ISSN: 0973-7510
E-ISSN: 2581-690X
Patients with malignancy are highly prone to infections by Extended spectrum beta-lactamases producing Enterobacteriaceae (ESBL-PE). Knowledge on local resistance profile and resistance genes is essential to decide empirical drug. Hence, the study aims to find the resistance profile and the resistance genes of ESBL-PE from cancer patients. 172 oxyimino-cephalosporins resistant Enterobacterial isolates from clinical specimens of cancer patients were obtained. Study isolates were speciated by conventional biochemical methods. Resistance to antibiotics was detected by disc diffusion method. Phenotypic detection of ESBLs was performed as stated in CLSI guidelines. Genotypic characterization of resistance determinants of ESBL-PE was done by PCR. Among 172 Enterobacterial isolates, 151 (87.7%) were ESBL producers. E. coli (67.5%) was the major species producing ESBL enzymes followed by K. pneumoniae (27.8%). Antibiotic susceptibility pattern showed lowest resistance to imipenem 11.2%, and netilmicin 13.9%. 72% of ESBL-PE was found to be Multidrug-resistant. Among ESBL genes, blaCTX M gp-1 (83.4%) was dominant followed by blaTEM (32.4%) and blaSHV (27.8%). 36% of the isolates were found to be positive for more than one ESBL gene. High level of plasmid encoding quinolone resistance genes (64.2%) was identified in ESBL-PE. Low levels of plasmid mediated AmpC gene (15.8%) and 16S rRNA genes (9.2%) were found in ESBL-PE. The predominant ESBL encoding gene belongs to blaCTX M group 1. High proportion of ESBL-PE was found to co-harbor PMQR genes. ESBL-PE had highest sensitivity for imipenem and netilmicin.
ESBL, Cancer, Enterobacteriaceae, Multidrug-resistance
Patients with malignancy are more susceptible to bacterial infections, as they are often immunocompromised due to malnutrition and various treatment modalities. Infection in cancer patients increases the hospital stay, financial burden, morbidity and mortality.1,2 Enterobacteriaceae accounts for approximately 65%-80% of Gram negative bacterial infections in cancer patients.3 Resistance in Enterobacteriaceae due to Extended spectrum β-lactamases (ESBLs) are reported increasingly and the organisms producing these enzymes has spread worldwide and considered as important pathogens in community and hospital settings.4,5 ESBLs producing Enterobacteriaceae (ESBL-PE) is a leading organism causing infection in cancer population and considered as major threat since it leads to serious illness in those patients.6
Wide usage of oxyimino-cephalosporins as therapeutic options for enterobacterial infections led to the appearance of ESBLs.5 ESBL enzymes hydrolyzes penicillins, cephalosporins except cephamycins. ESBL genes are encoded by plasmid that facilitates its dissemination among different Enterobacteriaceae species.7 The major types of ESBLs were from TEM, SHV, and CTX M families.8 blaCTX-M-15 gene has emerged all over the world, also common in nosocomial and community acquired infections.9 ESBL producing Enterobacteriaceae are often non-susceptible to fluoroquinolones, co-trimoxazole and aminoglycoside, due to coexistence of different resistance determinants on same plasmid and so exhibit the Multidrug-resistance phenotype and limits the treatment options.10 Although several studies of ESBL-PE species from hospital and community settings have been reported, very few data are available on ESBL-PE isolates from cancer population. The few reports on ESBL-PE are pertaining to bloodstream infections and there is hardly report on antibiotic resistance determinants of ESBL-PE from diverse infections in cancer patients. It is significant to identify the sensitivity pattern and resistance genes of Multidrug-resistant pathogen like ESBL-PE to aid appropriate choice of empirical therapy for infection in this high risk population.
Hence the present study was designed to find antibiotic susceptibility pattern and characterize the various antibiotic resistance genes of ESBL-PE isolates from cancer patients.
Study isolates
172 oxyimino-cephalosporins resistant Enterobacteriaceae were collected from Cancer care hospital, Chennai, Tamil Nadu, South India during the period commencing from February 2016 to May 2017. The collected isolates were obtained from various clinical specimens (urine, pus, sputum etc,) of cancer patients.
