ISSN: 0973-7510
E-ISSN: 2581-690X
This study aims to investigate the morphological and biochemical characteristics of thermophilic bacteria isolated from thermophilic biotopes in the Al-Jouf region of Saudi Arabia for the first time. Additionally, the production of thermostable bacterial enzymes (proteases, amylases, cellulases, gelatinases, and lipases) was investigated. Six main bacterial species were identified via 16S rRNA sequencing and phylogenetic analysis. Twenty water and soil samples were collected from several thermophilic sites in the Dumat Al-Jandal and Al-Qurayyat cities. In total, 28 bacterial strains were isolated and biochemically characterized. Most isolated strains showed high protease and amylase production. 46 and 28% of isolated strains showed the production of cellulase and gelatinase, respectively. However, lipase production was not observed in any isolated bacteria. Four Bacillus licheniformis and two Bacillus subtilis strains exhibited high GC content (55%). Our findings suggest thermophilic bacteria as a source of thermostable enzymes for pharmaceutical and industrial applications.
Hot Springs, Salt Marsh Soil, Bacillus licheniformis, Bacillus subtilis, Thermostable Enzymes
The increasing use of enzymes in food, chemical, and pharmaceutical industries has created a high demand for novel biocatalysts with advanced properties. To meet this growing demand, particularly of biocatalysts, it is crucial to develop more eco-friendly “green” chemistry approaches based on the “traditional” organic synthesis model.1 Advances in biotechnology, particularly in genetic and protein engineering, have enabled the production of enzymes with new activities that can adapt to various reaction conditions and are more favourable for industrial use.2,3
Such enzymes generally show good stability against changes in temperature, pH, and reaction medium composition as well as high reaction rates. These parameters are important for the industrial applications of enzymes.4
Thermophiles (optimal growth above 50°C) and hyperthermophiles (optimal growth above 80°C) are among the most studied microorganisms in extreme environments.5,6 Their natural biotopes vary and can be terrestrial (geothermal springs, desert and volcanic soils, oil drills, and compost) or marine (deep hydrothermal mountains). Extreme temperature, pH, and anoxic conditions are responsible for the high genomic and metabolic flexibility of thermophilic microbial communities colonising these hot environments, making the exploitation of such microbial proteins attractive for biotechnological applications.7-9
Proteases, lipases, and other hydrolases, such as cellulases, gelatinases, chitinases, and amylases, affect the stability of active enzymes of industrial interest at high temperatures.
Saudi Arabia is characterised by a semi-dry to dry desert climate with hot days and cold nights and extremely low annual rainfall, except the Aasir region in the southwestern parts of the Kingdom that receives an average precipitation of approximately 300 mm per year. The maximum temperature can reach 45°C in summer (NCM, 2023). Many hot springs contain hot natural sulphur water that is spread in several areas serving as tourist destinations, health retreats, and wellness centres in the Kingdom.10,11 Most studies have focused on hot springs in the southwestern regions of the Kingdom, such as the Gizan, Al-Lith, and Tabuk regions,10-14 however, the northern regions of the Kingdom, especially the Al-Jouf region, remain relatively unexplored.
In this study, we aimed to isolate thermophilic bacteria from different hot sites (hot springs, desert soil, lake water, and agricultural soil) in the Al-Jouf region and investigate their morphological, microscopic, and biochemical characteristics. Moreover, the ability of the isolated thermophilic bacteria to produce various enzymes, such as proteases, amylases, lipases, cellulases, and gelatinases, was studied. Molecular identification based on 16S rRNA sequencing was also performed for the identified six main bacteria.
Sampling and physico-chemical characterization
In this study, 20 water and soil samples were collected from different sites, especially the Dumat Al-Jandal and Al-Qurayyat cities, of the Al-Jouf region (29° 30′ 0″ N, 39° 30′ 0″ E). Samples were collected in sterile glass containers and transported to the laboratory for analysis (Table 1). Additionally, the temperature, pH, and electrical conductivity (EC) were determined. Temperature at the site was measured during sampling using a thermometer. The pH and EC of the water samples were directly measured using a pH meter (model WTW inoLab 7110, Germany) and conductivity meter (model CL 221, India). The soil samples were homogenised in distilled water (1w/5v), stirred for 2 h, and allowed to settle. Then, the pH and EC of the supernatants were determined.15
Isolation and characterization of the isolated thermophilic bacteria
For the isolation of thermophilic bacteria, 10 g of soil or 10 mL of water sample was placed in a sterile bottle containing 90 mL of physiological solution (9 g/L NaCl), and the mixture was vigorously stirred. Serial decimal dilutions were made and 100µL of the appropriate dilution was poured in triplicate on the nutrient agar (NA) medium (Techno Pharmchem, India) and incubated at 45°C for 48 h. Individual bacterial colonies from agar plates were selected and transferred to fresh NA plates. The strains were then streaked and transferred repeatedly to different plates to obtain pure colonies. Morphological characterisation of the isolated bacteria was performed via microscopic and macroscopic examinations, followed by gram staining. The ability of the isolated bacteria to assimilate substrates was analysed using the mannitol-motility, Kligler Hajna, and Simmon’s citrate media (Figure 1).16
Figure 1. Al Jouf Maps showing the studied sampling sites located at Al Qurayyet and Dumat Al Jandal cities
Thermophilic test
Thermophilic tests were performed by growing the isolated bacteria on NA at various temperatures (45, 50, 55, 60, 65 and 70°C) for 7 days.
