ISSN: 0973-7510
E-ISSN: 2581-690X
Bacteriophage, a predator virus of bacteria, is an abundant biological entity in the biosphere. With ultimate applications in medicine and biotechnology, new phages are extensively being isolated and characterized. The objective of the present study was to characterize lytic bacteriophage vB_Pae-PA14 infecting Pseudomonas aeruginosa ATCC 27853 that was isolated from seawater in Thailand. vB_Pae-PA14 was subjected to whole genome phylogenetic analysis, host range test, biofilm test and characterization. Results showed that the phage belonged to a group of N4-like viruses, could infect P. aeruginosa isolates including carbapenem-resistant P. aeruginosa. The burst size of vB_Pae-PA14 was 86 plaque-forming unit/infected cells. Also, the phage showed a greater ability to control planktonic P. aeruginosa cells than the biofilm cells. Phage could withstand physical stresses especially the high salt concentration. In brief, lytic bacteriophage vB_Pae-PA14 infecting P. aeruginosa was isolated and characterized, which might be useful in further bacteriophage lytic applications.
Bacteriophage, Lytic, Pseudomonas aeruginosa, N4-like viruses, Carbapenem-resistant Pseudomonas aeruginosa
Pseudomonas aeruginosa, a Gram-negative, aerobic bacteria are part of the normal flora of soil and water sources. It can cause infection in an immunocompromised patient due to broad-spectrum antibiotic resistance and diverse virulence factors including biofilm.1,2 It also contaminates and causes spoilage in food.3,4 These problems are a global issue not only limited to developing countries. Drugs and chemical sanitizers are mainly used to control P. aeruginosa.5 Alternatively, biological control of foodborne pathogens using bacteriophage has been evaluated.6 Furthermore, multidrug-resistant P. aeruginosa, especially carbapenem-resistant P. aeruginosa (CRPA) is one of the major issues to be a concern worldwide.7 Recently in the novel COVID-19 pandemic which antibiotics are highly likely overused to protect the patient, the forthcoming emergence of antibiotic resistant bacteria is inevitable.8 Hence, alternative treatment or preventive options are urgently required, with lytic bacteriophages being one of the possible candidates.
Bacteriophages (phages) have gained attention recently.9 Viruses infecting bacteria are abundant and play ecological roles.10,11 The previous review suggested that phages have their uses as vaccines and therapeutic agents.12 The recent breakthrough in phage therapy has been medical studies.13,14 Furthermore, the phage and host interaction might be exploited in biotechnology and other applications.15,16 Noteworthy is that phage genomes are diverse.17 Electron microscopy is one of the ways to systematically classify bacteriophages.18 Using molecular characterization, the phage genome might be de novo sequenced.19
Thai seawater is one of the possible ecological sources of phages but there has been no report on the isolation of Pseudomonas phage. In this work, seawater bacteriophage vB_Pae-PA14 was isolated using P. aeruginosa ATCC 27853 as a host. The genome sequence of the newly isolated vB_Pae-PA14 showed that it is an N4-like virus. The host range was examined, and the characterization was performed. In brief, this study provides information on the newly isolated phage in controlling P. aeruginosa.
This study was non-clinical, qualitative research on the bacteriophage isolate; and was performed at the Department of Microbiology, Faculty of Science, Bangkok, Thailand.
Strains and cultivation
Bacterial strains were stored in glycerol stock at -80˚C freezer at the Department of Microbiology, Faculty of Science until use. Cultures were streaked on tryptic soy agar (TSA; Himedia Pvt. Ltd., Mumbai, India) and cultivated in tryptic soy broth (TSB; Himedia Pvt. Ltd., Mumbai, India). P. aeruginosa ATCC 27853 was used as the host for bacteriophage isolation. Staphylococcus aureus ATCC 6538, Escherichia coli ATCC 25922, Klebsiella pneumoniae ATCC 13883, Listeria monocytogenes ATCC 7644 and E. coli DH5a (Life Technologies) were used as control. The P. aeruginosa isolate TN87 was obtained from the screening of P. aeruginosa in retail foods. Thirty-four (34) carbapenem-resistant P. aeruginosa (CRPA) strains were used in the study. P. aeruginosa isolates were cultured in TSB at 37˚C, 200 rpm using a rotary shaker. TSA was used in a double-layer agar (DLA) technique where the top layer contained 5 ml of 0.7% (w/v) agar.20
Isolation and purification of bacteriophages
Bacteriophage was isolated after enriching the water sample as described previously.21 To isolate bacteriophage, a seawater sample from Chonburi province, Thailand (12°36’29.1″N 100°55’51.5″E) was collected in January 2020. The water sample was mixed with overnight culture broth of P. aeruginosa ATCC 27853 and 10x TSB medium (45:0.5:5 ml) and incubated with shaking for 20 h. After enrichment, sample was centrifuged at 10,000×g for 10 min to pellet the host bacteria and the supernatant was passed through a 0.22 μm pore size polyethersulfone syringe filter (PURADISC 25 AS, Whatman) to remove host bacteria and other contaminants. The filtrate was mixed with the host and molten soft agar in the DLA method. The plates were incubated at 37˚C for 16 h and plaques appearance indicated the presence of phages. Distinct plaques were picked up and resuspended in 0.5 ml SM buffer (100 mM NaCl, 50 mM Tris pH 7.5, 8 mM MgSO4).22 Purification was performed at least 3 passages, using the described method to obtain a single plaque morphology on macroscopic observation. The phage titer was determined. The plaque diameter was measured (n=10) using a Vernier caliper.