Identification and speciation of study isolates
Isolation of the study isolates from the clinical specimens was done in hospital according to standard microbiological procedure. Only the pure culture isolates collected from the hospital were used in the study. All the 172 isolates were identified and speciated by using standard techniques such as Gram staining, studying colony morphology on MacConkey agar (Hi-Media Labs, Mumbai, India), oxidase test, catalase test, nitrate reduction, O/F test, TSI, IMViC ( Indole, Methyl Red, Voges Proskauer and Citrate), urease production, phenylalanine deaminase test, motility tests and sugar fermentation tests.
Antimicrobial susceptibility testing
Antibiotic resistance was detected by Kirby Bauer disc diffusion method and the zone diameters were interpreted according to the Clinical Laboratory Standard Institute (CLSI) break points.11 The following antibiotic discs were tested: Ampicillin (10μg), piperacillin (100μg), piperacillin/tazobactam (100/10μg), cefotaxime (30μg), ceftazidime (30μg), imipenem (10μg), meropenem (10μg), amikacin (30μg), gentamicin (10μg), netilmicin (30μg), ciprofloxacin (5μg), cotrimoxazole (1.25/23.75 μg), norfloxacin (10 μg) (Hi-Media Labs, Mumbai, India). The quality of antibiotic disc was determined using E.coli ATCC 25922 standard strain.
Phenotypic confirmation of ESBLs
Phenotypic confirmation of ESBL production was done by disc diffusion test as stated in CLSI guidelines.11 The ceftazidime (CAZ-30 μg) and Cefotaxime (CTX-30 μg), discs with and without clavulanic acid (10 μg) (Hi-Media Labs, Mumbai, India), were placed 30 mm apart on a lawn culture of the test isolate (0.5 McFarland opacity)and incubated for 24 hours at 37°C. Bacterial isolate was considered ESBL positive if ≥ 5mm increase in zone diameter of the clavulanic acid combination disc was observed than the disc without clavulanic acid.
Molecular detection of ESBL encoding genes and associated resistant genes
DNA extraction was carried out by boiling lysis method. Colonies of study isolates were inoculated in Luria-Bertani broth and incubated overnight. Then the broth was centrifuged and 200μl sterile water was added to the pellet; kept in dry bath at 100°C for 10 mins; immediately kept in -20°C deep freezer followed by centrifugation. Multiplex PCR detection of blaCTXM gp-1, blaCTXM gp-2, blaCTXM gp-8, blaCTXM gp-9, blaCTXM gp-25 genes was done.12 The presence of blaTEM and blaSHV were identified by the multiplex PCR according to the published methods.13 The other resistant genes such as plasmid-mediated AmpC genes, 16SrRNA methylase genes (armA, rmtB, rmtC) coding for aminoglycoside resistance and plasmid-mediated quinolone resistant (PMQR) genes (qnr A, B, S, aac (6’)-Ib-cr) were detected by PCR using previously described method (Table 1).14-17
Table (1):
ESBL genes primers.
| Gene | Primer | Amplicon size(bp) |
Reference |
|---|---|---|---|
| CTX –M Group I |
F: AAA AAT CAC TGC GCC AGT TC R: AGC TTA TTC ATC GCC ACG TT |
415 | Woodford et.al.,2005 |
| CTX–M Group2 |
F: CGA CGC TAC CCC TGC TAT T R:CCA GCG TCA GAT TTT TCA GG |
552 | |
| CTX–M Group9 |
F:CAA AGA GAG TGC AAC GGA TG R: ATT GGA AAG CGT TCA TCA CC |
205 | |
| CTX–M Group8 |
F: TCG CGT TAA GCG GAT GAT GC R: AAC CCA CGA TGT GGG TAG C |
666 | |
| CTX –M Group 25 |
F: GCA CGA TGA CAT TCG GG R: AAC CCA CGA TGT GGG TAG C |
327 | |
| blaTEM | F:TTTCGTGTCGCCCTTATTCC R:ATCGTTGTCAGAAGTAAGTTGG |
294 | Hassan MI et al.,2012 |
| blaSHV | F: CGCCTGTGTATTATCTCCCT R:CGAGTAGTCCACCAGATCCT |
404 |
[Table 1 contains the primers used to identify ESBLs determinants]
Statistics
GraphPad QuickCalcs were used to perform statistical analysis. Chi-square test was used to calculate associations between antibiotic resistance and its genes. A p-value of < 0.05 was considered statistically significant.