Protease activity
Next, the protease activity of the isolated bacteria was determined using NA containing 2.5% skimmed milk. The strains were streaked onto the medium and incubated at 45°C for 48 h. Transparent zones around the bacterial colonies confirmed the presence of protease enzyme.17
Amylase activity
Amylase-producing bacteria were screened using a starch agar medium. Isolated bacteria were streaked on the medium and incubated at 45°C for 48 h. After incubation, the plates were flooded with 1% freshly prepared iodine solution (2% I2 and 0.2% KI). The results were considered positive when a clear zone was formed around the bacterial colony.17
Cellulase activity
A pure bacterial culture was streaked on agar medium containing 1% (w/v) of carboxymethyl-cellulose and incubated at 45°C for 48 h. The plates were further incubated with the Congo red solution (1% w/v) for 30 min. Then, the Congo red solution was poured off, and the plates were flooded with 1 M NaCl for 15 min. Cellulose degradation was confirmed by the formation of a clear hydrolysis zone around the bacteria.18
Lipase activity
Lipase activity of the isolated bacteria was determined by inoculating the strains in NA supplemented with 1% Tween 80. After incubation at 45°C for 48 h, the appearance of a turbid halo surrounding the colonies confirmed lipase production.19
Gelatinase activity
Next, gelatinase production was detected by inoculating the isolated bacteria on NA supplemented with 1.5% gelatin. The plates were incubated at 45°C for 48 h, followed by refrigeration at 4°C for 30 min. Gelatin liquefaction was taken as a positive result.20
16S rRNA bacterial identification, GC content, and phylogenetic analysis
The six main isolated strains were molecularly identified via 16S rRNA sequencing. DNA was extracted from the bacterial strains using the QIAamp DNA Mini kit (Qiagen, Germany), with some modifications of the manufacturer’s guidelines. Briefly, 200µL of the bacterial cell suspension was incubated with 200µL of lysis buffer and 20µL of proteinase K at 56°C for 10 min. After incubation, 200µL of 100% ethanol was injected into the lysate. The samples were washed and centrifuged at 13000 rpm for 10 min. Nucleic acid was eluted with 100µL of elution buffer provided in the kit. For 16S rDNA amplification, the universal primer (AGAGTTTGATCMTGGCTCAG) and reverse primer (TACGGYTACCTTGTTACGACTT) were used in a 25-µL reaction containing 12.5µL of the Emerald Amp Max PCR Master Mix (Takara, Japan), in addition to 1µL of each primer of 20 pmol concentration, 5.5µL of water, and 5µL of DNA template. The reaction was conducted in an Applied Biosystems 2720 thermal cycler. The PCR program was as follows: primary denaturation at 94°C for 5 min, 35 amplification cycles of denaturation at 94°C for 0.5 min, annealing at 56°C for 1 min, extension at 72°C for 1.2 min, and extension at 72°C for 12 min.
Next, PCR products were separated via electrophoresis on 1% agarose gel (Applichem, Germany, GmbH) in 1× TBE buffer at room temperature with 5V/cm gradients. Each gel slot contained a 20µL load of the product. A Gelpilot100 bp plus ladder (Qiagen) was used to ascertain the fragment sizes. A gel documentation system (Alpha Innotech, Biometra) was used to photograph the gels, and computer software was used to analyse the data. A Single PCR product of 1500 bp was obtained. Purified PCR products were sequenced using the ABI 3730xl DNA Sequencer (Hitachi, Tokyo, Japan). The identified 16S rDNA nucleotide sequences were deposited on the European Nucleotide Archive, which assigned accession numbers to all the identified strains.
The ratio of guanine and cytosine was determined for the 16S rDNAs of the six main bacteria in addition to 12 similar sequences (thermophilic and non-thermophilic bacteria) collected from the GenBank database using the ACUA software.21 Phylogenetic analyses of the sequences of our isolates was performed, and they were aligned with the sequences of high similarity from GenBank using the MUSCLE Multiple Sequence Alignment Program with the ClustalX version 2.0 software.22 The similarity search program, BLASTn, was used to identify the homologous sequences from the National Center of Biotechnology Information (NCBI) nucleotide database, confirming species-level similarity with the isolate query sequence. The percentage of replicate trees is shown, in which related taxa were grouped together in the bootstrap test along the branches. The phylogenetic tree was plotted at a scale using branch lengths measured in the same units, with evolutionary distances used for phylogenetic tree inference. Furthermore, evolutionary distances were computed using the maximum composite likelihood method23 and expressed in units of the base substitution number for each site. The selected secondary structures for each isolate were downloaded in the Vienna file format from the Mfold server.24
Statistical analyses
The results are expressed as the mean ± standard deviation. Data were analysed using one-way analysis of variance followed by a Student t-test with the IBM SPSS 20 software. Statistical significance was set at p ≤ 0.05. All analyses were conducted in triplicate.
Physico-chemical characterization of the samples
The studied samples were collected from the Al-Jouf region, especially from the cities of Dumat Al-Jandal and Al-Qurayyat, during the winter period between November 2020 and February 2021 (Table 1). One of the most significant factors is temperature, which influences the activity and evolution of microorganisms. The hot springs samples collected from Al-Qurayyat represent a thermophilic environment (40-45°C). However, the samples from Dumat Al-Jandal were collected in the winter period (November 2020), when the temperature was moderate (24–27°C). The pH of all studied samples varied between neutrophile and alkalophile as well as the electrical conductivity varied from 0.75 to 12.85 dS/m (Table 2). Mineral ion and salt concentrations influence soil electrical conductivity.25 Furthermore, the salts marshes soils were characterized by high EC varied between 10.54 in Al-Qurayyat and 12.85 dS/m in Dumat Al-Jandal, while the pH was slightly alkaline (7.56). In addition, all the hot springs water samples showed comparability in pH (7.75–8.02) and EC (1.32–2) confirming the low salinity of this water. The lowest EC was observed in desert soil samples.
