies. Specifically, heteroresistance was observed in 10.46% (9/86) isolates of the FQ mono-resistant and 45.45% (5/11) of the SLID mono-resistant isolates and 16.16% (1/6) isolates of the XDR isolates. Heteroresistance phenotype in second line drug-resistant MTB isolates was reported in studies conducted in South Africa (18 isolates),30 Ukraine (7 isolates)31 and India (27 isolates).29 Heteroresistance phenomenon was very common among various drug-resistant MTB isolates, which is due to a sub-population of MTB strain evolving clonally under antibiotic selection pressure and owing to super-infection of drug-resistant and susceptible isolates. Clonal heterogeneity differs from polyclonal infection, i.e. clonal heterogeneity exhibits distinct Mycobacterial Interspersed Repetitive Unit-Variable Number Tandem Repeat (MIRU-VNTR) patterns at an individual locus, whereas polyclonal infection displays variations at multiple loci within the same sample.32
Treatment of a heteroresistant phenotype often has a negative impact on treatment outcomes. Furthermore, the challenge is compounded by the limitations in achieving rapid and precise phenotypic detection of heteroresistance. This challenge impairs efforts to prevent the selective expansion of drug-resistant MTB strains during therapeutic intervention.18,33 A heteroresistant phenotype was observed frequently in FQ drug-resistant isolates than with any other anti-tubercular drugs used for treatment. Research conducted in countries with high tuberculosis (TB) burdens has demonstrated that the percentage of heteroresistance was more common among FQ-resistant MTB isolates (14%-38%) than other anti-TB drugs.33 Standard phenotypic drug susceptibility tests often fail to identify heteroresistance, where a small subpopulation of cells within an otherwise susceptible strain exhibits resistance. Recent advancements in molecular diagnostics have enhanced heteroresistance detection, including the sloppy molecular beacon melting assay, digital PCR, and next-generation sequencing (NGS).23,34 Future research using NGS is needed to better understand heteroresistance for effective clinical management of drug-resistant MTB. FQs are the preferred choice for treating MTB isolates resistant to first-line drugs. As per the WHO global estimates, around 20.8% of first-line drug resistant isolates exhibited FQ resistance.11
According to the India TB report 2019, out of 59933 second-line LPAs performed, FQ resistance was noticed in 17797 (29.6%) of the cases. In the present study, FQ resistance was identified in 9.01% of MDR-TB cases, which aligns with findings from various other studies conducted in India, in which higher rates of FQ resistance have been reported, that is, 27.3%,13,26 28.87%,35 34.17%,36 30%,37 20.4%,38 34.2%,29 35%,39 57%40 and 39%.41 However, these figures are higher than other studies reported for Zimbabwe (2%),42 Pakistan (7%)43 and Delhi (7.5%).44 The elevated FQ resistance rates observed in those study regions could potentially be attributed to the frequent or routine administration of FQs and misuse of FQ due to over-the-counter availability without any prescription.8 The lower levels of FQ resistance observed in the study region could be attributed to less exposure to the MDR MTB isolates, better diagnostic laboratories, and on-time initiation of treatment protocol. Prompt identification of pre-XDR tuberculosis facilitates closer clinical monitoring, thereby aiding in the prevention of its progression to XDR-TB. This is crucial, as XDR-TB presents significant therapeutic challenges and is frequently associated with unfavorable treatment outcomes.