Distribution of study isolates
The source of the 172 oxyimino cephalosporin resistant enterobacterial isolates, are as follows 71.5% (123) from urine, followed by pus 12.7% (22), sputum 12.2% (21), blood 1.7% (3), wound 1.1% (2) and vaginal swab 0.5% (1). Of 172 oxyimino-cephalosporin resistant Enterobacteriaceae isolates, 67% (116) were from inpatients and 33% (56) were from outpatient’s samples. Among the 172 clinical isolates collected, 23.25% (40/172) were from hematological malignancy patients and 76.74% (132/172) were from solid organ tumor patients (Table 2).
Table (2):
Clinical characteristics of cancer patients infected by ESBL-PE.
Clinical Characteristics |
Percentage (n) N=151 |
|
|---|---|---|
Solid organ tumour |
Head and Neck cancer Lung Cancer Esophago-gastrointestinal cancer Colon and rectal cancer Hepatobiliary and pancreatic cancer Breast cancer Genitourinary cancer Gynecological cancer Others |
7.9 (12) 5.9 (9) 8.6 (13) 9.2 (14) 1.9 (3) 9.9 (15) 17.2 (26) 12.5 (19) 1.9 (3) |
Hematological malignancies |
Acute lymphoblastic leukemia Chronic lymphoblastic leukemia Hodgkin lymphoma Non- Hodgkin lymphoma Multiple myeloma |
6.6 (10) 1.3 (2) 0.6 (1) 7.9 (12) 7.9 (12) |
Age |
<15 15-60 >60 |
1.3 (2) 49 (74) 49.6 (75) |
Sex |
Male Female |
51.6 (78) 48.3 (73) |
Patient type |
Inpatient Outpateint |
64.2(97) 35.7(54) |
Table 2 shows the clinical details of the patients infected by ESBL-PE. n indicates no.of ESBL isolates
Of the 172 oxyimino-cephalosporin resistant Enterobacteriaceae isolates, 69.1% (n=119) was found to be Escherichia coli followed by Klebsiella pneumoniae 24.4% (n=42), Citrobacter koseri 3.4% (n=6), Enterobacter cloacae 1.7% (n=3) and Klebsiella oxytoca 1.1% (n=2) (Table 3).
Table (3):
Species distribution of ESBL producing isolates and its source.
| Species | Source (n=151) | Total | |||||
|---|---|---|---|---|---|---|---|
| Urine | Sputum | Pus | Blood | Wound swab | Vaginal swab | ||
| Escherichia coli | 81(71.6%) | 9(47.3%) | 10(71.4%) | 1(33.3%) | 0(0) | 1(100%) | 102(67.5%) |
| Klebsiella pneumoniae | 24(21%) | 9(47.3%) | 4(28.4%) | 2(66.6%) | 0(0) | 0(0) | 39(25.8%) |
| Citrobacter koseri | 4(3.5%) | 1(5.2% | 0(0) | 0(0) | 0(0) | 0(0) | 5(3.3%) |
| Enterobacter cloacae | 3(2.6%) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 3(1.9%) |
| Klebsiella oxytoca | 1(0.8%) | 0(0) | 0(0) | 0(0) | 1(100%) | 0(0) | 2(1.3%) |
| Total | 113 | 19 | 14 | 3 | 1 | 1 | 151 |
Table 3 shows the species distribution of ESBL-PE
Phenotypic detection of ESBLs
Among 172 oxyimino cephalosporin resistant Enterobacteriaceae, 140 (81.3%) isolates showed positive reaction for ESBL production phenotypically. Irrespective of phenotypic ESBL detection, all the 172 isolates were subjected to genotypic detection: 151/172 (87.7%) enterobacterial isolates were positive for ESBLs production genotypically. Among 151 ESBL-PE, E.coli (67.5% n=102) was found to be the predominant species followed by K.pneumoniae (25.8% n=39).
