ISSN: 0973-7510
E-ISSN: 2581-690X
The increasing prevalence of multidrug-resistant (MDR) bacteria highlights the urgent need for new antimicrobial agents. In this study, Bacillus species were isolated from mangrove sediments along the Red Sea coast in Jeddah, Saudi Arabia. A total of 30 isolates were screened for antibacterial activity against four MDR pathogens: Staphylococcus aureus, Escherichia coli, Klebsiella pneumoniae, and Pseudomonas aeruginosa. Eleven isolates designated JRSM (Jeddah Red Sea Mangrove) 1 to 11, demonstrated inhibitory effects, and four strains (JRSM 4, 6, 7, and 9) were selected for further investigation. The Minimum Inhibitory Concentration (MIC) and Minimum Bactericidal Concentration (MBC) assays confirmed their antimicrobial potential, particularly against S. aureus and P. aeruginosa. Molecular identification using 16S rRNA sequencing revealed the strains such as Bacillus mesophilus, Bacillus xiamenensis, Bacillus halotolerans, and Bacillus subtilis. All strains were Gram-positive and catalase-positive. These findings highlight the Red Sea mangrove sediments as a promising source of Bacillus spp. with potential applications in combating MDR and biofilm-associated infections. Further research is needed to optimize metabolite production and characterize the active compounds responsible for the observed antimicrobial activity.
Red Sea, Jeddah, Mangrove Sediments, Bacillus spp., Antimicrobial Activity, Antibiofilm, Multidrug-resistant Bacteria
Multidrug-Resistance (MDR) is described as the resistance of bacteria to commercial antimicrobial medications.1 Over the past few decades, MDR has increased the prevalence of microbial infections over the world.2 In Saudi Arabia, MDR infections have been a significant concern, with studies reporting that over 18% of the total MDR infection outbreaks in the Arabian Peninsula originated from the country, highlighting its high prevalence compared to neighboring regions.3 The cases of MDR in Saudi Arabia is high due to several factors, including the overuse and misuse of antibiotics in healthcare and agriculture, insufficient infection control measures, limited public awareness, the widespread use of broad-spectrum antimicrobials, and the influx of international travelers, which facilitates the spread of resistant pathogens. Additionally, environmental factors such as extreme climate conditions and socio-economic aspects, like reliance on expatriate labor in healthcare, contribute to the challenge by amplifying the spread and persistence of resistant strains.4 The rising prevalence of multidrug-resistant (MDR) microorganisms, driven by genetic adaptations such as efflux pumps, enzymatic deactivation, and altered target sites, create a serious worldly health issue by making the treatment of infections pathogens very complicated like Staphylococcus aureus, Escherichia coli, Klebsiella pneumoniae, and underscoring the urgent need for novel antimicrobial strategies and surveillance efforts.5
Escherichia coli is a common waterborne pathogen that can cause debilitating diarrhea due to the toxins it secretes.6 Skin infections caused by Staphylococcus aureus include various sores, such as impetigo, cellulitis, and staphylococcal scalded skin syndrome.7 Diseases of the respiratory tract, eyes, urinary tract, and skin are all caused by Pseudomonas aeruginosa.8 Extensive consolidation with hemorrhagic necrosis of the lungs is caused by Klebsiella pneumoniae.9 The development of antibiotic resistance in these microorganisms demonstrates the lack of effective long-term treatments, driven by the limited availability of antibacterial drugs and the frequent occurrence of cross-resistance between drug classes.10 Therefore, identifying potential resources for antimicrobials that are both more effective and sustainable is crucial.
Oceans account for 70 percent of the earth’s surface and contain 80 percent of all animal species on the planet. Due to the vast biological and chemical variety seen in marine habitats, these settings were thought to be significant producers of new bioactive chemicals.11 Maritime microorganisms are one of the most abundant natural product sources.12 The ecological condition of the Red Sea near Jeddah is marked by extreme environmental pressures, including high salinity levels, nutrient overloading from untreated wastewater, and significant pollutant accumulation such as heavy metals and organic contaminants, driven by rapid urbanization and industrialization; these factors not only alter water quality and sediment composition but also create harsh and fluctuating habitats that hypothetically could drive microbial diversity through adaptive evolution or, conversely, lead to the loss of sensitive microbial taxa due to the extreme and toxic conditions.13,14 Mangrove ecosystems are unique coastal environments that harbor a diverse range of microorganisms, including bacteria with potential biotechnological applications.15 These bacteria are frequently acclimated to extreme environmental circumstances, including elevated salinity, temperature variations, and low oxygen levels, rendering them a viable source of novel bioactive chemicals.16 These bacteria are recognized for their ability to synthesize a diverse range of secondary metabolites that include antibacterial and antibiofilm properties, which may be crucial in addressing multidrug-resistant diseases.17 The unique characteristics and capabilities of mangrove sediment bacteria make them focus on growing interest in microbial research and biotechnological innovations. The Red Sea, particularly along the coast of South Jeddah, is home to extensive mangrove forests that have not been extensively studied for their microbial diversity.
Marine bacteria are selected in biotechnology for antibiotic synthesis because of their distinctive metabolic pathways and capacity to generate novel bioactive chemicals absent in terrestrial microorganisms. The extreme and diverse environmental conditions of marine habitats, such as high pressure, salinity, and limited nutrients, drive marine bacteria to evolve distinctive secondary metabolites with potent antimicrobial properties.18 These metabolites include polyketides, lipopeptides, and alkaloids, which exhibit activity against multidrug-resistant pathogens, making marine bacteria a promising source for addressing the global antimicrobial resistance crisis.19 Additionally, their biosynthetic versatility and capacity for scalable production through fermentation processes enhance their applicability in sustainable drug development.
Marine bacteria, particularly species of the Bacillus genus, are well-known producers of antimicrobial compounds, making them promising sources for novel antibiotics. Bacillus species that are marine-derived produce a range of bioactive metabolites, such as polyketides, lipopeptides, and peptides that have antiviral, antifungal, and antibacterial qualities.20 For instance, Bacillus subtilis produces surfactant, a potent cyclic lipopeptide known for its biofilm-disrupting activity and effectiveness against Gram-positive bacteria.21 Commercially, antibiotics like bacitracin, produced by Bacillus licheniformis, are widely used to treat Gram-positive bacterial infections, while fengycin has gained attention for its antifungal properties.22
The robustness and distinctive metabolism of marine Bacillus, adapted to severe settings, augment their biosynthetic capability, making them crucial in the fight against diseases that are resistant to drugs.