To prepare a high titer phage sample, the phages were collected from confluent lysis plaques on a DLA plate. Three (3) ml of SM buffer was added to the surface of the top agar containing confluent lysis, and the plates were shaken on a rotary shaker at 100 rpm for 1 h. The solution was retrieved, centrifuged, and filtered. Purified phage with high titer was stored at 4˚C.
Host range determination
To check the phage specificity, a spot test was performed using the previously described protocol with modifications.23 Target bacteria-16 representative bacterial strains and 34 CRPA isolates-in liquid overnight culture were embedded onto TSA DLA and a spot test was performed. After the top agar had set, 5 μl of bacteriophage solution (109 plaque forming unit/ml (PFU/ml) and its 10-fold dilutions) was spotted over the bacterial lawn to evaluate the phage infectivity. After overnight incubation, clear lysis on the spot showed bacteriophage activity. SM buffer was used as a negative control.
Electron microscopy
The phage stock was adsorbed on 300 mesh carbon-coated copper grids and stained with 2% uranyl acetate for 1 min then air-dried. Electron microscopic analysis was performed at Scientific Equipment and Research Division, the Kasetsart University Research and Development Institute on Hitachi High-Technology HT7700 transmission electron microscope operated at 80 kV to determine the phage virion morphology.
Genomic extraction and analysis
Phage genomes were extracted by a column-based method after treatment of filtered phage with DNaseI using BioFACT Genomic DNA Prep Kit (Daejeon, South Korea) following manufacturer protocol. Shotgun library preparation and 500 Mb throughput analysis on HiSeq2500 100PE, including de novo assembly were performed at Macrogen Inc. (South Korea). Open reading frame prediction was analyzed on RASTtk pipeline24 and GeneMarkS,25 and then manually edited. Putative genes were searched on BLASTp.26 Transfer RNA sequences were searched using tRNAscan-SE.27 A genomic comparison was made on VICTOR platform28 using FASTME D6 formula. Branch support was inferred from 100 pseudo-bootstrap replicates each.
Bacterial challenge and One-step growth curve
Bacterial challenge assay and one-step growth curve analysis were performed as previously described procedures with modifications.29 For bacterial challenge assay, the active P. aeruginosa ATCC 27853 culture was separately infected with phage at a multiplicity of infection (MOI) of 0.01, 0.1, or 1. The OD600 was measured over a time course using GENESYS 20 spectrophotometer (Thermo Fisher Scientific). Experiments were performed in triplicate and the uninfected culture was used as control.
For the one-step growth curve analysis, phage in SM buffer was mixed with early log phase culture of P. aeruginosa ATCC 27853 at the MOI of 0.01 at room temperature (30°C ± 2°C) and allowed to adsorb for 10 min. The adsorption content was resuspended in fresh TSB medium and incubated at 37°C with shaking. Samples were taken at intervals from 20 to 90 min and directly plated on TSA DLA. Experiments were performed in triplicate. The growth curve between time and bacteriophage density was plotted. The burst size was calculated from the ratio of phage density at the burst period to that of the end of the latent period.