Antibiotic susceptibility pattern
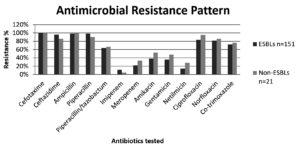
ESBL-PE isolates showed increased non-susceptibility to beta-lactam as well as non-beta-lactam drugs except for few antibiotics. Among the antibiotics tested, piperacillin showed resistance rate of 98.6% followed by ampicillin 98%, ciprofloxacin 83.4%, norfloxacin 81.4%, cotrimoxazole 72.1% and piperacillin/tazobactam 63.5%. Least resistance was observed for imipenem 11.2%, followed by netilmicin 13.9%, meropenem 21.8%, gentamicin 35.7%, and amikacin 38.4%. Multi-drug resistant phenotype (showing resistance to three or more classes of antibiotics) was observed in 72% of the ESBL-PE. Non-ESBL producing isolates showed highest resistance to fluoroquinolones and piperacillin. (Refer Figure)
Genotypic characterization of Esbl and other resistant genes
Among the ESBL genes detected, blaCTX M gp1 was the predominant ESBL type found in 83.4% (126/151) of the isolates followed by blaTEM 32.4% (49/151) and blaSHV 27.8% (42/151). 36% of the test isolates were found to be positive for more than one ESBLs encoding genes. The blaCTXM gp2, blaCTXM gp8, blaCTXM gp9, and blaCTXM gp25 genes were not found in any of the ESBL-PE. 64.2% of the ESBL producing strains harbored one or two PMQR genes, qnrA was not found on any of the isolates, 15.8% of the isolates were found to carry plasmid-mediated AmpC genes and 9.2% of the isolates were associated with 16SrRNA methylase gene (armA, rmtB). All the isolates were negative for rmtC gene (Refer Table 4). The statistical analysis revealed that the association between the antibiotic resistance and its determinants was not significant.
Table (4):
Resistance determinants of ESBL-PE.
| Species(n) | Other resistant determinants | |||
|---|---|---|---|---|
| ESBL genes(n) | AmpC (n) | PMQR(n) | 16SrRNA methylase(n) | |
| E.coli (102) | blaCTX-M gp-1(60) blaTEM-(10) blaSHV(3) blaCTX-M gp-1+blaTEM(17) blaCTX-M gp-1+blaSHV(10) blaCTX-M gp-1+blaTEM+blaSHV (2) |
CMY(19) | qnrB(8) qnrS(3) aac (6′)-Ib-cr (41) qnrB+aac(6′)-Ib-cr (13) qnrS+aac (6′)-Ib-cr (2) |
armA (5) rmtB (2) |
| K.pneumoniae(39) | blaCTX-M gp-1(9) blaTEM-(1) blaSHV(7) blaCTX-M gp-1+blaTEM(6) blaCTX-M gp-1+blaSHV(6) blaTEM+blaSHV (1) blaCTX-M gp-1+blaTEM+blaSHV (9) |
CMY (4) | qnrB(7) qnrS(1) aac (6′)-Ib-cr (7) qnrB+aac (6′)-Ib-cr (9) qnrS+aac (6′)-Ib-cr (1) |
armA (4) rmtB (3) |
| C.koseri(5) | blaCTX-M gp-1(3) blaSHV(1) blaCTX-M gp-1+blaTEM(1) |
CMY (1) | aac (6′)-Ib-cr (1) qnrB+aac (6′)-Ib-cr (1) |
– |
| E.cloacae(3) | blaCTX-M gp-1(1) blaTEM+blaSHV (1) blaCTX-M gp-1+blaTEM+blaSHV (1) |
– | qnrS(1) | – |
| K.oxytoca(2) | blaCTX-M gp-1(1) blaSHV(1) |
– | aac (6′)-Ib-cr (1) qnrB+aac (6′)-Ib-cr (1) |
– |
Table 4 shows the antibiotic resistance determinants of ESBL-PE (n represents No. of isolates).
In this study, it has been observed that 87.7% of the oxyimino-cephalosporin resistant enterobacterial isolates from cancer patients were carrying ESBL encoding genes. This is consistent with the study conducted in New Delhi which reports 80% prevalence of ESBLs among Gram negative infections in cancer patients.18 Similar report from western China observed that 72.8% of the nosocomial infections are caused by ESBL-PE in cancer patient.19 ESBL-PE has been frequently reported as a major cause of infections and it increases the mortality and morbidity rates among cancer patients. This is because of common usage of cephalosporin as first line drug for enterobacterial infections and colonization by ESBL producing organisms.
In this study, E. coli was the leading ESBL-PE isolate followed by K. pneumoniae and the chief sources of these isolates were urine samples. This is in accordance with the report of Barta et al., and Jiang et al.18,19 Earlier findings state that ESBL-PE is one of the leading pathogens causing bloodstream infections in cancer patients.20,21 This is contrary to the findings of our current study which showed only 2% of ESBL-PE isolates were from blood samples. The findings of the present study suggests that other sites of infections viz., urinary tract, respiratory tract, gastro-intestinal tract, surgical site, wound are also important sources of ESBL-PE infections. Although, there are several reports available for bloodstream infections in cancer patients, very few reports are available on ESBL-PE infections from other clinical sites.