Current findings indicate that bacteria isolated from mangrove sediments possess the ability to produce antibacterial compounds.23,24 Therefore, discovering new sources of antibacterial agents is crucial for developing alternative therapeutic strategies. In this study, we aim to isolate and characterize bacteria from mangrove sediments along the southern Red Sea coast of Jeddah, Saudi Arabia, an environment with extreme salinity and pollutant stress that has been largely unexplored for microbial diversity. These isolates were screened for their ability to produce antibacterial compounds against clinically relevant MDR pathogens. Unlike previous studies that focused on terrestrial or open marine Bacillus species, this work highlights the potential of Bacillus strains from a unique mangrove sediment niche, offering new insights into their antimicrobial capabilities and taxonomic diversity. This study contributes to the growing interest in harnessing extremophilic microbes from underexplored environments as sources of novel bioactive metabolites.
Chemicals and Materials
Marine sediment samples were collected from Red Sea mangroves in southern Jeddah, and their environmental parameters, like total dissolved solids, dissolved oxygen, salinity, temperature, and pH, were carefully measured using a pH meter and turbidimeter. Bacteria were isolated using two types of media: Glucose Yeast-Malt Extract Medium (GYM/GYM) and Starch Casein (SC) Medium purchased from HI-Media. Salts such as NaCl, KCl, CaCl2, MgCl2·6H2O, and CaCO3 were purchased from Merck as well as chemical, NaOH. Nystatin and Nalidixic Acid, agents were purchased from HI-Media. To identify bacterial isolates, Gram staining was performed using crystal violet, iodine, and safranin were bought from Merck. Additionally, the isolates were tested for catalase activity using hydrogen peroxide. Pathogenic bacteria, including Staphylococcus aureus, Escherichia coli, Klebsiella pneumoniae, and Pseudomonas aeruginosa were used for screening, along with Muller Hinton Broth and Agar (MHB and MHA) purchased from HI-Media.
Sediment collection
Sediment samples were collected from five distinct locations, identified through a randomized selection process utilizing Google Maps on June 5, 2023, from 2:37 to 3:17 pm Saudi Arabia Time. The procedure outlined by El-Kahawy et al.25 for sediment sampling involved scraping 15 cm from the area’s surface to gather sediment. The samples were then stored in a sterile, air-tight container within a chiller box. The evaluation of various environmental parameters, including total dissolved solids, temperature, dissolved oxygen, salinity, and pH, was conducted on the day of collection using specialized instruments: a pH Meter for acidity or alkalinity measurements, a Thermometer for temperature readings, a Salinity Meter for salt concentration, a Dissolved Oxygen Meter for oxygen levels, and a Total Dissolved Solids (TDS) Meter for the quantification of dissolved substances, respectively.
Media preparation
Glucose Yeast-Malt Extract Medium (GYM) and Starch Casein (SC) Medium. Both media types were prepared in a solution mimicking seawater composition, with concentrations of NaCl (23.5 g/L), KCl (0.66 g/L), CaCl2 (1.5 g/L), MgCl2·6H2O (10.6 g/L), Na2SO4 (3.9 g/L), NaHCO3 (0.19 g/L), Na2HPO4 (0.014 g/L), KBr (0.1 g/L), SrCl2 (0.04 g/L), and H3BO3 (0.03 g/L), with the addition of CaCO3 (0.2 g/L) for enhancement. The pH was adjusted to 8 with 1 M NaOH, and the media were sterilized by autoclaving at 121 °C for 15 minutes. Antifungal and antibacterial agents, Nystatin (25 mg/L) and Nalidixic Acid (20 mg/L), were added to the media, which were subsequently diluted to 70% with artificial seawater.26
Bacterial isolation
Following the procedure of Abdelmohsen et al.27 with minor adjustments, bacteria were isolated from mangrove sediments in the Red Sea. To ensure that no vegetative cellular forms were present, a one-gram mass from each sediment sample was heated to 60 °C for 30 minutes. Subsequently, ten milliliters of seawater were added to the treated samples, followed by their serial dilution in ratios of 1:10, 1:100, and 1:1000. This resultant mixture was then aseptically transferred onto agar-based culture media (either SCA or GYM medium) and underwent an incubation period ranging from 7 to 14 days at ambient temperature. Following incubation, individual colonies were isolated through streak plating onto identical media to ensure the procurement of pure cultures. After that, each separate colony was kept at a -80 °C temperature in a 30% glycerol reservoir until needed.
Preliminary test
The initial screening to assess the antimicrobial capabilities of cultures utilized the perpendicular streak technique as outlined by Singh et al.,28 targeting bacterial strains for example, Staphylococcus aureus, Klebsiella pneumoniae, Escherichia coli, and Pseudomonas aeruginosa. For this purpose, a solitary streak of each pure culture was applied centrally on the assay media plate. Following a four-day incubation period at 28 °C, the test organisms were streaked at a right angle to the strain streak. A subsequent incubation period of one to two days at 28 °C allowed for the evaluation of antagonistic activity between the cultures.
Gram staining
Gram staining was performed using the standard crystal violet-iodine method as described by Abdulrahman et al.29 followed by microscopic examination at 1000׳ magnification to determine whether isolates were Gram-positive or Gram-negative.
Catalase test
To find out whether the bacterial isolates produced the enzyme catalase,29 the catalase test was carried out. A sterile loop was used to transfer a small amount of bacterial colony from agar to a clean glass slide. A few drops of 3% hydrogen peroxide (H2O2) were added to the bacterial sample. The production of oxygen bubbles was observed within a few seconds. Quick bubbling was an indicator of a positive catalase response, which meant that the bacteria had created the enzyme necessary to decompose hydrogen peroxide into oxygen and water. The absence of bubbles indicated a negative catalase result.
Fermentation and extraction
Bacterial fermentation and metabolite extraction were performed according to Abdelmohsen et al.27 The preserved stock cultures were transferred onto various agar media and incubated at an ambient temperature of 28 °C. Fermentation processes were initiated by introducing a colony from each solid media into the corresponding broth media and maintaining the culture at 28 °C. Mycelial cakes were removed from broth by centrifugation at 3000 rpm for 10 minutes after fermentation. After that, the pellet was dissolved in methanol and the liquid on top was extracted using ethyl acetate. Thereafter, a rotary evaporator was used to concentrate the crude extracts. Next, to get the crude extracts ready for bioassay testing, they were reconstructed in DMSO.