Biofilm assay
Bacteriophage biofilm reduction assay was performed using a method adapted from the previously described protocol.30 The diluted P. aeruginosa ATCC 27853 cells (150 μl) were dispensed in wells of a flat bottom microplate with a blank medium as control. After 48 h incubation, wells were washed three times with phosphate-buffered saline (PBS) buffer pH 7.4. Phage stock was diluted in TSB medium and then dilutions containing 107, 105, 103, 10 and 1 PFU/150 μl were added to replicate wells (n=8) followed by 24 h incubation. Staining was performed by 0.1% (w/v) crystal violet solution, 30 min. After washing the unbound stain, the plate was air-dried and 150 μl of 30% (v/v) glacial acetic acid was added. The microplate was measured on a microplate reader (Thermo Multiskan GO; Thermo Fisher Scientific) at 570 nm. Experiments were performed in triplicate. The percent density was subtracted from background TSB and blank acetic acid and the percent of density relative to phage untreated wells was calculated.
Bacteriophage stability tests
Physical stability studies were tested at different levels of salinity, pH and temperature using protocols with modifications.31,32 Bacteriophage titer was adjusted to 106 PFU/ml in SM buffer and subjected to the following conditions. The pH stability was tested by incubating phages in pH-adjusted PBS for 60 min at 37°C followed by neutralization to pH 7.0. Temperature tolerance testing was conducted by incubating for 60 min. The boiling was carried out for 5 min and -20°C treatment was for 12 h. Salinity tolerance was tested by inoculating phage in distilled water containing varying NaCl concentrations for 60 min at 37°C. To determine the effect of chemicals to bacteriophage titers, sample was subjected to following treatments: 63% ethanol, 90% acetone, 50% chloroform, 0.09% sodium dodecyl sulfate (SDS), 0.06% sodium hypochlorite, 0.1% HgCl2 and 2.5% phenol. Commercial Multi-use disinfectant and Antiseptic disinfectant were diluted to obtain 0.1% active ingredient in sterile distilled water. The contact time is listed in Table 1. Following all treatments, dilutions of phage were plated on DLA immediately. Plaques are counted after overnight incubation. The percentage available was relative to availability in distilled water, at pH 7.0, at 4˚C and SM buffer for NaCl, pH, temperature and chemicals, respectively. Triplicate experiments were performed except in the chemical stability test that duplicates were performed.
Statistical analysis
Analysis of variance was performed in the data of the biofilm test and the stability test. Mean values ± SD were shown. Significance was considered at p-value < 0.05.
Pseudomonas phage vB_Pae-PA14 isolation and its morphological analysis
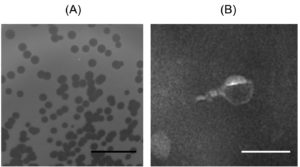
To isolate seawater bacteriophage, water was sampled from Chonburi province, Thailand. Phage was successfully isolated using P. aeruginosa ATCC 27853 as a host and named vB_Pae-PA14. The vB_Pae-PA14 appears with a clear plaque and plaque diameter of 1.5±0.1 mm on TSA DLA (Fig. 1A). When observed by transmission electron microscope, vB_PA14 has a short tail length morphology that can be classified as Podovirus (tail length < 40 nm) (n=4) (Fig. 1B).33
Fig. 1. Plaque morphology and morphological analysis of Pseudomonas phage vB_Pae-PA14
(A) Double-layer agar plating of vB_Pae-PA14 on TSA medium. The black scale bar indicates 1 cm.
(B) Virion morphology analysis by transmission electron microscope. vB_Pae-PA14 was observed at 25,000 times magnification, the white scale bar was 100 nm.
Host range analysis
Spot testing on different bacterial DLA plates was performed to investigate the host range and specificity of the vB_Pae-PA14. Sixteen bacterial strains in their log phase were embedded on DLA before 5 μl of phage suspensions were spotted. Spot testing showed that P. aeruginosa isolates, including the host and the food isolate TN87, were lyzed (Table 1). Furthermore, the result showed that this phage could form lysis on 21 CRPA isolates out of 34 tested (61%) (Supplementary Table 1). Results suggested that the vB_Pae-PA14 had a narrow host range and could form lysis on P. aeruginosa isolates in vitro.
Table (1):
Host range of vB_Pae-PA14.