Table (1):
Samples collection and references of isolated bacteria
| Sample location | Samples | References of isolated bacteria | |
|---|---|---|---|
| Number | Quality | ||
| Doumat Al -Jandal location | |||
| Lake | 2 | Water | W1; W2 |
| Agricultural soil | 3 | Soil | DS1-; DS1-2; DS2-1; DS2-2; DS3 |
| Salt marshes soil | 3 | Salt soil | SP1-1; SP1-2; SP2-1; SP2-2; SP2-3; SP2-4; SP3-1; SP3-2 |
| Desert soil | 3 | Hot soil | SS1; SS2; SS3-1; SS3-2 |
| Al- Qurayyat location | |||
| Salt marshes soil | 2 | Salt soil | WH1 |
| Ain Hawas | 2 | Hot soil | KWG1; KWG2; KWG3 |
| Ain Hawas | 3 | Hot springs | GWG1; GWG2; GWG3 |
| Ain Kaaf | 1 | Hot springs | SKS1 |
| Ain Garqar | 1 | Hot springs | SGS1 |
| Total | 20 | 28 | |
Table (2):
Physico-chemical characterization of collected samples
Sample |
Temperature (°C) |
pH |
EC (dS/m) |
|---|---|---|---|
Dumat al-Jandal city |
|||
Water lake |
24 |
7.60 |
3.42 |
Agricultural soil |
27 |
7.22 |
1.80 |
Salt marsh soil |
27 |
6.89 |
12.85 |
Desert soil |
27 |
7.06 |
0.75 |
Al –Qurayyat city |
|||
Hot springs water (Ain-Hwass) |
42 |
7.85 |
1.32 |
Hot springs water (Ain-Kaf) |
44 |
8.02 |
2.00 |
Hot springs water (Ain-GarGar) |
40 |
7.75 |
1.85 |
Salt marsh soil |
22 |
7.03 |
10.54 |
EC: Electrical conductivity.
Isolation of thermophilic bacteria
Twenty-eight thermophilic bacteria were isolated from 20 samples collected from various sites in the Al-Jouf region as illustrated in Table 1. The results revealed a relatively low biodiversity of thermophilic bacteria in the collected samples. This may be due to the increase in stress conditions in the original medium.26 Indeed, the negative effects of high salt content in salt marsh soils and low organic matter content in desert soils decrease the biodiversity of bacteria.27
Bacterial strain isolation focused on the morphological differences (macroscopic characterisation) of the isolates on NA media. All isolated bacteria were macroscopically, microscopically, and biochemically characterised.
Morphological and biochemical characterization of the isolated thermophilic bacteria
All thermophilic isolates were identified macroscopically, microscopically, and biochemically (Tables 3 and 4). The majority of the strains showed a white colour on the NA. Similar results were reported by Ifandi and Alwi26 and Masi et al.,28 who reported the dominant white and yellow colours of thermophilic bacteria. The majority had round colonies with an irregular form, elevated elevation, rough surface, and translucent opacity. The size of the colonies varied significantly depending on the bacterial species used. All isolated bacteria revealed a positive Gram stain with a rod shape, confirming Bacillus as the genus of all the isolated bacteria.
Table (3):
Macroscopic and microscopic identification of thermophilic bacteria
| Bacteria | Macroscopic observation | Microscopic observation | Identification | |||||
|---|---|---|---|---|---|---|---|---|
| Color | Shaped colony | Form | surface | Opacity | Form | Gram stain | ||
| W1 | white | Round | Irregular | rough | translucent | Rod | + | Bacillus sp. |
| W2 | white | Round | Irregular | Smooth | translucent | Rod | + | Bacillus sp. |
| DS1-1 | white | Round | regular | Smooth | Opaque | Rod | + | Bacillus sp. |
| DS1-1 | creamy | Round | regular | rough | translucent | Rod | + | Bacillus sp. |
| DS2-1 | white | Round | Round | rough | translucent | Rod | + | Bacillus sp. |
| DS2-2 | creamy | Round | regular | rough | translucent | Rod | + | Bacillus sp. |
| DS3 | white | Round | Irregular | Smooth | Opaque | Rod | + | Bacillus sp. |
| SP1-1 | white | Round | Irregular | rough | translucent | Rod | + | Bacillus sp. |
| SP1-2 | creamy | Round | regular | Smooth | Opaque | Rod | + | Bacillus sp. |
| SP2-1 | white | Round | Round | Smooth | Opaque | Rod | + | Bacillus sp. |
| SP2-2 | white | Round | regular | rough | translucent | Rod | + | Bacillus sp. |
| SP2-3 | white | Round | Round | rough | translucent | Rod | + | Bacillus sp. |
| SP2-4 | white | Round | regular | Smooth | translucent | Rod | + | Bacillus sp. |
| SP3-1 | white | Round | Irregular | Smooth | Translucent | Rod | + | Bacillus sp. |
| SP3-2 | creamy | Round | regular | rough | Translucent | Rod | + | Bacillus sp. |
| SS1 | white | Round | Irregular | Round | translucent | Rod | + | Bacillus sp. |
| SS2 | white | Round | regular | rough | Opaque | Rod | + | Bacillus sp. |
| SS3-1 | Creamy | Round | Irregular | rough | translucent | Rod | + | Bacillus sp. |
| SS3-2 | white | Round | regular | Round | Opaque | Rod | + | Bacillus sp. |
| WH1 | white | Round | Irregular | Smooth | translucent | Rod | + | Bacillus sp. |
| GWK1 | white | Round | Irregular | rough | translucent | Rod | + | Bacillus sp. |
| GWK2 | white | Round | regular | Smooth | Opaque | Rod | + | Bacillus sp. |
| GWK3 | white | Round | Irregular | rough | Opaque | Rod | + | Bacillus sp. |
| GWG1 | white | Round | Irregular | rough | translucent | Rod | + | Bacillus sp. |
| GWG2 | creamy | Round | regular | Smooth | Opaque | Rod | + | Bacillus sp. |