Minimum Inhibition Concentration (MIC)
Assessment of MIC using Priyanto et al.30 method was modified in this research. Pathogenic bacteria were grown overnight in Muller Hinton Agar (MHA) medium at 37 degrees Celsius. A bacterial culture was introduced to a 0.85% physiological saline solution and its density was compared to the 0.50 McFarland Standard. About 200 µL of the culture was pipetted from the physiological solution and diluted in 20 mL of 0.85% physiological saline. 96-well plates were filled with approximately 100 µL of Muller Hinton Broth medium, and crude extracts with concentrations of 2500, 1250, 625, 312.5, 156.25, 78.13, 39.06, and 19.53 µg/mL were combined. About 100 L of microbial culture in a physiological solution was added. Plates containing cultures and test compounds were incubated overnight at 37 degrees Celsius and 200 rpm with shaking. Clear well was compared to Cefalexin and Ampicillin antibiotics from the regular supply. The experiment was conducted duplicate.
Minimum Bactericidal Concentration (MBC)
Assessment of MIC using Priyanto et al.30 method, around 20 µL of MIC-concentration clear well solution was pipetted onto the MHA plate. Before observation, the agar plate was incubated overnight at 37 degrees Celsius. The concentration of the well where no bacterial growth was seen on the plate served as the MBC value. The experiment was done in duplicate.
Microbial identification
The bacterial isolates were cultivated in the proper growth media and their genomic DNA was extracted following the manufacturer’s protocol. Microgen, located in Seoul, South Korea, was consulted to analyse the concentration, quality and sequencing of 16S rRNA gene for bacterial identification.
The 16S rRNA gene was amplified utilizing the conventional primers 785F 5′ (GGA TTA GAT ACC CTG GTA) 3′, 27F 5′ (AGA GTT TGA TCM TGG CTC AG) 3′, 907R 5′ (CCG TCA ATT CMT TTR AGT TT) 3′, and 1492R 5′ (TAC GGY TAC CTT GTT ACG ACT T) 3′. The PCR cycling protocol comprised an initial denaturation at 94 °C for 5 minutes, succeeded by 35 cycles of 94 °C for 30 seconds, 55 °C for 30 seconds, and 72 °C for 1 minute, concluding with a final extension at 72 °C for 10 minutes. Gel electrophoresis was used to confirm the amplified PCR products, and a PCR purification kit was used to purify them. Purified products were sequenced using Sanger sequencing in both forward and reverse directions. Following a quality control analysis in Chromas software that removed low-quality areas, the resultant sequences were matched to the NCBI database using BLAST to identify which bacteria were present.
Phylogenetic analysis was conducted using ClustalW for sequence alignment and MEGA software (version X) to construct phylogenetic trees with the neighbor-joining method. The results were summarized in a report detailing the identified bacterial species, percentage similarity, and phylogenetic relationships.
Statistical analysis
Averages with standard deviations from three independent samples were used to compile all the data. Further analysis was established by use of a one-way Analysis of Variance (ANOVA). For statistical purposes, a p-value below 0.05 was considered significant.
Bacteria Isolation
Table 1 provides a detailed account of the environmental parameters observed in mangrove sediments from five distinct sampling sites, labelled JSM 1 to JSM 5, along the southern coastline of the Red Sea near Jeddah. This study seeks to explore the complex environmental interactions within this critical mangrove ecosystem, offering valuable insights essential for its conservation and comprehension.
Table (1):
Chemical indicators assessed during the collection of sediment samples reflect the environmental state
| Region | Location | Chemical Condition | ||||
|---|---|---|---|---|---|---|
| pH | Temp. (Celsius) | Salinity | Dissolved Oxygen mg/L) | Total Dissolved Solids (g/L) | ||
| JSM 1 | 21°15’57.7″N 39°07’41.5″E | 8.1 | 37.64 | 39.88 | 42.56 | 39.14 |
| JSM 2 | 21°15’56.8″N 39°07’42.7″E | 8.1 | 36.53 | 39.99 | 7.63 | 39.19 |
| JSM 3 | 21°15’55.2″N 39°07’43.6″E | 8.6 | 36.53 | 39.99 | 7.61 | 39.19 |
| JSM 4 | 21°15’49.0″N 39°07’44.0″E | 8.5 | 37.30 | 39.60 | 39.38 | 38.88 |
| JSM 5 | 21°15’44.4″N 39°07’45.0″E | 8.6 | 37.23 | 39.69 | 8.39 | 38.97 |
Key environmental parameters, including pH, temperature, salinity, dissolved oxygen, and total dissolved solids, were meticulously recorded. The pH ranged from 8.1 to 8.6, indicating a slightly alkaline environment conducive to mangrove ecosystems, which often require specific pH conditions for optimal growth. Temperature readings averaged around 30 °C, reflecting the warm coastal waters typical of the region and aligning with the thermal adaptations of mangroves. Salinity levels, just under 40, highlighted the Red Sea’s naturally high salinity, a defining factor in shaping its unique biodiversity. Dissolved oxygen levels varied significantly across sites, ranging from a robust 42.56 mg/L to a lower 7.62 mg/L, suggesting differences in biological activity or water flow at each sampling point. The total dissolved solids, approximately 39 g/L, further illustrated the mineral-rich composition of the water, a critical factor in determining the habitat’s suitability for mangrove ecosystems.
This study analysed chemical parameters, including pH, temperature, salinity, dissolved oxygen, and total dissolved solids, to establish their relationship with the diversity and abundance of bacteria in different mangrove sediments. This approach provides important insights into how distinct environmental factors influence microbial communities within this specialized ecosystem.31,32 The differences in pH and temperature across the sampling locations could account for the variations in bacterial populations, as bacteria tend to thrive within specific pH and temperature ranges.33 Likewise, the distinct salinity levels of the Red Sea likely favor salt-tolerant bacterial species, influencing the structure and composition of the microbial community.34 Additionally, oxygen availability is a key factor in shaping the dominance of aerobic or anaerobic bacteria, highlighting the dynamic and adaptive characteristics of these ecosystems.35 The measurement of total dissolved solids, reflecting nutrient availability, further aligns with the potential for greater bacterial diversity and abundance in nutrient-rich locations.36
By incorporating data from these five sampling locations, the study offers a detailed overview of the environmental conditions in the mangrove ecosystem along South Jeddah’s Red Sea coast. This baseline information was vital for preparing media that closely mimicked the natural conditions of the mangrove sediments in laboratory settings for bacterial isolation.