In this report, ESBL production was phenotypically negative in 7% of PCR positive ESBL isolates. A surveillance study from India reported 40% PCR positive ESBL isolates were undetectable phenotypically. Production of multiple β-lactamases can mask the inhibitory effect of clavulanic acid and may make ESBLs phenotypically undetectable.22
The antibiotic susceptibility testing of the current study revealed high level of non-susceptibility to non-beta-lactam drugs. ESBL-PE isolates showed higher resistance to fluoroquinolones and co-trimoxazole as previously reported by Aldrazi et al., and Mahamat et al.23,24 Resistance to other classes of antibiotics among ESBL-PE is worrisome because it further limits the empirical therapeutic options. ESBL-PE showed less resistance to aminoglycosides particularly netilmicin. The resistance rates of amikacin was 38% and of gentamicin 36% unlike other reports. A study from Spain reported 13% of gentamicin resistance and nil amikacin resistance among ESBL-PE which is much lower than the current study.25 Another study has reported 70.2% of gentamicin resistance, 67% of netilmicin resistance and 18% of amikacin resistance in ESBL-PE which is contrary to the current report.24 These variations may be due to different geographical locations and study period.
Carbapenems are the generally preferred drugs for the treatment of ESBL-PE infection and it has lower failure rate in treating these infections compared to cephalosporin and fluoroquinolones.26 In the current study, ESBL-PE showed high susceptibility to carbapenems. They showed higher susceptibility to imipenem than meropenem which is in concordance with previous report from India.22 Even low rate of carbapenem resistance among ESBL-PE is of major concern and hence it should be used with caution. In the current study, increased non-susceptibility was observed for ampicillin, piperacillin, and 3rd generation cephalosporins and this is due to the production of ESBLs and other betalactam hydrolyzing enzymes.
In the current study, the predominant ESBL encoding gene among the enterobacterial isolates is blaCTXM group1 (83.4%) followed by TEM (37.4%) and SHV (27.8%). Several studies have reported similar findings of high levels of blaCTXM genes among Enterobacteriaceae.24 A study from India by Ensor et al., has reported high rate of blaCTXM (73%) among cephalosporin resistant Enterobacteriaceae.27 ESBL genes blaCTX-M15 belongs to blaCTX-M group1 and has disseminated worldwide and is widely reported as predominant ESBL gene among Enterobacteriaceae.9 In the present report, blaTEM was found in 37.4% ESBL-PE isolates; contrary to previous reports from India. A study by Jena et al, reported high proportion of blaTEM 96.42% in ESBL-PE27 and another study by Ray et al., has also reported blaTEM as the most common ESBL among E. coli and K. pneumoniae.22 blaSHV gene was detected in less proportion (27.8%) in the current study and this is similar to the earlier study from India which documented lower rates of blaSHV (6% and 21%) among E. coli and K. pneumoniae isolates.22 Co-occurrence of ESBL genes were observed in the present study and the same has been reported in the earlier studies.22,24,28
Findings of the current study demonstrate the co-harboring of various resistant determinants by ESBL-PE. Co-existence of plasmid-mediated AmpC and ESBL genes were found in 15.8% of the isolates and this is similar to the study by Rizi et al., who reported co-production of AmpC in 16.7% of ESBL isolates.29 Even in the present study, they exhibit lower resistance to aminoglycoside antibiotics, 9.2% of ESBL-PE was found to carry 16SrRNA methylase gene which is lower than the previous report by Ayad et al.30 Our study shows high prevalence of fluoroquinolone resistant determinants among ESBL-PE isolates. The rate of co-existence of PMQR genes are qnrB–25.8%, qnrS- 5.2% and aac6’1b-cr gene is 50.3% and it is similar to the previous report.31 Since all these resistant determinants are located on the mobile genetic elements, it can be easily acquired by the ESBL-PE facilitating the Multidrug-resistant phenotype, consequently making the treatment more difficult. Hence, it is critical to know the local resistance pattern and prevalent resistance genes of this Multidrug-resistant ESBL-PE as improper empirical therapy may lead to severe sepsis and poor outcome in cancer patients.
The study has certain limitations. The study data was from single centre, hence it may not depict epidemiology of different geographical locations. More clinical details of the cancer patients such as previous exposure to antibiotics and other co-morbidities are unavailable.