| GWG3 | White | Round | Irregular | Smooth | translucent | Rod | + | Bacillus sp. |
| SKS1 | Creamy | Round | regular | rough | Opaque | Rod | + | Bacillus sp. |
| SGS1 | white | Round | Irregular | Smooth | translucent | Rod | + | Bacillus sp. |
Table (4):
Biochemical identification of thermophilic bacteria
Bacteria |
Glucose |
Lactose |
Mannitol |
Citrate |
Motility |
H2S |
Gas |
|---|---|---|---|---|---|---|---|
W1 |
+ |
– |
+ |
+ |
+ |
– |
+ |
W2 |
+ |
– |
+ |
+ |
+ |
– |
– |
DS1-1 |
+ |
– |
+ |
+ |
+ |
– |
– |
DS1-2 |
+ |
– |
+ |
+ |
+ |
– |
– |
DS2-1 |
+ |
– |
+ |
+ |
+ |
– |
– |
DS2-2 |
+ |
– |
+ |
+ |
+ |
– |
– |
DS3 |
+ |
– |
+ |
+ |
+ |
– |
+ |
SP1-1 |
+ |
– |
+ |
+ |
+ |
– |
|
SP1-2 |
+ |
– |
+ |
+ |
+ |
– |
– |
SP2-1 |
+ |
– |
+ |
+ |
+ |
– |
– |
SP2-2 |
+ |
– |
+ |
+ |
+ |
– |
+ |
SP2-3 |
+ |
– |
+ |
+ |
+ |
– |
– |
SP2-4 |
+ |
– |
+ |
+ |
+ |
– |
– |
SP3-1 |
+ |
– |
+ |
+ |
+ |
– |
– |
SP3-2 |
+ |
– |
+ |
+ |
+ |
– |
– |
SS1 |
+ |
– |
+ |
+ |
+ |
– |
– |
SS2 |
+ |
– |
+ |
+ |
+ |
– |
– |
SS3-1 |
+ |
– |
+ |
+ |
+ |
– |
– |
SS3-2 |
+ |
– |
+ |
+ |
+ |
– |
– |
WH1 |
+ |
– |
+ |
+ |
+ |
– |
– |
GWK1 |
+ |
– |
+ |
+ |
+ |
– |
– |
GWK2 |
+ |
– |
+ |
+ |
+ |
– |
– |
GWK3 |
+ |
– |
+ |
+ |
+ |
– |
– |
GWG1 |
+ |
– |
+ |
+ |
+ |
– |
– |
GWG2 |
+ |
– |
+ |
+ |
+ |
– |
– |
GWG3 |
+ |
– |
+ |
+ |
+ |
– |
– |
SKS1 |
+ |
– |
+ |
+ |
+ |
– |
– |
SGS1 |
+ |
– |
+ |
+ |
+ |
– |
– |
+:positive enzyme activity; -: negative enzyme activity
The biochemical characteristics of the isolated thermophilic bacteria were determined by studying the ability of the strains to hydrolyse glucose, lactose, mannitol, and citrate substrates, and thus the production of H2S and gas, as well as the motility of the strains. The results showed that all isolated bacteria were glucose-, citrate-, and mannitol-positive but lactose- and H2S-negative. Notably, only three strains (W1, DS3, and SP2-2) produced gas, and all strains were motile.
Bacillus genus was isolated from all the studied thermophilic sites, which could explain its ability to move and be resistant to harsh environmental conditions.29 Different studies have found Bacillus dominance at all the thermophilic sites. Aanniz et al.30 revealed that approximately 98% of bacilli were recovered from hot springs in Morocco. Maugeri et al.31 isolated 87 thermophilic and spore-forming bacteria from the Aeolin Islands in Italy. Malkawi and Al-Omari32 and Abou-Shanab33 reported that Bacillus was the dominant bacterium in Jordanian hot springs. In addition, Khiyami et al.12 identified 15 thermoaerobic bacteria from the Jazan and Al-Lith geothermal springs, where Bacillus licheniformis, Bacillus cereus, Bacillus thermoamylovorans, Enterobacter sp, and Pseudomonas aeruginosa were the dominant strains.
Thermo-tolerance test of the isolated thermophilic bacteria
Thermo-tolerance of the isolated bacteria was screened in various temperatures from 45 to 75°C (Table 5). All isolated bacteria showed a high growth in 45, 50, and 55°C. However, this growth decreased slightly from 60 to 75°C (Table 5). Only 3 isolates could sustain the temperature of 75°C: SP1-1 and SP2-4 isolated from salt marshes soil of Dumat Al-Jandal and GWK1 was isolated from hot water springs in Ain-Kaf at Al-Qurayyat city. This could be attributed to the current temperature of the hot spring.
Table (5):
Thermophilic test of isolated bacteria
| Isolate | Temperature °C | |||||
|---|---|---|---|---|---|---|
| 45 | 50 | 55 | 60 | 65 | 75 | |
| W1 | +++ | +++ | +++ | ++ | – | – |
| W2 | +++ | +++ | +++ | ++ | + | – |
| DS1-1 | +++ | +++ | +++ | ++ | – | – |
| DS1-2 | +++ | +++ | ++ | + | – | – |
| DS2-1 | +++ | +++ | +++ | ++ | – | – |
| DS2-2 | +++ | +++ | ++ | ++ | + | – |
| DS3 | +++ | +++ | +++ | ++ | – | – |
| SP1-1 | +++ | +++ | +++ | +++ | ++ | + |
| SP1-2 | +++ | +++ | ++ | ++ | – | – |
| SP2-1 | +++ | +++ | + | + | – | – |
| SP2-2 | +++ | +++ | +++ | ++ | + | – |
| SP2-3 | +++ | +++ | +++ | – | – | – |
| SP2-4 | +++ | +++ | +++ | +++ | ++ | + |
| SP3-1 | +++ | +++ | ++ | ++ | + | – |
| SP3-2 | +++ | +++ | ++ | + | + | – |
| SS1 | +++ | +++ | ++ | ++ | + | – |
| SS2 | +++ | +++ | +++ | – | – | – |
| SS3-1 | +++ | +++ | +++ | – | – | – |
| SS3-2 | +++ | +++ | +++ | – | – | – |
| WH1 | +++ | +++ | + | – | – | – |
| GWK1 | +++ | +++ | +++ | +++ | ++ | + |
| GWK2 | +++ | +++ | +++ | + | + | – |
| GWK3 | +++ | +++ | ++ | + | – | – |
| GWG1 | +++ | +++ | ++ | + | – | – |
| GWG2 | +++ | +++ | ++ | – | – | – |
| GWG3 | +++ | +++ | +++ | + | + | – |
| SKS1 | +++ | +++ | +++ | + | – | – |
| SGS1 | +++ | +++ | +++ | + | – | – |
-: No growth; +: weak growth; ++: medium growth; +++: high growth
Panda et al.17 mentioned that the thermophilic bacteria from hot springs live and survive in temperatures around 60°C. The optimum growth temperature of three Bacillus strains isolated by Sarhan and Alamrri,13 from Jazan Hot Springs in Saudi Arabia were 45 and 50°C. In the same case, Ifandi and Alwi26 reported that the average temperature of Bora hot spring (Indonesia) is around 50°C, with pH that reaches 6.8–8.5.