In the research conducted within the Jeddah mangrove area, sediment samples were collected from five distinct sites, labeled Jeddah Sea Mangrove 1 to 5 (JSM 1 to JSM 5), to isolate the total bacterial community.
Bacterial isolation utilized two media types, GYM and SCA, selected for their effectiveness in supporting a diverse range of microbial growth. The use of GYM and SCA media for isolating marine bacteria from sediment samples highlights the adaptability and nutritional preferences of these microorganisms. These media were selected for their ability to support a wide range of microbial growth, catering to the diverse metabolic needs of marine bacteria. The incorporation of artificial seawater, simulating the natural salinity of the Red Sea, ensured that the isolates were cultured under conditions resembling their native environment, preserving their ecological characteristics. This methodology revealed variations in microbial diversity across the mangrove ecosystem, emphasizing the critical ecological roles of these bacteria in nutrient cycling and ecosystem stability.37-39
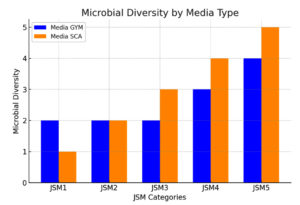
Initial results from the five locations showed varying bacterial counts. At JSM 1, three colonies were observed on GYM media compared to two on SCA, suggesting slightly higher bacterial diversity in GYM for this site. In contrast, JSM 2 and JSM 3 exhibited an even distribution, with two colonies on GYM and three on SCA, indicating a possible preference for SCA at these sites. Sampling points JSM 4 and JSM 5 demonstrated greater bacterial diversity overall, with most colonies flourishing on SCA, highlighting the media’s suitability for cultivating bacteria from these locations. Figure 1 illustrates the total colony counts for each sampling point, grown on specific media.
Figure 1. Microbial diversity of each sampling point. All counts were based on consistent replicate values, resulting in zero error bars
This methodical approach to bacterial isolation underscores the rich microbial diversity present in mangrove sediments, emphasizing the critical ecological roles these microorganisms play in the coastal environment. Variations in bacterial colony counts across different sampling locations and media provide important insights into the intricate interactions and environmental influences shaping microbial communities in the Jeddah Red Sea mangroves. Consequently, 30 colonies were chosen for antibacterial screening.
Antibacterial activity
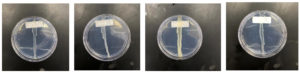
In this study, we looked at individual bacterial colonies to see if they could produce substances that stop drug-resistant bacteria from growing. We used a specific method to test the bacteria’s ability to fight against bacteria resistant to multiple drugs. Our initial tests found that 11 out of 30 (Labeled as Jeddah Red Sea Mangrove 1 to 11, JRSM 1 to 11) bacterial colonies showed promising results, meaning they might have compounds that could act against these pathogenic bacteria. Figure 2 shows the results of the Perpendicular Test for Chosen Active Bacteria.
MIC and MBC results
To dig deeper, we conducted further tests, known as Minimum Inhibitory Concentration (MIC) and Minimum Bactericidal Concentration (MBC) tests, on these 11 colonies. These tests helped us figure out the smallest amount of the substance needed to stop the MDR bacteria from growing and against them. This step is crucial because it gives us a result of how effective these bacterial compounds could be in treating infections caused by drug-resistant bacteria. Table 2 shows MIC and MBC results of 11 bacteria extracts against chosen pathogenic bacteria.
Table (2):
MIC and MBC results of bacteria extracts against four pathogenic bacteria
| Isolate Codes | Media | MIC (μg/mL) | MBC (μg/mL) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| E.coli | S. aureus | P. aeruginosa | K. pneumoniae | E.coli | S. aureus | P. aeruginosa | K. pneumoniae | ||
| JRSM 1 | GYM | >5000 | 625 | 625 | >5000 | >5000 | >1250 | 625 | >5000 |
| JRSM 2 | GYM | >5000 | >5000 | >5000 | >5000 | >5000 | >5000 | >5000 | >5000 |
| JRSM 3 | GYM | 2500 | 5000 | 2500 | – | 5000 | >5000 | 5000 | >5000 |
| JRSM 4 | GYM | 5000 | 156,25 | 312,5 | 2500 | >5000 | 312,5 | 625 | >5000 |
| JRSM 5 | GYM | 2500 | 2500 | 2500 | 2500 | >5000 | >5000 | 2500 | >5000 |
| JRSM 6 | GYM | 2500 | 312,5 | 312,5 | 2500 | >5000 | 625 | 312,5 | >5000 |
| JRSM 7 | SC | >5000 | 78,125 | 156,25 | >5000 | >5000 | 312,5 | 312,5 | >5000 |
| JRSM 8 | SC | >5000 | >5000 | 2500 | >5000 | >5000 | >5000 | >2500 | >5000 |
| JRSM 9 | SC | >5000 | 156,25 | 5000 | >5000 | >5000 | 625 | 5000 | >5000 |
| JRSM 10 | SC | >5000 | >5000 | >5000 | >5000 | >5000 | >5000 | >5000 | >5000 |
| JRSM 11 | SC | >5000 | >5000 | >5000 | >5000 | >5000 | >5000 | >5000 | >5000 |
| DMSO | – | >5000 | >5000 | >5000 | >5000 | >5000 | >5000 | >5000 | >5000 |
| Cefalexin | – | >5000 | 2,44 | 4,88 | >5000 | >5000 | 9,70 | 4,88 | >5000 |
| Ampicillin | – | >5000 | 2,44 | 2,44 | >5000 | >5000 | 2,44 | 2,44 | >5000 |
In this study, four bacterial strains were selected for further analysis: two cultured on GYM media and two on SCA-based on their superior performance in MIC and MBC tests. The antimicrobial activity of extracts from JRSM 4, 6, 7, and 9 was evaluated against a panel of multidrug-resistant bacteria, including Escherichia coli, Staphylococcus aureus, Pseudomonas aeruginosa, and Klebsiella pneumoniae. The results revealed varying degrees of efficacy among the extracts.
JRSM 4 demonstrated MIC values of 5000 µg/mL and 2500 µg/mL against E. coli and K. pneumoniae, respectively, with lower MICs of 156.25 µg/mL for S. aureus and 312.5 µg/mL for P. aeruginosa. Its MBC values followed a similar trend, indicating a moderate level of antimicrobial activity. JRSM 6 showed MIC values of 2500 µg/mL for E. coli and K. pneumoniae, while S. aureus and P. aeruginosa exhibited MICs of 312.5 µg/mL, with MBC values reflecting similar or slightly enhanced bactericidal effects.