To the best of our knowledge, this is the first report regarding the antibiotic resistance determinants of ESBL-PE from cancer patients from our geographical location. The study data showed high prevalence of ESBLs among Enterobacteriaceae from cancer patients. The predominant ESBL encoding gene was blaCTX M group 1. High proportion of ESBL-PE was found to co-harbor PMQR determinants and co-existence of AmpC and 16SrRNA methylase genes were also observed. Most of the ESBL-PE isolates showed Multidrug-resistant phenotype with few treatment options. Thus, the judicious usage of antibiotics and surveillance program should be implemented for cancer patients to prevent spread of ESBL-PE. Imipenem and netilmicin have highest sensitivity for ESBL-PE and can be considered for the treatment in our region. Further studies on prevailing resistant pattern from different regions of India are required to affirm the appropriate empirical therapeutic options.
ACKNOWLEDGMENTS
The authors would like to thank VS Hospitals, Chetpet, Chennai, Tamil Nadu for providing the isolates for the study.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
The study was partially funded by Senior Research Fellowship awarded by Indian Council of Medical Research (ICMR).
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
Not applicable.
- Zembower TR. Epidemiology of infections in cancer patients. Cancer Treat Res. 2014:43-89.
Crossref - Girmenia C, Menichetti F. Current epidemiology and prevention of infectious complications in cancer patients. Eur Oncol Haematol. 2011;07(04):270.
Crossref - Saghir S, Faiz M, Saleem M, Younus A, Aziz H. Characterization and anti-microbial susceptibility of gram-negative bacteria isolated from bloodstream infections of cancer patients on chemotherapy in Pakistan. Indian J Med Microbiol. 2009;27(4):341-347.
Crossref - Rupp ME, Fey PD. Extended spectrum beta-lactamase (ESBL)-producing Enterobacteriaceae considerations for diagnosis, prevention and drug treatment. Drugs. 2003;63(4):353-365.
Crossref - Chaudhary U, Aggarwal R. Extended spectrum β-lactamases (ESBL) – an emerging threat to clinical therapeutics. Indian J Med Microbiol. 2004;22(2):75-80.
Crossref - Tohamy S, Aboshanab K, El-Mahallawy H, El-Ansary MR, Afifi S. Prevalence of multidrug-resistant gram-negative pathogens isolated from febrile neutropenic cancer patients with bloodstream infections in Egypt and new synergistic antibiotic combinations. Infect Drug Resist. 2018;11:791-803.
Crossref - Chandramohan L, Revell PA. Prevalence and molecular characterization of extended-spectrum-β-lactamase-producing Enterobacteriaceae in a pediatric patient population. Antimicrob Agents Chemother. 2012;56(9):4765-4770.
Crossref - Bradford PA. Extended-spectrum β-lactamases in the 21st century: Characterization, epidemiology, and detection of this important resistance threat. Clin Microbiol Rev. 2001;14(4):933-951.
Crossref - Canton R, Novais A, Valverde A, et al. Prevalence and spread of extended-spectrum β-lactamase-producing Enterobacteriaceae in Europe. Clin Microbiol Infect. 2008;14(Suppl 1):144-153.
Crossref - Paterson DL, Bonomo RA. Extended-spectrum β-lactamases: A clinical update. Clin Microbiol Rev. 2005;18(4):657-686.
Crossref - Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fourth Informational Supplement. CLSI document. M100-S24 (ISBN 1-56238-897-5[Print]; ISBN 1-56238-898-3.
- Woodford N, Fagan EJ, Ellington MJ. Multiplex PCR for rapid detection of genes encoding CTX-M extended-spectrum β-lactamases. J Antimicrob Chemother. 2005;57(1):154-155.
Crossref - Hassan MI, Alkharsah KR, Alzahrani AJ, Obeid OE, Khamis AH, Diab A. Detection of extended spectrum beta-lactamases-producing isolates and effect of AMPC overlapping. J Infect Dev Ctries. 2013;7(8):618-629.
Crossref - Perez-Perez F. Javier, Hanson ND. Detection of plasmid-mediated AMPC β-lactamase genes in clinical isolates by using multiplex PCR. J Clin Microbiol. 2002;40(6):2153-2162.
Crossref - Doi Y, Arakawa Y. 16S ribosomal RNA methylation: Emerging resistance mechanism against aminoglycosides. Clin Infect Dis. 2007;45(1):88-94.