The ability of thermophilic bacteria to grow at high temperatures is due to their higher G+C content than that of mesophilic organisms,34 especially their membrane lipid chemical stability.35 Sprott et al.36 reported that the growth temperature increases from 45 to 65°C decrease the diether lipids (archaeol based lipids) from 80 to 20% and increase the standard caldarchaeol-based and cyclic archaeol-based lipids increase from 10 to 40%.
The hot spring environment in Saudi Arabia has attracted the attention of several researchers. Kalil37 screened 30 isolates from hot springs in Jazan. All isolated showed a growth temperature ranging from 50 to 70°C.
As part of the same study, Sarhan and Alamrri13 isolated 28 thermophilic bacteria from Jazan hot water springs at optimal growth temperatures of 45–50°C and pH values of 6.4.
Production of extracellular thermostable enzymes
The bacteria isolated from different sites in the Al-Jouf region were screened for extracellular thermostable enzymes production: protease, lipase, amylase, cellulase, and gelatinase (Table 6). Among 28 isolates, 23 isolates produced protease (82%); 19 isolates produced amylase (68%), 13 isolates produced cellulase (46%), 8 isolates produced gelatinase (28%). No lipase activity was observed for any of the isolates. In addition, two isolates (7%) (SP1-1 and GWK1) combined the production of four extracellular enzymes, seven isolates (25%) produced three tested enzymes, 11 isolates (40%) produced two enzymes, seven bacteria (25%) produced one extracellular enzyme, and one bacterium (WH1) was inactive on all substrates (Table 6).
Table (6):
Enzymatic activity produced by isolated thermophilic bacteria
Protease |
Amylase |
Cellulase |
Gelatinase |
Lipase |
|
|---|---|---|---|---|---|
W1 |
+ |
– |
+ |
– |
– |
W2 |
+ |
+ |
+ |
– |
– |
DS1-1 |
+ |
– |
– |
+ |
– |
DS1-2 |
+ |
+ |
– |
– |
– |
DS2-1 |
+ |
– |
+ |
+ |
– |
DS2-2 |
+ |
– |
– |
– |
– |
DS3 |
+ |
+ |
+ |
– |
– |
SP1-1 |
+ |
+ |
+ |
+ |
– |
SP1-2 |
+ |
– |
+ |
– |
– |
SP2-1 |
+ |
+ |
– |
– |
– |
SP2-2 |
+ |
+ |
– |
+ |
– |
SP2-3 |
+ |
+ |
– |
– |
– |
SP2-4 |
+ |
+ |
+ |
– |
– |
SP3-1 |
+ |
+ |
– |
– |
– |
SP3-2 |
– |
+ |
+ |
– |
– |
SS1 |
+ |
– |
+ |
– |
– |
SS2 |
+ |
+ |
– |
+ |
– |
SS3-1 |
+ |
+ |
– |
+ |
– |
SS3-2 |
– |
+ |
– |
– |
– |
WH1 |
– |
– |
– |
– |
– |
GWK1 |
+ |
+ |
+ |
+ |
– |
GWK2 |
+ |
+ |
+ |
– |
– |
GWK3 |
– |
+ |
– |
+ |
– |
GWG1 |
+ |
+ |
+ |
– |
– |
GWG2 |
+ |
– |
– |
– |
– |
GWG3 |
+ |
+ |
+ |
– |
– |
SKS1 |
+ |
– |
– |
– |
– |
SGS1 |
– |
+ |
– |
– |
– |
+: enzymes production; – non enzymes production
Thermophilic bacteria are gaining industrial importance through the production of enzymes with a high potential to function under extreme conditions, such as pH, temperature, and pressure. This confirmed that these bacteria could have special genetic and physiological mechanisms capable of using different available carbon sources via the production of enzymes, and provide them with the ability to adapt and grow in severe conditions and uptake available nutrients.38
Thermostable enzymes, such as lipases, proteases, cellulases, and amylases, have important industrial value. Specifically, proteases constitute 60% of the global enzyme market and are used in various industries, including the food, detergent, leather, medical diagnostics, and cosmetic industries.39-41 Amylases, which are starch hydrolases, have been utilised in the manufacture of syrups containing high fructose, starch fermentation to ethanol, and starch-processing wastewater treatment.42 In addition, lipases are widely used in food, detergents, bio-resolution pharmaceuticals, the perfumery industry, cosmetics, agrochemicals, and bioremediation.43 Cellulases are also used to convert cellulosic materials into useful products.44
Extracellular enzyme production by thermophilic bacteria is of great interest to researchers. Thirteen thermophilic bacteria have been isolated from hot water springs in the Kingdom of Saudi Arabia.37 Biochemically, all the isolates were positive for lipase, 11 had amylase activity, and three were cellulase-positive. Benammar et al.45 studied thermophilic bacterial diversity in eight hot springs located in Algeria. The authors mentioned that 65.87% of the isolated strains presented five types of studied extracellular enzyme activities at least and 6.48% of strains combined all tested enzymes (pectinase, amylase, cellulase, lipase, esculinase, protease, gelatinase, nuclease, and lecithinase). The same authors showed that Aneurinibacillus, Aeribacillus, and Anoxybacillus exhibited high enzyme activity. Recently, Masi et al.28 isolated 102 thermophilic strains from the Addis Ababa landfill site (2023). Among the bacteria analysed, 35 were cellulases, 22 were amylases, 17 were proteases, and 9 were lipases.