Interestingly, JRSM 7 exhibited strong activity with lower MIC values of 78.125 µg/mL for S. aureus and 156.25 µg/mL for P. aeruginosa, and consistent MBC values of 312.5 µg/mL for both, highlighting its significant antimicrobial potential against these strains. However, it was less effective against E. coli and K. pneumoniae. JRSM 9 showed an MIC of 156.25 µg/mL against S. aureus but a much higher MIC of 5000 µg/mL for P. aeruginosa, with corresponding MBC values indicating variable effectiveness. The lack of MIC and MBC data for E. coli and K. pneumoniae for JRSM 9 limited a more comprehensive assessment of its antimicrobial properties.
Bacteria identification 16S rRNA sequencing and phylogenetic tree
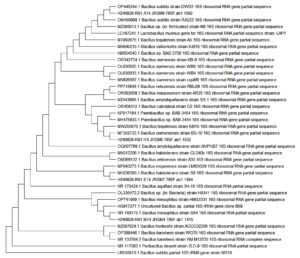
The bacterial identification and sequencing results from Figure 3, Table 3 provide critical insights into the identification of the bacterial isolates JRSM 4, JRSM 6, JRSM 7, and JRSM 9, each corresponding to specific Bacillus species, and their potential role in antibacterial and antibiofilm activities. The sequencing was performed using primers 785F 5′ (GGA TTA GAT ACC CTG GTA) 3′, 27F 5′ (AGA GTT TGA TCM TGG CTC AG) 3′, 907R 5′ (CCG TCA ATT CMT TTR AGT TT) 3′, and 1492R 5′ (TAC GGY TAC CTT GTT ACG ACT T) 3′, which are standard primers for bacterial 16S rRNA gene amplification.
Figure 3. Phylogenetic Trees of Bacillus Species Identified from Red Sea Sediment Mangroves in South Jeddah
Table (3):
Identification and Characteristics of Bacillus Species Isolated from Red Sea Sediment Mangroves in South Jedda
Sample Code |
Samples Name |
Media |
Morphology |
Gram Staining |
Catalase |
Identified Bacteria |
Accession Number |
Percentage (%) |
|---|---|---|---|---|---|---|---|---|
ISP2A12 |
JRSM 4 |
GYM |
Orange color, rod shape |
Positive |
Positive |
Bacillus mesophilus |
NR_149175.1 |
99.42 |
ISP2A10 |
JRSM 6 |
GYM |
White color, rod shape |
Positive |
Positive |
Bacillus xiamenensis |
MF359733.1 |
99.57 |
B5SCA |
JRSM 7 |
SC |
White color, rod shape |
Positive |
Positive |
Bacillus halotoleran |
MN512206.1 |
99.86 |
B1SCA |
JRSM 9 |
SC |
Yellow color, rod shape |
Positive |
Positive |
Bacillus subtilis |
OP445244.1 |
99.57 |
JRSM 4 was identified as Bacillus mesophilus with a 99.42% similarity and showed as Gram-positvie bacteria with catalase-positive. The identification of Bacillus mesophilus suggests its involvement in the observed antibacterial and antibiofilm properties, contributing to the diversity of bioactive compounds found in the mangrove sediment isolates. JRSM 6, identified as Bacillus xiamenensis, Gram-positive bacteria and catalase-positive with a 99.57% match, is known for its bioactive compound production, highlighting its likely role in the antibiofilm activities observed against pathogenic bacteria. Colony JRSM 7 was identified as Bacillus halotolerant with catalase and Gram-positive, with a 99.86% similarity, a species recognized for its antifungal properties and non-pathogenic nature. This identification supports the potential of JRSM 7 in developing treatments targeting fungal and bacterial pathogens, aligning with the study’s objectives. The fourth colony, JRSM 9 was identified as Bacillus substilis with a 99.57% match to known sequences, indicating its capacity to produce bioactive compounds that could exacerbate infections, based on Gram staining it was Gram-positive and catalase-positive.
Sequencing data revealed that the colony labeled JRSM 4 was identified as Bacillus mesophilus. This Gram-positive, rod-shaped bacterium is mesophilic, thriving in moderate temperature ranges of approximately 20 °C to 45 °C.40 It plays a crucial role in ecosystems by decomposing organic matter in water and soil environments.41 A distinguishing characteristic of Bacillus mesophilus is its ability to form endospores, allowing it to withstand harsh environmental conditions, including extreme temperatures, desiccation, and exposure to UV radiation.42 These spores remain dormant until favorable conditions allow them to germinate.41 B. mesophilus is metabolically versatile, capable of utilizing a wide range of organic compounds for growth. It also produces extracellular enzymes, such as amylases and proteases, which facilitate the breakdown of complex carbohydrates and proteins.43 This capability has significant industrial applications, particularly in bioremediation and the synthesis of bioactive compounds. Phylogenetically, Bacillus mesophilus shares similarities with other Bacillus species but is distinguishable based on genetic markers, as verified through 16S rRNA gene analysis.
JRSM 6 was identified as Bacillus xiamenensis, a Gram-positive, aerobic, rod-shaped bacterium initially isolated from the intestinal tract of a flathead mullet in Xiamen, China. It is part of the Bacillus genus, renowned for its ecological diversity and adaptability to a wide range of environments.44 Similar to other species within the Bacillus genus, Bacillus xiamenensis has the ability to form endospores, enabling it to endure extreme environmental conditions such as high temperatures, desiccation, and chemical exposure.41
Bacillus xiamenensis is distinguished by its production of various extracellular enzymes, such as amylases and proteases, which are industrially significant for their ability to degrade complex organic molecules. These enzymes position B. xiamenensis as a valuable candidate for biotechnological applications, particularly in bioremediation, food processing, and waste management industries.42 Additionally, Bacillus xiamenensis has exhibited antifungal activity against Fusarium oxysporum, a capability that provides a competitive edge in natural environments and highlights its potential for biocontrol applications.45
Phylogenetically, Bacillus xiamenensis is closely related to other species in the Bacillus genus, including Bacillus subtilis.46 Additionally, studies have demonstrated its plant growth-promoting abilities, making it a promising biofertilizer candidate for agricultural use. This is attributed to its capacity to solubilize phosphate and produce substances that enhance plant growth.47
JRSM 7 was identified as Bacillus halotolerans, a Gram-positive, rod-shaped bacterium recognized for its ability to thrive in high-salinity environments, classifying it as a halotolerant species.48 The buildup of osmotic balance-supporting solutes-including proline, glycine betaine, and trehalose are responsible for its salt tolerance.49 The buildup of osmotic balance-supporting solutes-including proline, glycine betaine, and trehalose-are responsible for its salt tolerance.41 This ability to form endospores is a common trait within the Bacillus genus, allowing the organism to survive in harsh and unfavourable environmental conditions.