Test strain |
vB_Pae-PA14 lysis |
|---|---|
Acinetobacter baumannii |
– |
Bacillus cereus |
– |
Bacillus subtilis |
– |
Escherichia coli ATCC 25922 |
– |
E. coli DH5α |
– |
E. coli O157:H7 |
– |
Klebsiella pneumoniae ATCC 13883 |
– |
Listeria innocua |
– |
Listeria monocytogenes ATCC 7644 |
– |
Proteus mirabilis |
– |
Pseudomonas aeruginosa ATCC 27853 (host) |
+ |
P. aeruginosa food isolate TN87 |
+ |
Pseudomonas fluorescens |
– |
Staphylococcus aureus ATCC 6538 |
– |
Staphylococcus aureus MRSA |
– |
Streptococcus epidermidis |
– |
+: lysis observed, -: no lysis observed
Genomic analysis
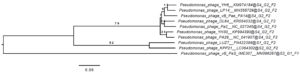
To look into genomic identity, the genome of vB_Pae-PA14 was sequenced and submitted to Genbank (accession number MZ229661). The double-stranded DNA genome contains 73,404 bp, 55.1% GC with predicted 92 open reading frames. tRNA sequence was not found in the genome. The phylogenetic tree of the whole genome sequence analyzed on the VICTOR platform (Fig. 2) showed that the vB_Pae-PA14 is closely related to Pseudomonas phage YH6 and belongs to the same clade with Pseudomonas phage Pa2, LP14 and PA26, while different from Pseudomonas phage KPP21. The comparison of deducted amino acid sequence of major capsid protein showed that the vB_Pae-PA14 belongs to the N4 type major capsid protein group. Furthermore, the DNA sequence of the predicted amino acid sequence of open reading frame number 32 also showed no similarity to sequences listed in the NCBI database. No putative sequence of integrase or virulence gene was found.
Fig. 2. Phylogenetic analysis of whole vB_Pae-PA14 bacteriophage genome
The genome sequence of vB_Pae-PA14 was compared to other deposited Pseudomonas phage genomes in GenBank. Phylogenomic Genome BLAST Distance Phylogeny (GBDP) trees inferred using D6 formula yielding average support of 34%. The numbers above branches are GBDP pseudo-bootstrap support values from 100 replications. The branch lengths of the resulting VICTOR trees are scaled in terms of the respective distance formula used. Two clusters are visualized.
Bacterial challenge assay and one-step growth curve of vB_Pae-PA14
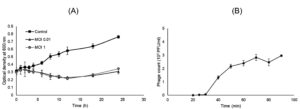
To determine the bacteriolytic activity, a bacterial challenge assay was conducted. The growth inhibition of P. aeruginosa ATCC 27853 occurred at different MOIs tested (Fig. 3A). MOI of 0.01, 1 and 0.1 (chart not shown) offered a similar inhibition pattern up to 12 h post-challenge compared to the control experiment (no phages added). After that, a gradual increase in the optical density of both MOI 0.01 and 1 treated culture was observed. The result suggests the strong and persisting lytic activity of vB_Pae-PA14, that only MOI 0.01 was required to inhibit the culture of P. aeruginosa. And vB_Pae-PA14 was at least stable for 12 h at 37°C in tested conditions.
The one-step growth kinetics of vB_Pae-PA14 showed a latent period of 25 min, a burst period at 70 min, and a burst size of 86 PFU/infected cell when infecting P. aeruginosa ATCC 27853 (Fig. 3B).
Fig. 3. Bacterial challenge assay and one-step growth curve analysis of vB_Pae-PA14
(A) Bacterial challenge of vB_Pae-PA14 on a culture of Pseudomonas aeruginosa ATCC 27853 in TSB medium with 10 mM CaCl2 at 37°C.
(B) One-step growth curve analysis of vB_Pae-PA14 on a culture of Pseudomonas aeruginosa ATCC 27853 in TSB medium at 37°C with agitation. Error bar = ± SD, MOI = multiplicity of infection and PFU = plaque forming units.
Biofilm reduction assay
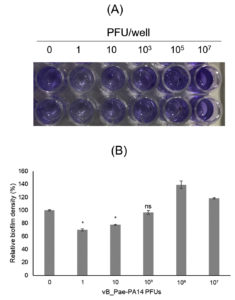
Biofilm formation is one of P. aeruginosa virulence factors.34 To test the biofilm inhibition activity of vB_Pae-PA14, after the biofilm of P. aeruginosa ATCC 27853 was formed, phage dilutions were added and incubated for 24 h. After removal of planktonic cells and multiple washings, biofilms were stained with crystal violet. The assessment of biofilm biomass was performed by measuring the well content at 570 nm. Fig. 4 showed that vB_Pae-PA14 at 1 and 10 PFU markedly reduces 30% and 23%, respectively, of formed biofilm compared to the control treatment. When the higher titers were used; however, biofilm was not eliminated. The 107 PFU treatment resulted in an increase of 18% biofilm and 105 PFU phages resulted in 39% increased biofilm. The 48 h phage with incubation has an effect that biofilm cannot be eliminated by phage treatments, showing a similar density to control (result not shown).
Fig. 4. Biofilm inhibition test
(A) Photograph of a microplate containing stained and dissolved biofilm of Pseudomonas aeruginosa ATCC 27853.