Molecular identification of the isolated bacteria
Genomic DNAs of six thermophilic bacteria of Bacillus genus (W1, SP1-1, SP2-4, SS2, GWG3, and GWK1) were extracted, and their 16S rDNAs were subjected to PCR amplification. The products of PCR were separated via electrophoresis on 1% agarose gel. PCR showed six bands of the expected size of 16S rDNA (1485 bp; Figure 2).
Figure 2. Electrophoresis of 16S rDNA PCR products on 1% agarose gel of six isolated bacteria. A ~1485 bp DNA fragment amplified using 16S primer pair, L= DNA marker (1 kb), N= Negative control, P= Positive control, Lanes 1-4 represent PCR products of B. licheniformis isolates and Lanes 5-6 represent PCR products of B. subtilis isolates
Polymerase chain reaction (PCR) and sequencing techniques have been used to differentiate the isolated species. Nucleotide sequences were obtained and the size of the nuclear 16S rRNA genes ranged between 1414–1458 bp for the different isolated species. After sequencing, the 16S rDNA sequences obtained from the six thermophilic bacteria were blasted against the GenBank database. The accession numbers of all isolated strains are presented in Table 7.
Table (7):
Molecular identification and Accession number of six isolated thermophilic bacteria
Strain |
Identification |
Accession No. |
% ID |
Isolation |
|---|---|---|---|---|
W1 |
Bacillus licheniformis |
MZ419876 |
100% |
Lake water – Dumat Aljandal |
SS2 |
Bacillus licheniformis |
MZ419878 |
100% |
Desert soil – Dumat Al Jandal |
SP2-4 |
Bacillus licheniformis |
MZ419877 |
100% |
Salt marshes soil- Dumat Al Jandal |
GWG3 |
Bacillus licheniformis |
MZ419880 |
100% |
Hot srings – Ain Hawas |
GWK1 |
Bacillus subtilis |
MZ419881 |
100% |
Hot soil- Ain Hawas |
SP1-1 |
Bacillus subtilis |
MZ419879 |
100% |
Agricultural soil- Dumat Al Jandal |
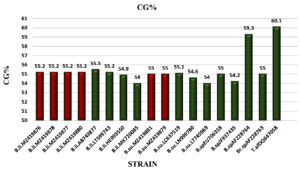
GC percentages of the Bacillus strains were compared to those of other bacterial strains (mesophilic and thermophilic) collected from GenBank. The obtained data showed a CG% of 55.2 for all B. licheniformis isolates and 55 for two B. subtilis isolates. However, the high CG contents of 59.3 and 60.1% was recorded respectively to hyperthermophilic bacteria collected from GenBank, Bacillus sp. (AF228764) and Thermosipho africanus (DQ647058; Figure 3). Panda et al.17 isolated thermophilic Bacillus sp. from hot spring of Tarabalo, Odisha, India, with GC content equal to 54.78%. In addition to the ability of thermophilic bacteria to survive at high temperatures, the authors speculated that high GC content may contribute to high survival rates. The same authors suggested that microorganisms isolated from soil samples had a higher GC content than those isolated from aquatic media. However, Wu et al.46 reported that a higher GC% is not the only reason for microorganisms to survive at extreme temperatures.
Figure 3. GC percentage of identified Bacillus strains and other strains collected from Genbank. The red bar indicates the GC content of our isolates
Phylogenetic analysis and identification of sequence clusters
Several phylogenetic analyses were conducted for the nuclear 16S rDNA sequence data. Phylogenetic analyses of the sequences of our isolates were aligned with sequences of high similarity from GenBank using the MUSCLE Program (ClustalX version 2.0) for distance, phylogenetic, and neighbour-joining analyses.
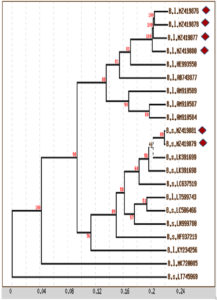
Complete gene sequences, including informative sites, were used for distance analysis. Distance analysis was performed for the 16S rRNA gene sequences of the six main species and 15 reference sequences collected from the NCBI database. The distance matrix and phylogenetic tree with bootstrap values (cluster algorithm) based on 16S rRNA gene sequence alignment of the isolated species revealed that closely related isolates of B. licheniformis were monophylogenetic, with an average distance of 0.020 and percent identity of 100. The same monophylogenetic relationship was recorded among B. subtilis isolates, with an average distance of 0.001 and percent identity of 100. In addition, the relationship between B. licheniformis and B. subtilis isolates was monophylogenetic, with an average distance of 0.097 and percent identity of 98 (Figure 4). The phylogeny of B. licheniformis isolates proved that all our isolates were closely related to each other but were polyphylogenetic to other B. licheniformis isolates (HE993550 and AB743877), with a percent identity of 81. Generally, isolates from the same location were monophylogenetic, whereas those from different locations were polyphylogenetic.
Figure 4. Phylogenetic tree with bootstrap values (Cluster Algorism) basing on 16S rRNA gene sequences alignment of isolated species (Diamond shapes represent our isolates and remaining are reference sequences; B.s.: Bacillus subtilis; B.l. Bacillus licheniformis)
A neighbour-joining phylogenetic tree was plotted at scale, with branch lengths in the same units measured as the evolutionary distances used for the inference of the phylogenetic tree (Figure 4). Based on the alignment of 16S rRNA gene sequences among the isolated species, the results were similar to those obtained via distance and phylogenetic tree analyses, which revealed a high similarity in the phylogenetic relationship between our isolated Bacillus species, where they were monophylogenetic, but other isolates were polyphylogenetic.
rRNA secondary structure prediction
Nuclear 16S rRNA, transcribed from 16S rDNA, is a single strand with many complementary (paired) sequence stems and single (unpaired) stranded loops. The stems, formed by complementary nucleotides, along with the resulting loops, form a unique structure referred to as the RNA secondary structure.