AGs (amikacin, kanamycin) and capreomycin are the main SLID employed in the control of MDR and XDR MTB. These SLIDs are parenterally administered along with other SLDs. The emergence of MTB isolates in resistance to SLIDs is widely considered a grave concern to public health. According to the India TB report 2019, out of 59933 second-line LPAs performed, SLID resistance was observed in 880 (1.5%) cases. In our study, 1.15% of MDR cases were resistant to at least one of the SLIDs. The figures reported for MTB resistance to SLIDs in India were 4.52%,36 2.13%,35 0.5%-35.9%,37 0.8%,45 1.8%,38 8.1%29 and 1.1%.41
According to the India TB report 2019, out of 59933 second-line LPAs performed, XDR phenotype was detected in 3794 (6.3%) cases. In our study, 0.63% MTB isolates showed the XDR phenotype. These findings are consistent with reports from Thailand (0.38%),46 Iran (0.2%),47 and Ethiopia (0.6%).8 Several other studies conducted in India have shown a higher percentage of XDR cases: 5.53%,36 5.88%,35 4.7%,37 3.7%,38 8.6%,29 and 11.4%.41 Furthermore, higher rates of XDR TB was reported in countries like Sao Paulo, Brazil (9%),48 Ukraine (35%),31 Botswana (5%).49 The 2020 Global Tuberculosis Report stated that 9.0% of all MDR-TB cases were classified as XDR-TB. 12,350 XDR TB cases were reported as of 2019, of which, Europe (8560) had the highest cases followed by South East Asia (2,444), Africa (618), Western Pacific (517), The Americas (138) and the Eastern Mediterranean with the lowest number of XDR cases (73).1
High-level FQ resistance in MTB strains is primarily attributed to SNPs within the QRDR region of the gyrA gene. Conversely, mutations in gyrB generally confer lower FQ resistance, while gyrA mutations are more prevalent than those in gyrB.8,17 In our study, among 86 FQ-resistant isolates, mutations in gyrA were observed in 77.90% (67/86) isolates, and those in gyrB were observed in 22.09% (19/86) isolates. Amino acid substitutions in the QRDR region were most frequently observed at positions 90, 91, and 94, with D94G and A90V emerging as the predominant mutations. These alterations are associated with high-level resistance to FQ. D94G mutation confers MTB isolate resistance against levofloxacin and even fourth-generation moxifloxacin, while A90V mutation makes levofloxacin ineffective. Apart from D94G and A90V, other mutations such as S91P, D94A and D94N/Y were also observed in a few MTB isolates, which confer various levels of resistance to different FQs. Among XDR-TB cases, FQ resistance was correlated with the following amino acid substitutions within the gyrA gene: D94A, S91P, A90V, and D94A. Concurrently, resistance to SLIDs was linked to specific genetic alterations: The G1484T and A1401G mutations in the rrs gene, as well as the C-14t variant in the eis promoter region.
Notably, all observed mutations within the gyrB gene remained uncharacterized. SLID resistance is mainly linked to rrs gene mutations affecting 16S rRNA, leading to high-level aminoglycoside resistance, and eis promoter mutations, which contribute to low-level kanamycin resistance.8,17 The most frequently detected amino acid alterations occur at positions 1401 (A1401G), 1402 (C1402T), and 1484 (G1484T) of the rrs gene. In this study, the G1484T mutation within the rrs gene was identified in 63.63% of SLID-resistant isolates (7 out of 11), while the A1401G mutation was detected in two isolates, and two isolates exhibited uncharacterized mutations. A single isolate exhibiting the XDR phenotype harbored the C-14T mutation in the eis promoter region.