In terms of metabolic capabilities, Bacillus halotolerans is highly versatile. Its ability to use a broad variety of organic substances as carbon and energy sources makes it able to thrive in environments with minimal nutrients.42 This bacterium also produces various extracellular enzymes, including proteases, amylases, and lipases, which are of biotechnological interest due to their potential applications in industries such as food processing, detergent formulation, and biofuel production.43 The high salt tolerance of these enzymes further enhances their utility in industrial processes that operate under saline conditions, where traditional enzymes may not function efficiently.48
In biotechnology, Bacillus halotolerans has shown promise in several applications, including bioremediation, where it is used to degrade organic pollutants in saline environments.50 Moreover, this bacterium has potential use in agriculture as a biofertilizer, particularly in saline soils, where it promotes plant growth by solubilizing phosphates and producing plant growth-promoting hormones.51
Bacillus halotolerans also exhibits notable antimicrobial activity, making it a potential candidate for biocontrol applications in agriculture and medicine. Studies have shown that this species produces antimicrobial peptides (AMPs) that inhibit the growth of various pathogenic bacteria, including Staphylococcus aureus, Escherichia coli, and Pseudomonas aeruginosa.52 These AMPs, such as bacillomycin and fengycin, are part of a larger class of lipopeptides known for their ability to disrupt bacterial cell membranes.53 Additionally, B. halotolerans has been shown to produce antimicrobial compounds that are effective against fungal pathogens, such as Fusarium oxysporum, making it a potential biocontrol agent in plant protection.54
JRSM 9 was identified as Bacillus subtilis, a Gram-positive, aerobic, rod-shaped bacterium that is widely studied due to its robust physiology.55 It exhibits remarkable environmental resilience, capable of surviving in extreme conditions by forming endospores.56 This bacterium is non-pathogenic and is found naturally in soil and plant environments.57 One of the key characteristics of B. subtilis is its ability to produce a wide range of secondary metabolites, including enzymes and antimicrobial compounds.58
B. substilis is employed in various industrial applications, including the production of enzymes like amylases and proteases, which are used in food, pharmaceutical, and agricultural industries.22 Additionally, B. subtilis is known for its ability to produce antimicrobial peptides such as subtilin and bacitracin, which have broad-spectrum activity against Gram-positive bacteria.56 These antimicrobial properties make B. subtilis a significant producer in biocontrol, where it is used to suppress plant pathogens and reduce the need for chemical pesticides in agriculture.59 Varius such useful microbes may also be isolated for petroleum bioremediations from red sea and Saudi soils.60
This research needs more exploration for optimization of fermentation conditions to get optimum results in metabolic production. Altering parameters such as pH, temperature, nutrient composition, and aeration during fermentation may increase the yield of active metabolites, potentially enhancing all extract’s antibiofilm activity.
This study successfully identified and characterized four Bacillus strains from mangrove sediments along the Red Sea coast of Jeddah, Saudi Arabia. These isolates exhibited antimicrobial activity against multiple clinically relevant multidrug-resistant (MDR) pathogens. Molecular analysis confirmed their identity as Bacillus mesophilus, Bacillus xiamenensis, Bacillus halotolerans, and Bacillus subtilis, all of which are known producers of bioactive compounds. Bacillus mesophilus (JRSM 4) demonstrated MIC values of 156.25 µg/mL against Staphylococcus aureus, 312.5 µg/mL against Pseudomonas aeruginosa, and 5000 µg/mL against both Escherichia coli and Klebsiella pneumoniae. Bacillus xiamenensis (JRSM 6) showed MIC values of 312.5 µg/mL against S. aureus and P. aeruginosa and 2500 µg/mL against E. coli and K. pneumoniae. Bacillus halotolerans (JRSM 7) displayed MIC values of 78.125 µg/mL against S. aureus and 156.25 µg/mL against P. aeruginosa, with MBC values of 312.5 µg/mL for both. Bacillus subtilis (JRSM 9) had MIC values of 156.25 µg/mL against S. aureus and 5000 µg/mL against P. aeruginosa. The findings highlight the potential of Red Sea mangrove ecosystems as a valuable reservoir of antimicrobial-producing bacteria. Future work should focus on optimizing fermentation conditions, isolating specific bioactive compounds, and evaluating their efficacy through in vivo and biofilm-based studies. Further genomic analysis may also reveal biosynthetic gene clusters responsible for antimicrobial activity, opening new avenues for drug discovery and environmental biotechnology.
ACKNOWLEDGMENTS
The authors acknowledge the Deanship of Scientific Research (DSR), King Abdulaziz University, Jeddah, Saudi Arabia, for the financial and technical support.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
AA conceptualized the project. FNH performed the experiments. MAZ, AT, AAA, RHA, HNA, MAZ, AAA, NAA and RMA assisted in writing, reviewing and editing the manuscript. All authors read and approved the final manuscript for publication.
FUNDING
This work was supported by the Deanship of Scientific Research (DSR), King Abdulaziz University, Jeddah, Saudi Arabia, under grant number KEP-34-130-41.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
Not applicable.
- Tanwar J, Das S, Fatima Z, Hameed S. Multidrug resistance: An emerging crisis. Interdiscip Perspect Infect Dis. 2014;2014(1):541340.
Crossref - Wartu JR, Butt AQ, Suleiman U, et al. Multidrug resistance by microorganisms: a review. Sci World J. 2019;14(4):49-56.
- Aly MM, Abu Alsoud NM, Elrobh MS, Al Johani SM, Balkhy HH. High prevalence of the PER-1 gene among carbapenem-resistant Acinetobacter baumannii in Riyadh, Saudi Arabia. Eur J Clin Microbiol Infect Dis. 2016;35(11):1759-1766.
Crossref - Borgio JF, Rasdan AS, Sonbol B, Alhamid G, Almandil NB, Azeez SA. Emerging Status of Multidrug-Resistant Bacteria and Fungi in the Arabian Peninsula. 2021;10(11):1144.