The stem and fold positions were recorded for each isolated species, and a distinct secondary structure was drawn using Mfold (Figure 5). The rRNA secondary structure indicated a degree of similarity between closely related isolates of B. licheniformis, and the structure elucidated the monophylogenetic relationships among them. The same monophylogenetic relationships were elucidated among B. subtilis isolates using RNA secondary structures. The isolates related to B. licheniformis were monophylogenetic, but polyphylogenetic to other isolates of B. subtilis.
To date, thermal ecosystems in the Al-Jouf region of Saudi Arabia have not been adequately studied from a microbiological perspective. Hot springs, salt marsh soil, and desert soil are good sources to isolate thermophilic organisms. Therefore, in this study, we successfully isolated 28 thermophilic bacteria from 20 hot samples of various locations, especially the Dumat Al-Jandal and Al-Qurayyat cities, in the Al-Jouf region. Morphological and biochemical analyses revealed Bacillus as the dominant genus in the collected samples. Notably, most isolated thermophilic strains showed high production of thermostable enzymes, mainly proteases, amylases, and cellulases. These enzymes exhibit a high potential for various industrial applications. Overall, this study suggests the potential biotechnological and bioprocess applications of thermostable enzymes isolated from thermophilic bacteria.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
SSA performed methodology and investigated the study. MSA performed molecular analysis. RJ supervised the study. SAS and RJ wrote the manuscript. RJ reviewed and edited the manuscript. All authors read and approved the final manuscript for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
This article does not contain any studies on human participants or animals performed by any of the authors.
- Pathak AP, Rathod MG, MahaboleMP, Khairnar RS. Enhanced catalytic activity of Bacillus aryabhattai P1 protease by modulation with nanoactivator. Heliyon 2020;6:Article e04053.
Crossref - Liu X, Kokare C. Microbial Enzymes of use in industry. In Biotechnology of Microbial Enzymes, Elsevier, 2017; 267–298.
Crossref - Esclapez-Espliego JM, Bautista-Saiz V, Torregrosa-Crespo J, Luque AV, Camacho-Carrasco ML, Pire C, Bonete MJ, Martínez-Espinosa RM. Extremophile Enzymes and Biotechnology. In R. Durvasula and D.S. Rao eds. Extremophiles: from Biology to Biotechnology. CRC Press, 2018: p. 227-242.
- Kumar S, Arumugam N, Permaul K, Singh S. Thermostable enzymes and their industrial applications. Chapter 5. In Microbial Biotechnology, CRC Press: Taylor and Francis Group, Boca Raton, FL 33487-2742, 2016;115–162.
Crossref - Wagner ID, Wiegel J. Diversity of Thermophilic Anaerobes. Ann NY Acad Sci. 2008;11:1-43.
Crossref - Taha GH, El-ShatouryEH, Tolba ST, Ibrahim MK. Potential enzyme activity of thermophilic bacteria from hot spring in Egypt. Afr J Biol Sci. 2020;16:197-206.
Crossref - Badhai J, Ghosh TS, Das SK. Taxonomic and functional characteristics of microbial communities and their correlation with physicochemical properties of four geothermal springs In Odisha, India. Front Microbiol. 2015;6:Article 1166.
Crossref - DumornéK, Córdova DC, Astorga-Eló M, Renganathan P. Extremozymes: a potential source for industrial applications. J Microbiol Biotechnol. 2017;27:649-659.
Crossref - Mohammad BT, Al DaghistaniHI, Jaouani A, Abdel-Latif S, Kennes C. Isolation and characterization of thermophilic bacteria from Jordanian hot springs: Bacillus licheniformis and Thermomonas hydrothermalis isolates as potential producers of thermostable enzymes. Internet J Microbiol. 2017;Article ID 6943952:1-12.
Crossref - El-Gayar K, Al Abboud M, Essa AMM, Characterization of Thermophilic Bacteria Isolated from two Hot Springs in Jazan, Saudi Arabia. J Pure App. Microbiol. 2017;11(2):1-9.
Crossref - Alrumman SA, Mostafa YS, Al-Qahtani STS, Sahlabji T, Taha TH. Antimicrobial activity and GC-MS analysis of bioactive constituents of thermophilic bacteria isolated from Saudi hot springs. Arab J Sci Eng. 2019;44:75–85.
Crossref - Khiyami MA, Serour EA, Shehata MM, Bahklia AH. Thermo-aerobic bacteria from geothermal springs in Saudi Arabia. Afr J Biotechnol. 2012;11:4053-4062.
Crossref - Sarhan MA, Alamri S. Characterization and identification of moderate thermophilic bacteria isolated from Jazan hot springs in Saudi Arabia. Egypt Acad J Biolo Sci. 2014;6:67-75.
Crossref - AlSediy K, Ashy R, Al-fassi F, and Al-Judaibi A. Microbial diversity and abundance in the hot springs on the west coast of Saudi Arabia as a potential source of novel industrial products. Microb Biosyst. 2022;7: 8-17.
Crossref - Japanese Standards Association JIS Handbook. Wastewater Treatment Japanese Standards Association Tokyo. 1995.
- Aslam M, Nasim FUH, Ruhi R, Murad H, Ejaz S, Choudhary MS, Mustafa G, Ashraf M, Rehman J. Isolation and Characteristics of Biotechnologically Important Antagonistic Thermophilic Bacteria from Rhizosphere of Haloxylon salicornicum. Pol J Microbiol. 2018;67:49–58.
Crossref - Panda MK, Sahu MK, Tayung K. Isolation and characterization of a thermophilic Bacillus sp. with protease activity isolated from hot spring of Tarabalo, Odisha, India. Iran J Microbiol. 2013;5:159–165. http://ijm.tums.ac.ir
- Yanmis D, Adiguzel A. Molecular typing of thermophilic bacilli isolated from different hot springs of Turkey. Res J Biotech. 2014;9:83–88.