Another significant finding from our study is that the higher number of unknown or un-characterized mutations. In this study, uncharacterized genetic alterations were identified within specific genes of MTB isolates. Specifically, indeterminate mutations were observed in the gyrA gene of 9 isolates, gyrB gene of 19 isolates, and in the rrs gene of 3 isolates. Isolates exhibiting uncharacterized mutations, as determined by the assay used, are defined by the concurrent absence of both the wild-type (WT) and corresponding mutant (MUT) bands in the relevant genetic target region. This indicates a deviation from the expected banding patterns associated with both susceptible and known resistant strains.21 Intriguingly, the majority of these gene mutations were determined in FQ mono resistant MTB isolates, and the occurrence of unknown mutations implies that drug resistance is not due to mutations reported widely in the genic region incorporated in the GenoType MTBDRsl VER 2.0.49 These mutations could likely be rare mutations and further in-depth characterization of these unknown mutations through phenotypic DST and/or gyrA and gyrB sequencing is warranted to identify the novel and emerging second-line drug resistance mutations. Furthermore, studying the relationship between these unknown mutations and treatment outcomes would enable the formulation of personalized treatment. In the recent past, FQ resistance reported in drug-sensitive MTB isolates has been of great concern.26
This investigation yielded crucial data regarding the prevalence and mutation patterns associated with second-line drug resistance within a cohort of first-line drug-resistant tuberculosis cases in Andhra Pradesh, India, by employing the GenoType MTBDRsl VER 2.0. This study represents a novel contribution, being the first to provide such detailed molecular epidemiological data for this specific geographic area. Our findings underscore the occurrence of FQ and SLID mono-resistant isolates, and XDR MTB isolates, highlighting the urgent need for improved surveillance and focused treatment approaches. The study’s results emphasize the importance of further comparative analyses with traditional drug susceptibility testing (DST) methods, such as LJ solid DST or MGIT, to refine and personalize treatment regimens for the effective clinical management of MDR TB.
ACKNOWLEDGMENTS
The consumables and equipment for the GenoType MTBDRsl VER 2.0 assay were generously provided free of cost by the Damien Foundation India Trust for the management of patients with multidrug-resistant tuberculosis (MDR-TB) under the Programmatic Management of Drug-Resistant TB (PMDT). The authors gratefully acknowledge the technical support provided by the administrative and laboratory staff of the Damien Foundation’s TB Research Center, Nellore. The authors also extend their sincere thanks to the District TB Officer for their continuous support.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
VGPP, BSR and USA conceptualized the study. VGPP, BSR, USA, KNM and MSK applied methodology. VGPP, BSR, USA, KNM, MSK, KN and KSV performed formal analysis. VGPP, BSR, USA, KNM, MSK, KN and KSV performed data curation. VGPP, BSR, USA, KNM, MR, BWB, KN, BVSR, SRA, KSV and MSK wrote the original draft. VGPP, BSR, USA, KNM, MR, BWB, KN, BVSR, SRA, KSV and MSK wrote, reviewed and edited the manuscript.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
This study was approved by the Institutional Ethics Committee, SVS Medical College, Mahabubnagar, Telangana, India (SVSMC/IEC Approval N.07/2020).
- World Health Organization (WHO). Global Tuberculosis Report 2020. https://iris.who.int/bitstream/handle/10665/336069/9789240013131-eng.pdf?sequence=1. Accessed August 2, 2025.
- Mabhula A, Singh V. Drug-resistance in Mycobacterium tuberculosis: where we stand. Med Chem Comm. 2019;10(8):1342-1360.
Crossref - Centers for Disease Control and Prevention (CDC). CDC Tuberculosis (TB); Treating Tuberculosis. https://www.cdc.gov/tb/treatment/index.html. Accessed August 2, 2025.
- World Health Organization. WHO announces updated definitions of extensively drug-resistant tuberculosis. Published January 27, 2021. https://www.who.int/news/item/27-01-2021-who-announces-updated-definitions-of-extensively-drug-resistant-tuberculosis. Accessed August 2, 2025
- Dixit R, Mohan E, Gupta A, Patni T. Pattern and characteristics of mutations conferring resistance to second line drugs in Mycobacterium tuberculosis isolates of pulmonary and extrapulmonary TB samples. Indian J Tuberc. 2024;71(Suppl):S37-S43.
Crossref - Nehru VJ, Vandakunnel MJ, Brammacharry U, et al. Risk assessment and transmission of fluoroquinolone resistance in drug-resistant pulmonary tuberculosis: a retrospective genomic epidemiology study. Sci Rep. 2024;14(1):19719.