(B) Relative biofilm density of different vB_Pae-PA14 PFUs treatments. PFU = plaque forming units, error bar = ± SD, * = statistical significance was reported at p < 0.05 and ns = non significant.
Stability test
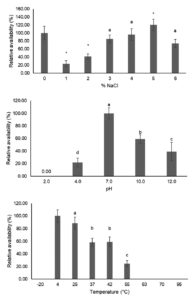
The stability of phage is one of the concerns regarding further applications. Three parameters of vB_Pae-PA14 stability (salinity, pH and temperature) were investigated. The result is shown in Fig. 5. The vB_Pae-PA14 could tolerate various concentrations of NaCl, up to 6%. The phage density in 3% and 4% NaCl was not different from that of the control. When tested with different pH levels, phage was viable from ranges of pH 4.0 to pH 12.0 with a significant titer decrease. The availability in pH 4.0, pH 10.0 and pH 12.0 were 21%, 58% and 39%, respectively. An increasing temperature beyond 25°C notably effects by decreasing the phage density and temperature above 63°C could inactivate vB_Pae-PA14. Beyond that, phage was not detected even in the undiluted samples. Moreover, it was not stable at -20°C in SM buffer without preservatives added. For chemical tolerance, residual vB_Pae-PA14 were found in 0.09% SDS and commercial disinfectants tested (Table 2). Phage was not able to tolerate other solvents and disinfectants tested.
Table (2):
Chemical stability of vB_Pae-PA14.
Chemicals |
Contact time (min) |
Relative availability (%) |
|---|---|---|
63% ethanol |
60 |
N.D. |
90% acetone |
60 |
N.D. |
50% chloroform |
90 |
N.D. |
0.09% SDS |
10 |
20.7 |
0.06% sodium hypochlorite |
10 |
N.D. |
Multi-use disinfectant (0.1% 4-chloro-3-methylphenol) |
10 |
29.6 |
Antiseptic disinfectant (0.1% chloroxylenol) |
10 |
16.2 |
0.1% HgCl2 |
10 |
N.D. |
2.5% phenol |
10 |
N.D. |
Mean from independent replicate (n=2) is shown, N.D.: not detected
Fig. 5. Stability of vB_Pae-PA14 in different conditions
Percentage available was relative to availability in distilled water, at pH 7.0 and at 4˚C for NaCl concentration, pH and temperature, respectively. Error bar = ± SD, * = statistical significance was reported at p < 0.05 and different letters indicate significant differences in means.
P. aeruginosa can be found in many ecological sources from soil to water. It is considered as pathogenic bacteria in humans causing a variety of infections and resistance to multiple antimicrobials. Its bacteriophage is highly likely to be found wherever the host is present. There have been reports of marine Pseudomonas phage, but in Thailand, this has not been reported. In the current study, Pseudomonas phage vB_Pae-PA14 was isolated from seawater sampled in Chonburi province, Thailand by using P. aeruginosa ATCC 27853 as a host for isolation of phage.
Morphological analysis showed that vB_Pae-PA14 is a Podovirus that forms a clear plaque on the DLA plate. The host range of vB_Pae-PA14 was limited to P. aeruginosa strains. The phage could infect one P. aeruginosa food isolate and 61% of CRPA isolates but no activity was observed with other bacteria. It is possible that this phage isolate could be effective in controlling clinical P. aeruginosa isolates, especially CRPA. The genome was sequenced, and the virus was found to be an N4-like virus. The phage belonging to genus N4-like viruses could be useful as a therapeutic agent.35 vB_Pae-PA14 is highly related to the branch containing Pseudomonas phage Pa2 and YH6, the gene arrangement showed a highly conserved genome of N4-like viruses isolated from various locations.36 Moreover, none of the harmful genes such as toxin production or lysogenization were found. This genomic property of vB_Pae-PA14 would not prohibit the potential of therapeutic use.
Biofilm of P. aeruginosa ATCC 27853 was inhibited when lower titers of vB_Pae-PA14 were used. However, when higher titers were used the bacterial biofilm level was increased. The reasons might be that P. aeruginosa cells are stressed and becoming more difficult to eliminate when formed into biofilm or had assembled into higher-order biofilm structure37 thus tolerant to phages. Beyond 24 h incubation, phages might unstable and lose the activity, failing to disrupt the biofilm. Planktonic cell killing in microplate was not tested in this study, but it might be inferred from the result of bacterial challenge assay that vB_Pae-PA14 is more effective in controlling the unadhered cell more than the preformed biofilm. Overall, it can be suggested that vB_Pae-PA14 should be added to bacteria before they can bind to a surface or use in combination with other phages as a cocktail to eliminate preformed biofilm.