Crossref - Kashif M, Ahmad A, Khan MJ, et al. Current Research in Microbiology The Emerging Prospects of Global Anti Microbial Resistance/: Pros and Cons; Chapter 6. https://openaccessebooks.com/current-research-in-microbiology/the-emerging-prospects-of-global-anti-microbial-resistance-pros-and-cons.pdf
- Odonkor ST, Addo KK. Prevalence of Multidrug-Resistant Escherichia coli Isolated from Drinking Water Sources. Int J Microbiol. 2018;2018(1):7204013.
Crossref - Panda SK, Das R, Lavigne R, Luyten W. Indian medicinal plant extracts to control multidrug-resistant S. aureus, including in biofilms. South Afr J Bot. 2020;128:283-291.
Crossref - Aloush V, Navon-Venezia S, Seigman-Igra Y, Cabili S, Carmeli Y. Multidrug-resistant Pseudomonas aeruginosa: Risk factors and clinical impact. Antimicrob Agents Chemother. 2006;50(1):43-48.
Crossref - Farhadi M, Ahanjan M, Goli HR, Haghshenas MR, Gholami M. High frequency of multidrug-resistant (MDR) Klebsiella pneumoniae harboring several β-lactamase and integron genes collected from several hospitals in the north of Iran. Ann Clin Microbiol Antimicrob. 2021;20(1):70.
Crossref - Catalano A, Iacopetta D, Ceramella J, et al. Multidrug Resistance (MDR): A Widespread Phenomenon in Pharmacological Therapies. Molecules. 2022;27(3):616.
Crossref - Blunt JW, Carroll AR, Copp BR, Davis RA, Keyzers RA, Prinsep MR. Natural Product Reports. 2018;35(1):8-53.
Crossref - Genilloud O. Actinomycetes: still a source of novel antibiotics. Nat Prod Rep. 2017;34(10):1203-1232.
Crossref - Al-Farawati R. Environmental conditions of the coastal waters of Southern corinche, Jeddah, eastern Red Sea: Physico-chemical approach. Aust J Basic Appl Sci. 2010;4(8):3324-3337.
- Bantan RA, Ghandour IM, El-Kahawy RM, et al. Environmental assessment of toxic heavy metals in bottom sediments of the Sharm Obhur, Jeddah, Saudi Arabia. Mar Pollut Bull. 2024;205:116675.
Crossref - Kathiresan K, Bingham BL. Biology of mangroves and mangrove ecosystems. Adv Mar Biol. 2001;40:81-251.
Crossref - Basak P, Pramanik A, Sengupta S, et al. Bacterial diversity assessment of pristine mangrove microbial community from Dhulibhashani, Sundarbans using 16S rRNA gene tag sequencing. Genomics Data. 2016;7:76-78.
Crossref - Ye JJ, Zou RJ, Zhou DD, et al. Insights into the phylogenetic diversity, biological activities, and biosynthetic potential of mangrove rhizosphere Actinobacteria from Hainan Island. Front Microbiol. 2023;14:1157601.
Crossref - Subramani R, Aalbersberg W. Marine actinomycetes: an ongoing source of novel bioactive metabolites. Microbiol Res. 2012;167(10):571-580.
Crossref - Barzkar N, Sukhikh S, Babich O. Study of marine microorganism metabolites: new resources for bioactive natural products. Front Microbiol. 2023;14:1285902.
Crossref - Mondol MAM, Shahidullah Tareq F, Kim JH, et al. New antimicrobial compounds from a marine-derived Bacillus sp. J Antibiot. 2013;66(2):89-95.
Crossref - Beltran-Gracia E, Macedo-Raygoza G, Villafana-Rojas J, et al. Production of Lipopeptides by Fermentation Processes: Endophytic Bacteria, Fermentation Strategies and Easy Methods for Bacterial Selection. Ferment Process. 2017.
Crossref - Abriouel H, Franz CMAP, Ben Omar N, Galvez A. Diversity and applications of Bacillus bacteriocins. FEMS Microbiol Rev. 2011;35(1):201-232.
Crossref - Dias ACF, Andreote FD, Dini-Andreote F, et al. Diversity and biotechnological potential of culturable bacteria from Brazilian mangrove sediment. World J Microbiol Biotechnol. 2009;25(7):1305-1311.
Crossref - Sangkanu S, Rukachaisirikul V, Suriyachadkun C, Phongpaichit S. Evaluation of antibacterial potential of mangrove sediment-derived actinomycetes. Microb Pathog. 2017;112:303-312.
Crossref - El-Kahawy R, El-Shafeiy M, Helal SA, Aboul-Ela N, El-Wahab MA. Morphological deformities of benthic foraminifera in response to nearshore pollution of the Red Sea, Egypt. Environ Monit Assess. 2018;190(5):312.
Crossref - Alghamdi MA, Ayed L, Aljarad MR, Altayeb HN, Abbes S, Chaieb K. Whole genome sequencing analysis and Box-Behnken design for the optimization of the decolourization of mixture textile dyes by halotolerant microbial consortium. Microbiol Res. 2023;276:127481.
Crossref - Abdelmohsen UR, Yang C, Horn H, Hajjar D, Ravasi T, Hentschel U. Actinomycetes from red sea sponges: Sources for chemical and phylogenetic diversity. Mar Drugs. 2014;12(5):2771-2789.
Crossref - Singh V, Haque S, Singh H, et al. Isolation, Screening and Identification of Novel Isolates of Actinomycetes from India for Antimicrobial Applications. 2016;7:1921.
Crossref - Abdulrahman I, Jamal MT, Pugazhendi A, et al. Antibacterial and antibiofilm activity of extracts from sponge-associated bacterial endophytes. Prep Biochem Biotechnol. 2023;53(9):1143-1153.
Crossref - Priyanto JA, Prastya ME, Sinarawadi GS, et al. The antibacterial and antibiofilm potential of Paederia foetida Linn. leaves extract. J Appl Pharm Sci. 2022;12(10):117-124.
Crossref - Zhu D, Sethupathy S, Gao L, et al. Microbial diversity and community structure in deep-sea sediments of South Indian Ocean. Environ Sci Pollut Res. 2022;29(30):45793-45807.
Crossref - Pinnaka AK, Tanuku NRS. Marine Microbial Diversity for Sustainable Development. In: Satyanarayana, T., Johri, B., Das, S. (eds) Microbial Diversity in Ecosystem Sustainability and Biotechnological Applications. Springer, Singapore.