Crossref - Nath H, Ryoo S. First- and Second-Line Drugs and Drug Resistance. In: Mahboub B, ed. Tuberculosis – Current Issues in Diagnosis and Management. InTech. 2013.
Crossref - Shibabaw A, Gelaw B, Gebreyes W, Robinson R, Wang SH, Tessema B. The burden of pre-extensively and extensively drug-resistant tuberculosis among MDR-TB patients in the Amhara region, Ethiopia. PLoS ONE. 2020;15(2):e0229040.
Crossref - Rao M, Wollenberg K, Harris M, et al. Lineage classification and antitubercular drug resistance surveillance of Mycobacterium tuberculosis by whole-genome sequencing in Southern India. Microbiol Spectr. 2023;11(5):e0453122.
Crossref - World Health Organization. Global Tuberculosis Report 2018. Accessed August 2, 2025. https://iris.who.int/bitstream/handle/10665/274453/9789241565646-eng.pdf
- World Health Organization. Global Tuberculosis Report 2019.; 2019. https://iris.who.int/bitstream/handle/10665/329368/9789241565714-eng.pdf?sequence=19. Accessed August 2, 2025
- Utpat KV, Rajpurohit R, Desai U. Prevalence of pre-extensively drug-resistant tuberculosis (Pre XDR-TB) and extensively drug-resistant tuberculosis (XDR-TB) among extra pulmonary (EP) multidrug resistant tuberculosis (MDR-TB) at a tertiary care center in Mumbai in pre Bedaquiline (BDQ) era. Lung India. 2023;40(1):19-23.
Crossref - Central TB Division of India. India TB Report 2019.; 2019. https://www.tbdiah.org/resources/publications/india-tb-report-2019/. Accessed August 2, 2025.
- Singh N, Singh AK, Kumar S, Chaudhary A, Mishra A, Singh NP. Prevalence and Genetic Profiling of Second-line Drug Resistant Tuberculosis at the Tertiary Care Center of Northern India. Curr Microbiol. 2025;82(4):176.
Crossref - Gopalaswamy R, Palani N, Viswanathan D, et al. Resistance Profiles to Second-Line Anti-Tuberculosis Drugs and Their Treatment Outcomes: A Three-Year Retrospective Analysis from South India. Medicina (Mex). 2023;59(6):1005.
Crossref - Brossier F, Guindo D, Pham A, et al. Performance of the New Version (v2.0) of the GenoType MTBDR sl Test for Detection of Resistance to Second-Line Drugs in Multidrug-Resistant Mycobacterium tuberculosis Complex Strains. J Clin Microbiol. 2016;54(6):1573-1580.
Crossref - Rufai SB, Umay K, Singh PK, Singh S. Performance of Genotype MTBDRsl V2.0 over the Genotype MTBDRsl V1 for detection of second line drug resistance: An Indian perspective. PLoS ONE. 2020;15(3):e0229419.
Crossref - Nguyen TNA, Anton-Le Berre V, Banuls AL, Nguyen TVA. Molecular Diagnosis of Drug-Resistant Tuberculosis; A Literature Review. Front Microbiol. 2019;10:794.
Crossref - Misra R, Das P, Nath A, Neyaz Z. Detection of extensive drug resistance by the Xpert MTB/XDR assay in multidrug resistant tuberculosis cases at a tertiary care centre in northern India, and therapeutic decision making for the six-month BPaLM regimen. J Clin Tuberc Mycobact Dis. 2025;39:100520.
Crossref - Reta MA, Maningi NE, Fourie PB. Patterns and profiles of drug resistance-conferring mutations in Mycobacterium tuberculosis genotypes isolated from tuberculosis-suspected attendees of spiritual holy water sites in Northwest Ethiopia. Front Public Health. 2024;12:1356826.
Crossref - Polu GP, Mohammad SJ, Kota NMK, Karumanchi D, Allam US. Analysis of drug resistance mutations in pulmonary Mycobacterium tuberculosis isolates in the Southern coastal region of Andhra Pradesh, India. Braz J Infect Dis. 2019;23(5):281-290.