Phage vB_Pae-PA14 was active at physiological temperature, comparable to another report.38 However, it was inactivated at 63°C which would limit its use in a high-temperature environment such as in compost,39 but might be applicable to surfaces as an anti-P. aeruginosa agent. This data showed that vB_Pae-PA14 can be inactivated at using pasteurizing temperature after usage. Phage vB_Pae-PA14 showed the highest activity at pH 7.0 and reduced availability was observed at a pH range from 4.0 to 12.0. Ranges of pH stability are essential for phage to find use in different applications. For therapeutic uses, a phage must be stable between pH 6.0 and 8.0.40 For NaCl tolerance, vB_Pae-PA14 could withstand the NaCl concentration as high as 6% with the highest density at 5% NaCl. The low salt concentration (1%); however, resulted in the lowest density of phage. This is consistent with the level of salinity in seawater that phage was isolated from. This phage might find its application in controlling P. aeruginosa in salt-fermented foods. To test tolerance to chemicals in various conditions, phages were subjected to solvents and disinfectants with contact time listed by the manufacturer. Most of the chemicals tested have reduced phage to a not-detectable level. A fraction of phage can tolerate incubation with SDS, a general detergent. Differently, a part of vB_Pae-PA14 seemed to endure after treating with both formulae of disinfectant. The fact that it is a non-enveloped virus, so the contact time might need to be increased to completely disrupt it.
Pseudomonas phage has many applications. In agricultural biocontrol, commercialized phages that is stable to variable physical and chemical stresses are available.41 P. aeruginosa belongs to the ESKAPE group (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp.) that are pathogens associated with hospital infection and exhibit virulence along with a drug resistance phenotype.42 As prolonged use of antibiotics has generated drug-resistant strains, perhaps the most important application of a phage is for the elimination of antibiotic-resistant or multidrug-resistant P. aeruginosa.8,43,44 After in vitro characterization, animal models have been used for studies.45,46 For phage therapy in humans, as an alternative to using chemical drugs, is an ultimate goal and studies have suggested phage in treatments and prophylaxis, in this era of emerging antibiotic resistance.13,47-50
In summary, newly seawater isolate vB_Pae-PA14 was a member of N4-like viruses. Furthermore, the virus can inhibit one food isolate P. aeruginosa and CRPA isolates. This lytic phage is stable in physiological conditions as well as high salinity. Also, it can effectively reduce the number of P. aeruginosa planktonic cells. Isolated phage is presented as a candidate to control bacterial activity, and might have its application in the future. Further experimentation such as confirming the phage activity in food matrices and the performance on drug-resistant isolates are required to obtain more information for use of newly isolated phages in controlling P. aeruginosa infections.
Additional file: Additional Table 1.
ACKNOWLEDGMENTS
J Soonthornsima, K Chanworawit and N Choychod provided assistance. J Chuerduangphui assisted with proofreading an earlier version of the manuscript.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
This work was supported by the Kasetsart University Research and Development Institute (KURDI) and the Faculty of Science, Kasetsart University, Bangkok, Thailand (Grant number 8.63).
ETHICS STATEMENT
Not applicable.
AVAILABILITY OF DATA
The sequence of bacteriophage genome was available in the NCBI GenBank, accession number MZ229661.
- Streeter K, Katouli M. Pseudomonas aeruginosa: a review of their pathogenesis and prevalence in clinical settings and the environment. Infect Epidemiol Med. 2016;2:25-32.
Crossref - Chegini Z, Khoshbayan A, Moghadam MT, Farahani I, Jazireian P, Shariati A. Bacteriophage therapy against Pseudomonas aeruginosa biofilms: a review. Ann Clin Microbiol Antimicrob.2020;19(1):45.
Crossref - Raposo A, Perez E, de Faria CT, Ferrus MA, Carrascosa C. Food spoilage by Pseudomonas spp.-An overview. Foodborne Pathogens and Antibiotic Resistance.2017:41-71.
Crossref - Quintieri L, Fanelli F, Caputo L. Antibiotic resistant Pseudomonas spp. spoilers in fresh dairy products: An underestimated risk and the control strategies. Foods. 2019;8(9):372.
Crossref - Meesilp N, Mesil N. Effect of microbial sanitizers for reducing biofilm formation of Staphylococcus aureus and Pseudomonas aeruginosa on stainless steel by cultivation with UHT milk. Food Sci Biotechnol. 2018;28(1):289-296.