Crossref - Wu J-J, Ko W-C, Tsai S-H, Yan J-J. Prevalence of plasmid-mediated quinolone resistance determinants QNRA, qnrb, and qnrs among clinical isolates of Enterobacter cloacae in a Taiwanese hospital. Antimicrob Agents Chemother. 2007;51(4):1223-1227.
Crossref - Wareham DW, Umoren I, Khanna P, Gordon NC. Allele-specific polymerase chain reaction (PCR) for rapid detection of the AAC(6′)-IB-CR quinolone resistance gene. Int J Antimicrob Agents. 2010;36(5):476-477.
Crossref - Batra U, Goyal P, Jain P, et al. Epidemiology and resistance pattern of bacterial isolates among cancer patients in a Tertiary Care Oncology Centre in North India. Indian J Cancer.2016;53(3):448-451.
Crossref - Jiang A-M, Shi X, Liu N, et al. Nosocomial infections due to multidrug-resistant bacteria in cancer patients: A six-year retrospective study of an oncology center in Western China. BMC Infect Dis. 2020;20(1).
Crossref - Abbasi Montazeri E, Khosravi AD, Saki M, Sirous M, Keikhaei B, Seyed-Mohammadi S. prevalence of extended-spectrum beta-lactamase-producing enterobacteriaceae causing bloodstream infections in cancer patients from southwest of iran. Infect Drug Resist. 2020;13:1319-1326.
Crossref - Ha YE, Kang C-I, Cha MK, et al. Epidemiology and clinical outcomes of bloodstream infections caused by extended-spectrum β-lactamase-producing Escherichia coli in patients with cancer. Int J Antimicrob Agents. 2013;42(5):403-409.
Crossref - Ray P, Gautam V, Thakur A, et al. Molecular characterization of extended-spectrum β-lactamases among clinical isolates of Escherichia coli & Klebsiella pneumoniae: A multi-centric study from Tertiary Care Hospitals in India. Indian J Med Res. 2019;149(2):208.
Crossref - Aldrazi FA, Rabaan AA, Alsuliman SA, et al. ESBL expression and antibiotic resistance patterns in a hospital in Saudi Arabia: Do Healthcare staff have the whole picture? J Infect Public Health. 2020;13(5):759-766.
Crossref - Ouchar Mahamat O, Lounnas M, Hide M, et al. High prevalence and characterization of extended-spectrum ß-lactamase producing Enterobacteriaceae in Chadian Hospitals. BMC Infect Dis. 2019;19(1):205.
Crossref - Gudiol C, Tubau F, Calatayud L, et al. Bacteraemia due to multidrug-resistant gram-negative bacilli in cancer patients: Risk factors, antibiotic therapy and outcomes. J Antimicrob Chemother. 2010;66(3):657-663.
Crossref - Rodriguez-Bano J, Gutierrez-Gutierrez B, Machuca I, Pascual A. Treatment of infections caused by extended-spectrum-beta-lactamase-, AMPC-, and carbapenemase-producing Enterobacteriaceae. Clin Microbiol Rev. 2018;31(2).
Crossref - Ensor VM, Shahid M, Evans JT, Hawkey PM. Occurrence, prevalence and genetic environment of CTX-M -lactamases in Enterobacteriaceae from Indian Hospitals. J Antimicrob Chemother. 2006;58(6):1260-1263.
Crossref - Jena J, Debata NK, Sahoo RK, Gaur M, Subudhi E. Molecular characterization of extended spectrum β-lactamase-producing Enterobacteriaceae strains isolated from a tertiary care hospital. Microbial Pathogenesis. 2018;115:112-116.
Crossref - Rizi KS, Mosavat A, Youssefi M, et al. High prevalence of blaCMY AMPC beta-lactamase in ESBL co-producing Escherichia coli and Klebsiella spp. clinical isolates in the northeast of Iran. J Glob Antimicrob Resist. 2020;22:477-482.
Crossref - Ayad A, Drissi M, de Curraize C, et al. Occurence of arma and rmtb aminoglycoside resistance 16s rrna methylases in extended-spectrum β-lactamases producing Escherichia coli in Algerian hospitals. Front Microbiol. 2016;7:1409.
Crossref - Azargun R, Sadeghi MR, Soroush Barhaghi MH, et al. The prevalence of plasmid-mediated quinolone resistance and ESBL-production in enterobacteriaceaeisolated from urinary tract infections. Infect Drug Resist. 2018;11:1007-1014.
Crossref
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.