Crossref - Reddy R, Alvarez-Uria G. Molecular Epidemiology of Rifampicin Resistance in Mycobacterium tuberculosis Using the GeneXpert MTB/RIF Assay from a Rural Setting in India. J Pathog. 2017;2017(1):1-5.
Crossref - Bhanushali A, Atre S, Nair P, et al. Whole-genome sequencing of clinical isolates from tuberculosis patients in India: real-world data indicates a high proportion of pre-XDR cases. Microbiol Spectr. 2024;12(5):e0277023.
Crossref - Kent PT, Kubica GP. Public Health Mycobacteriology: A Guide for the Level III Laboratory. Centers for Disease Control, Atlanta.1985.
- Hain Lifescience GmbH. GenoType MTBDRsl Ver 2.0: Instructions for Use. Published 2015. https://www.hainlifescience.de/include_datei/kundenmodule/packungsbeilage/download.php?id=2020. Accessed August 2, 2025
- Sharma R, Singh BK, Kumar P, Ramachandran R, Jorwal P. Presence of Fluoroquinolone mono-resistance among drug-sensitive Mycobacterium tuberculosis isolates: An alarming trend and implications. Clin Epidemiol Glob Health. 2019;7(3):363-366.
Crossref - Chatterjee S, Poonawala H, Jain Y. Drug-resistant tuberculosis: is India ready for the challenge? BMJ Glob Health. 2018;3(4):e000971.
Crossref - Migliori GB, Tiberi S, Zumla A, et al. MDR/XDR-TB management of patients and contacts: Challenges facing the new decade. The 2020 clinical update by the Global Tuberculosis Network. Int J Infect Dis. 2020;92(Suppl):S15-S25.
Crossref - Sethi S, Agarwal P, Khaneja R, et al. Second-line Drug Resistance Characterization in Mycobacterium tuberculosis by Genotype MTBDRsl Assay. J Epidemiol Glob Health. 2020;10(1):42.
Crossref - Gardee Y, Dreyer AW, Koornhof HJ, et al. Evaluation of the GenoType MTBDR sl Version 2.0 Assay for Second-Line Drug Resistance Detection of Mycobacterium tuberculosis Isolates in South Africa. J Clin Microbiol. 2017;55(3):791-800.
Crossref - Daum LT, Konstantynovska OS, Solodiankin OS, et al. Next-Generation Sequencing for Characterizing Drug Resistance-Conferring Mycobacterium tuberculosis Genes from Clinical Isolates in the Ukraine. J Clin Microbiol. 2018;56(6):e00009-18.
Crossref - Kamakoli MK, Sadegh HR, Farmanfarmaei G, et al. Evaluation of the impact of polyclonal infection and heteroresistance on treatment of tuberculosis patients. Sci Rep. 2017;7(1):41410.
Crossref - Rigouts L, Miotto P, Schats M, et al. Fluoroquinolone heteroresistance in Mycobacterium tuberculosis: detection by genotypic and phenotypic assays in experimentally mixed populations. Sci Rep. 2019;9(1):11760.
Crossref - Chakravorty S, Kothari H, Aladegbami B, et al. Rapid, High-Throughput Detection of Rifampin Resistance and Heteroresistance in Mycobacterium tuberculosis by Use of Sloppy Molecular Beacon Melting Temperature Coding. J Clin Microbiol. 2012;50(7):2194-2202.
Crossref - Sidiq Z, Hanif M, Chopra KK, Khanna A, Jadhav I, Dwivedi KK. Second-line drug susceptibilities of multidrug- and rifampicin-resistant Mycobacterium tuberculosis isolates in Delhi. Biomed Biotechnol Res J. 2019;3(2):87.