Crossref - Yaradoddi JS, Kontro MH, Ganachari S V, et al. Actinobacteria in Marine Environments. In: Yaradoddi, J.S., Kontro, M.H., Ganachari, S.V. (eds) Actinobacteria. Rhizosphere Biology. Springer, Singapore.
Crossref - Jiang H, Huang Q, Deng S, Dong H, Yu B. Planktonic actinobacterial diversity along a salinity gradient of a river and five lakes on the Tibetan Plateau. Extremophiles. 2010;14(4):367-376.
Crossref - Banti DC, Tsali A, Mitrakas M, Samaras P. The Dissolved Oxygen Effect on the Controlled Growth of Filamentous Microorganisms in Membrane Bioreactors. 2020;2(1):39.
Crossref - Weber-Scannell PK, Duffy LK. Effects of total dissolved solids on aquatic organisms: A review of literature and recommendation for salmonid species. Am J Environ Sci. 2007;3(1):1-6.
Crossref - Laid B, Kamel K, Mouloud G, et al. Effects of Plant Growth Promoting Rhizobacteria (PGPR) on In Vitro Bread Wheat (Triticum aestivum L.) Growth Parameters and Biological Control Mechanisms. Adv Microbiol. 2016;06(09):677-690.
Crossref - Bull AT, Ward AC, Goodfellow M. Search and Discovery Strategies for Biotechnology: the Paradigm Shift. Microbiol Mol Biol Rev. 2000;64(3):573-606.
Crossref - Jensen PR, Fenical W. Marine bacterial diversity as a resource for novel microbial products. J Ind Microbiol Biotechnol. 1996;17(5-6):346-351.
Crossref - Todar K. Bacillus and Related Endospore Forming Bacteria. Todar’s Online Textbook of Bacteriology; 2012. https://www.scribd.com/document/374847410/Todars-Online-Textbook-of-Bacteriology-pdf
- Nicholson WL, Munakata N, Horneck G, Melosh HJ, Setlow P. Resistance of Bacillus endospores to extreme terrestrial and extraterrestrial environments. Microbiol Mol Biol Rev. 2000;64(3):548-572.
Crossref - Priest FG. Extracellular enzyme synthesis in the genus Bacillus. Bacteriol Rev. 1977;41(3):711-753.
Crossref - Haki GD, Rakshit SK. Developments in industrially important thermostable enzymes: a review. Bioresour Technol. 2003;89(1):17-34.
Crossref - Lai Q, Liu Y, Shao Z. Bacillus xiamenensis sp. nov., isolated from intestinal tract contents of a flathead mullet (Mugil cephalus). Antonie Van Leeuwenhoek. 2014;105(1):99-107.
Crossref - Liaqat I, Mubin M, Chaudhry MA, et al. Multienzyme and Antibacterial Potential of Bacteria Isolated from gut of Asian Honey Bee (Apis cerana Indica), Lahore Using Culture Dependent Method. Braz Arch Biol Technol. 2021;64.
Crossref - Xu D, Cote JC. Phylogenetic relationships between Bacillus species and related genera inferred from comparison of 32 end 16S rDNA and 52 end 16S-23S ITS nucleotide sequences. Int J Syst Evol Microbiol. 2003;53(3):695-704.
Crossref - Barros-Rodríguez A, Pacheco P, Peñas-Corte M, et al. Comparative Study of Bacillus-Based Plant Biofertilizers: A Proposed Index. Biology. 2024;13(9):668. Published 2024.
Crossref - Ventosa A, Nieto JJ, Oren A. Biology of moderately halophilic aerobic bacteria. Microbiol Mol Biol Rev. 1998;62(2):504-544.
Crossref - Oren A. Diversity of halophilic microorganisms: environments, phylogeny, physiology, and applications. J Ind Microbiol Biotechnol. 2002;28(1):56-63.
Crossref - Margesin R, Schinner F. Bioremediation (natural attenuation and biostimulation) of diesel-oil-contaminated soil in an alpine glacier skiing area. Appl Environ Microbiol. 2001;67(7):3127-3133.
Crossref - Chen YP, Rekha PD, Arun AB, Shen FT, Lai W-A, Young CC. Phosphate solubilizing bacteria from subtropical soil and their tricalcium phosphate solubilizing abilities. Appl Soil Ecol. 2006;34(1):33-41.
Crossref - Sumi CD, Yang BW, Yeo I-C, Hahm YT. Antimicrobial peptides of the genus Bacillus: a new era for antibiotics. Can J Microbiol. 2015;61(2):93-103.
Crossref - Ongena M, Jacques P. Bacillus lipopeptides: versatile weapons for plant disease biocontrol. Trends Microbiol. 2008;16(3):115-125.
Crossref - Slama H Ben, Cherif-Silini H, Bouket AC, et al. Screening for Fusarium Antagonistic Bacteria From Contrasting Niches Designated the Endophyte Bacillus halotolerans as Plant Warden Against Fusarium. Front Microbiol. 2018;9:3236.
Crossref - Earl AM, Losick R, Kolter R. Ecology and genomics of Bacillus subtilis. Trends Microbiol. 2008;16(6):269-275.
Crossref - Stein T. Bacillus subtilis antibiotics: structures, syntheses and specific functions. Mol Microbiol. 2005;56(4):845-857.
Crossref - Reva ON, Swanevelder DZH, Mwita LA, et al. Genetic, Epigenetic and Phenotypic Diversity of Four Bacillus velezensis Strains Used for Plant Protection or as Probiotics. Front Microbiol. 2019;10:2610.
Crossref - Awais M, Pervez A, Yaqub A, Shah MM. Production of antimicrobial metabolites by Bacillus subtilis immobilized in polyacrylamide gel. Pak J Zool. 2010;42(3):267-275.
- Sagar A, Yadav SS, Sayyed RZ, Sharma S, Ramteke PW. Bacillus subtilis: A Multifarious Plant Growth Promoter, Biocontrol Agent, and Bioalleviator of Abiotic Stress. In: Islam, M.T., Rahman, M., Pandey, P. (eds) Bacilli in Agrobiotechnology. Bacilli in Climate Resilient Agriculture and Bioprospecting. Springer, Cham..
Crossref - Ahmad A, Baothman OA, Nadeem MS, Ahmad V. Biodesulfurizing Microbes in the Petroleum Refinery Areas of Saudi Arabia. J Pure Appl Microbiol. 2023;17(3):1737-1747.
Crossref
© The Author(s) 2025. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.