Crossref - Kazi M, Annapure US. Bacteriophage biocontrol of foodborne pathogens. J Food Sci Technol. 2016;53(3):1355-1362.
Crossref - Nordmann P, Poirel L. Epidemiology and diagnostics of carbapenem resistance in Gram-negative bacteria. Clin Infect Dis. 2019;69(Suppl 7):S521-s528.
Crossref - Wojewodzic MW. Bacteriophages could be a potential game changer in the trajectory of coronavirus disease (COVID-19). Phage. 2020;1(2):60-65.
Crossref - Duzgunes N, Sessevmez M, Yildirim M. Bacteriophage therapy of bacterial infections: The rediscovered frontier. Pharmaceuticals. 2021;14(1):34.
Crossref - Clokie MR, Millard AD, Letarov AV, Heaphy S. Phages in nature. Bacteriophage. 2011;1(1):31-45.
Crossref - Weitz JS, Poisot T, Meyer JR, et al. Phage-bacteria infection networks. Trends Microbiol. 2013;21(2):82-91.
Crossref - Clark JR, March JB. Bacteriophages and biotechnology: vaccines, gene therapy and antibacterials. Trends Biotechnol. 2006;24(5):212-218.
Crossref - LaVergne S, Hamilton T, Biswas B, Kumaraswamy M, Schooley RT, Wooten D. Phage therapy for a multidrug-resistant Acinetobacter baumannii craniectomy site infection. Open Forum Infect Dis. 2018;5(4):ofy064-ofy064.
Crossref - Schooley RT, Biswas B, Gill JJ, et al. Development and use of personalized bacteriophage-based therapeutic cocktails to treat a patient with a disseminated resistant Acinetobacter baumannii infection. Antimicrob Agents Chemother. 2017;61(10):e00954-17.
Crossref - Stone E, Campbell K, Grant I, McAuliffe O. Understanding and exploiting phage-host interactions. Viruses. 2019;11(6):567.
Crossref - Harada LK, Silva EC, Campos WF, et al. Biotechnological applications of bacteriophages: State of the art. Microbiol Res. 2018;212-213:38-58.
Crossref - Deveau H, Labrie SJ, Chopin M-C, Moineau S. Biodiversity and Classification of Lactococcal Phages. Appl Environ Microbiol. 2006;72(6):4338-4346.
Crossref - Ackermann HW. Phage classification and characterization. Methods Mol Biol. 2009;501:127-140.
Crossref - Hatfull GF, Hendrix RW. Bacteriophages and their genomes. Curr Opin Virol. 2011;1(4):298-303.
Crossref - Kropinski AM, Mazzocco A, Waddell TE, Lingohr E, Johnson RP. Enumeration of bacteriophages by double agar overlay plaque assay. Methods Mol Biol.. 2009;501:69-76.
Crossref - Bao H, Zhang H, Wang R. Isolation and characterization of bacteriophages of Salmonella enterica serovar Pullorum. Poult Sci. 2011;90(10):2370-2377.
Crossref - Born Y, Knecht LE, Eigenmann M, Bolliger M, Klumpp J, Fieseler L. A major-capsid-protein-based multiplex PCR assay for rapid identification of selected virulent bacteriophage types. Arch Virol. 2019;164(3):819-830.
Crossref - Jensen KC, Hair BB, Wienclaw TM, et al. Isolation and host range of bacteriophage with lytic activity against methicillin-resistant Staphylococcus aureus and potential use as a fomite decontaminant. PLoS One. 2015;10(7):e0131714.
Crossref - Brettin T, Davis JJ, Disz T, et al. RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci Rep. 2015;5:8365.
Crossref - Besemer J, Lomsadze A, Borodovsky M. GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Research. 2001;29(12):2607-2618.
Crossref - Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403-410.
Crossref - Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Research. 1997;25(5):955-964.
Crossref - Meier-Kolthoff JP, Goker M. VICTOR: genome-based phylogeny and classification of prokaryotic viruses. Bioinformatics. 2017;33(21):3396-3404.
Crossref - Chang Y, Shin H, Lee J-H, Park CJ, Paik S-Y, Ryu S. Isolation and genome characterization of the virulent Staphylococcus aureus bacteriophage SA97. Viruses. 2015;7(10):5225-5242.
Crossref - Gomaa S, Serry F, Abdellatif H, Abbas H. Elimination of multidrug-resistant Proteus mirabilis biofilms using bacteriophages. Arch Virol. 2019;164(9):2265-2275.