Crossref - Sharma AK, Gupta N, Kala DK, et al. A study on pattern of resistance to second line anti tubercular drugs among multi drug resistant tuberculosis patients. Indian J Tuberc. 2018;65(3):233-236.
Crossref - Rufai SB, Singh J, Kumar P, Mathur P, Singh S. Association of gyrA and rrs gene mutations detected by MTBDRsl V1 on Mycobacterium tuberculosis strains of diverse genetic background from India. Sci Rep. 2018;8(1):9295.
Crossref - Chaubey J, Shrivastava D, Pawar S, Singh B, Sharma R, Sinha S. High degree of fluoroquinolone resistance among extrapulmonary tuberculosis patients at a tertiary care center in North India. Int J Mycobacteriol. 2020;9(3):309.
Crossref - Dalal A, Pawaskar A, Das M, et al. Resistance Patterns among Multidrug-Resistant Tuberculosis Patients in Greater Metropolitan Mumbai: Trends over Time. PLoS ONE. 2015;10(1):e0116798.
Crossref - Mirza IA, Khan FA, Khan KA, Satti L, Ghafoor T, Fayyaz M. Extensively and pre-extensively drug resistant tuberculosis in clinical isolates of multi-drug resistant tuberculosis using classical second line drugs (levofloxacin and amikacin). J Coll Physicians Surg Pak. 2015;25(5):337-341.
- Singhal P, Dixit P, Singh P, Jaiswal I, Singh M, Jain A. A study on pre-XDR & XDR tuberculosis & their prevalent genotypes in clinical isolates of Mycobacterium tuberculosis in north India. Indian J Med Res. 2016;143(3):341-347.
Crossref - Sagonda T, Mupfumi L, Manzou R, et al. Prevalence of Extensively Drug Resistant Tuberculosis among Archived Multidrug Resistant Tuberculosis Isolates in Zimbabwe. Tuberc Res Treat. 2014;2014(1):1-8.
Crossref - Rao NA, Irfan M, Soomro MM, Mehfooz Z. Drug resistance pattern in multidrug resistance pulmonary tuberculosis patients. J Coll Physicians Surg Pak. 2010;20(4):262-265.
- Porwal C, Kaushik A, Makkar N, et al. Incidence and Risk Factors for Extensively Drug-Resistant Tuberculosis in Delhi Region. PLoS ONE. 2013;8(2):e55299.
Crossref - Sharma R, Sharma SK, Singh BK, Mittal A, Kumar P. High degree of fluoroquinolone resistance among pulmonary tuberculosis patients in New Delhi, India. Indian J Med Res. 2019;149(1):62-66.
Crossref - Reechaipichitkul W, Tubtim S, Chaimanee P. Drug susceptibility patterns of Mycobacterium tuberculosis and clinical outcomes of drug-resistant tuberculosis at Srinagarind Hospital, a tertiary care center in northeastern Thailand. Southeast Asian J Trop Med Public Health. 2011;42(5):1154-1162.
- Tasbiti AH, Yari S, Ghanei M, Shokrgozar MA, Fateh A, Bahrmand A. Low Levels of Extensively Drug-resistant Tuberculosis among Multidrug Resistant Tuberculosis Isolates and Their Relationship to Risk Factors: Surveillance in Tehran, Iran; 2006 to 2014. Osong Public Health Res Perspect. 2017;8(2):116-123.
Crossref - Matsui T, Pinhata JMW, Rabello MCdS, et al. Frequency of first and second-line drug resistance-associated mutations among resistant Mycobacterium tuberculosis clinical isolates from Sדo Paulo, Brazil. Mem Inst Oswaldo Cruz. 2020;115:e200055.
Crossref - Mogashoa T, Melamu P, Derendinger B, et al. Detection of Second Line Drug Resistance among Drug Resistant Mycobacterium Tuberculosis Isolates in Botswana. Pathogens. 2019;8(4):208.
Crossref
© The Author(s) 2025. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.