- Kumar D, Kumar L, Nagar S, Raina C, Parshad R. Gupta VK. Screening, isolation and production of lipase/esterase producing Bacillus sp. strain DVL2 and its potential evaluation in esterification and resolution reactions. Arch Appl Sci Res. 2012;4:1763-1770. http://scholarsresearchlibrary.com/archive.html
- Forbes BA, Sahm DF, Weissfeld AS. Bailey and Scott’s diagnostic microbiology, Elsevier Mosby, Missouri, Mo, USA, 12 edition. 2007:p.1031.
- Umashankar V, Arunkumar V, Dorairaj S. ACUA: A software tool for automated codon usage analysis. Bioinformation 2007;2:62-63.
Crossref - Larkin MA, Blackshields G, Brown NP, Chenna R, Mcgettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics 2007;23:2947–2948.
Crossref - Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739.
Crossref - Meyer IM, Miklos I. SimulFold: Simultaneously inferring rna structures including pseudoknots, alignments, and trees using a Bayesian MCMC framework. PLoS Comput Biol. 2007;3:e149.
Crossref - Artiola JF, Walworth JL, Musil SA, Crimmins MA. Soil and land pollution, in: Environmental and Pollution Science, Academic Press, 2019, pp. 219–235.
- Ifandi S, Alwi M. Isolation of thermophilic bacteria from bora hot springs in central Sulawesi. Biosaintifika 2018;10:291-297
Crossref - Ventosa A, Nieto JJ, Oren A. Biology of moderately halophilic aerobic bacteria. Microbiol Mol Biol Rev. 1998;62:504-44.
Crossref - Masi C, Tebiso A, Kumar KVS. Isolation and characterization of potential multiple extracellular enzyme-producing bacteria from waste dumping area in Addis Ababa. Heliyon 2023;9:e12645.
Crossref - Connor N, Sikorski J, Rooney AP, Kopac S, Koeppel AF, Burger A, Cole SG, Perry EB, Krizanc D, Field NC. Ecology of speciation in the genus Bacillus. App Environ Microbiol. 2010;76: 1349–1358.
Crossref - Aanniz T, Ouadghiri M, Melloul M, Swings J, Elfahime E, Ibijbijen J, Ismaili M, Amar M. Thermophilic bacteria in Moroccan hot springs, salt marshes and desert soils. Braz J Microbiol. 2015;46(2):443–453.
Crossref - Maugeri TL, Gugliandolo C, Caccamo D, Stackebrandt E. A polyphasic taxonomic study of thermophilic bacilli from shallow, marine vents. Syst Appl Microbiol.2001;24:572–587.
Crossref - Malkawi HI, Al-Omari MN. Culture-Dependent and culture-independent approaches to study the bacterial and archaeal diversity from Jordanian hot springs. Afr J Microbiol Res. 2010;4:923–932. http://www.academicjournals.org/ajmr
- Abou-Shanab R. Characterization and 16S rDNA identification of thermo-tolerant bacteria isolated from hot springs. J Appl Sci Res. 2007;3:994–1000.
- Galtier N, Lobry JR. Relationships between Genomic G+C content, RNA secondary structures, and optimal growth temperature in Prokaryotes. J Mol Evol. 1997;44:632–636.
Crossref - Koga Y. Thermal adaptation of the archaeal and bacterial lipid membranes. Archaea 2012:Article ID 789652, 6p.
Crossref - Sprott GD, Meloche M, Richards JC. Proportions of diether, macrocyclic diether, and tetraether lipids in Methanococcus jannaschii grown at different temperatures. J Bacteriol. 1991;173:3907–3910.
Crossref - Khalil A. Screening and characterization of thermophilic bacteria (lipase cellulose and amylase producers) from hot springs in Saudi Arabia.J Food Agric Environ. 2011;9:672-675. www.world-food.net
- Baltaci MO, Genc B, Arslan S, Adiguzel G, Adiguzel A. Isolation and characterization of thermophilic bacteria from geothermal areas in Turkey and preliminary research on biotechnologically enzyme production. Geomicrobiol. 2017;34:53–62.
Crossref - Gupta R, Beg QK, Khan S, Chauhan B. An Overview on fermentation, downstream processing and properties of microbial alkaline proteases. App Microbiol Biotechnol. 2002;60:381–395.
Crossref - Laxman RS, Sonawane AP, More SV, Rao BS, Rele MV, Jogdand VV. Optimization and scale up of production of alkaline protease from Conidiobolus coronatus. Process Biochem. 2005;40:3152–3158.
Crossref - Yadav P, Korpole S, Prasad GS, Sahni G, Maharjan J, Sreerama L, Bhattarai T. Morphological, enzymatic screening, and phylogenetic analysis of thermophilic bacilli isolated from five hot springs of Myagdi, Nepal. J Appl Biol Biotechnol. 2018;6:1-8.
Crossref - Aiyer PV. Amylases and their applications. Afr J Biotechnol. 2005;4 (13):1525–1529. http://www.academicjournals.org/AJB
- Hasan F, Shah AA, Hameed A. Industrial applications of microbial lipases. Enzyme Microb Technol. 2006;39:235–251.
Crossref - Kuhad RC, Gupta R, Singh A. Microbial cellulases and their industrial applications. Enzyme Res. 2011;Article ID 280696:1-10.
Crossref - Benammar L, İnan Bektaş K, Menasria T, Beldüz AO, Güler HI, Bedaida IK, Gonzalez JM, Ayachi A. Diversity and enzymatic potential of thermophilic bacteria associated with terrestrial hot springs in Algeria. Braz J Microbiol. 2020;51:1987–2007.
Crossref - Wu H, Zhang Z, Hu S, Yu J. On the molecular mechanism of GC content variation among eubacterial genomes. Biol Direct 2012;7(2):1-16. http://www.biology-direct.com/content/7/1/2
© The Author(s) 2024. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.