Crossref - Phumkhachorn P, Rattanachaikunsopon P. Isolation and partial characterization of a bacteriophage infecting the shrimp pathogen Vibrio harveyi. African J Microbiol Res. 2010;4(16):1794-1800.
- Jurczak-Kurek A, Gasior T, Nejman-Falenczyk B, et al. Biodiversity of bacteriophages: morphological and biological properties of a large group of phages isolated from urban sewage. Sci Rep. 2016;6(1):34338.
Crossref - Ackermann H-W. Tailed Bacteriophages: The Order Caudovirales. Advances in Virus Research. Academic Press; 1998;51:135-201.
Crossref - Alonso B, Fernandez-Barat L, Di Domenico EG, et al. Characterization of the virulence of Pseudomonas aeruginosa strains causing ventilator-associated pneumonia. BMC Infect Dis. 2020;20(1):909.
Crossref - McVay CS, Velasquez M, Fralick JA. Phage therapy of Pseudomonas aeruginosa infection in a mouse burn wound model. Antimicrob Agents Chemother. 2007;51(6):1934-1938.
Crossref - Ceyssens P-J, Brabban A, Rogge L, et al. Molecular and physiological analysis of three Pseudomonas aeruginosa phages belonging to the “N4-like viruses”. Virology. 2010;405(1):26-30.
Crossref - Secor PR, Sweere JM, Michaels LA, et al. Filamentous bacteriophage promote biofilm assembly and function. Cell Host Microbe. 2015;18(5):549-559.
Crossref - Guo Y, Chen P, Lin Z, Wang T. Characterization of two Pseudomonas aeruginosa viruses vB_PaeM_SCUT-S1 and vB_PaeM_SCUT-S2. Viruses. 2019;11(4):318.
Crossref - Amgarten D, Martins LF, Lombardi KC, et al. Three novel Pseudomonas phages isolated from composting provide insights into the evolution and diversity of tailed phages. BMC genomics. 2017;18(1):346.
Crossref - Yang H, Liang L, Lin S, Jia S. Isolation and characterization of a virulent bacteriophage AB1 of Acinetobacter baumannii. BMC Microbiol. 2010;10:131.
Crossref - Pinheiro LAM, Pereira C, Frazao C, Balcao VM, Almeida A. Efficiency of phage φ6 for biocontrol of Pseudomonas syringae pv. syringae: An in vitro preliminary study. Microorganisms. 2019;7(9):286.
Crossref - Mulani MS, Kamble EE, Kumkar SN, Tawre MS, Pardesi KR. Emerging strategies to combat ESKAPE pathogens in the era of antimicrobial resistance: A review. Front Microbiol. 2019;10:539.
Crossref - Chan BK, Sistrom M, Wertz JE, Kortright KE, Narayan D, Turner PE. Phage selection restores antibiotic sensitivity in MDR Pseudomonas aeruginosa. Sci Rep. 2016;6:26717.
Crossref - Al-Wrafy F, Brzozowska E, Gorska S, Drab M, Strus M, Gamian A. Identification and characterization of phage protein and its activity against two strains of multidrug-resistant Pseudomonas aeruginosa. Sci Rep. 2019;9(1):13487.
Crossref - Cafora M, Deflorian G, Forti F, et al. Phage therapy against Pseudomonas aeruginosa infections in a cystic fibrosis zebrafish model. Sci Rep. 2019;9(1):1527.
Crossref - Raz A, Serrano A, Hernandez A, Euler CW, Fischetti VA. Isolation of phage lysins that effectively kill Pseudomonas aeruginosa in mouse models of lung and skin infection. Antimicrob Agents Chemother. 2019;63(7).
Crossref - Oechslin F, Piccardi P, Mancini S, et al. Synergistic interaction between phage therapy and antibiotics clears Pseudomonas aeruginosa infection in endocarditis and reduces virulence. J Infect Dis. 2017;215(5):703-712.
- Waters EM, Neill DR, Kaman B, et al. Phage therapy is highly effective against chronic lung infections with Pseudomonas aeruginosa. Thorax. 2017;72(7):666-667.
Crossref - Chan BK, Turner PE, Kim S, Mojibian HR, Elefteriades JA, Narayan D. Phage treatment of an aortic graft infected with Pseudomonas aeruginosa. Evol Med Public Health. 2018;2018(1):60-66.
Crossref - Reuter M, Kruger DH. Approaches to optimize therapeutic bacteriophage and bacteriophage-derived products to combat bacterial infections. Virus Genes. 2020;56(2):136-149.
Crossref
© The Author(s) 2021. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.