ISSN: 0973-7510
E-ISSN: 2581-690X
Among patients admitted to the intensive care unit (ICU) the most common cause of infection is lower respiratory tract infections, which account for 4.4% of hospital admissions. Among the non-fermenters, Pseudomonas aeruginosa and Acinetobacter calcoaceticus-baumannii complex, cause many types of nosocomial infections. Ventilator-associated pneumonia is the most common emerging infection among them. The goal of this study was to isolate and speciate non-fermenting Gram-negative organisms from respiratory samples of ICU patients and to detect antimicrobial susceptibility patterns of the isolated non-fermenters. This cross-sectional study was conducted at the Institute of Microbiology in collaboration with Medical Intensive Care units in Rajiv Gandhi Government General Hospital. A total of 200 patients who satisfied the inclusion criteria were included in the study conducted between March 2019 and March 2020. Culture, sensitivity, and anti-microbial susceptibility tests were performed for the respiratory samples collected as per standard protocols. Pseudomonas aeruginosa (49%) was the most commonly isolated non-fermenter followed by Acinetobacter baumannii (24.3%). Among Pseudomonas aeruginosa isolates, the highest percentage were ESBL producers (44.4%). Carbapenem resistance among Pseudomonas isolates was 33%. The study showed increased isolation of MDR non-fermenters from the ICU causing Ventilator-associated pneumonia (VAP). To prevent VAP caused by these MDR pathogens, clinicians should follow strict infection control practices, use invasive devices on a short-term basis, and use antibiotics judiciously.
Non-fermenters, Ventilator Associated Pneumonia(VAP), Pseudomonas aeruginosa, Acinetobacter baumannii
Among patients admitted to the intensive care unit (ICU), the most common cause of infection is lower respiratory tract infections, which account for 4.4% of hospital admissions and occur in 10-25% of all intensive care patients, leading to high overall mortality (22.71%).1
Gram-negative followed by Gram-positive organisms are usually implicated in the etiologies of respiratory tract infections in ICU patients. Multi-drug resistant non-fermenting Gram-negative organisms1 are commonly the most notorious among them.
Non-fermenting Gram-negative organisms are ubiquitous, prefer moist environments, and in hospital ICUs can be isolated from nebulizers, dialysate fluids, saline, catheters, and other devices.2 Among the emerging infections wide variety of nosocomial infections such as bloodstream infections, ventilator-associated pneumonia (VAP), urinary tract infections, and wound infections1 are caused by non-fermenters such as Pseudomonas aeruginosa, Acinetobacter calcoaceticus-baumannii complex, and Stenotrophomonas maltophilia.
Pseudomonas aeruginosa is a common pathogen present in hospital ICUs involved in causing various life-threatening infections due to various innate and acquired drug resistance.3 Metallo beta-lactamase-producing Pseudomonas has been documented with increasing mortality rates in ICU patients.4 Beta lactamases which cause carbapenem hydrolysis, with elevated carbapenem MICs, belong to molecular classes A, B, and D.5 Acinetobacter species is an emerging opportunistic infection in ICUs. There is an increasing trend of aminoglycoside resistance and carbapenems resistance in nosocomial outbreaks.6,7
Pseudomonas resists a wide range of antipseudomonal agents8 and develops resistance via both intrinsic and extrinsic mechanisms. Intrinsic genes cause over-expression of efflux pumps (mexAB, mexCD, mexEF, and mexXY), development of chromosomal hyper-Amp C producers, and loss of porins (opr D). ESBL (blaSHV, blaTEM, blaVEB, blaPER, and blaOXA types) carbapenems (blaGES, blaKPC, blaIMP, blaSPM, blaVIM and blaNDM) are the extrinsic genes.
The mortality rate of Pseudomonas aeruginosa causing pneumonia is up to 70%.8 The present study focuses on the isolation, speciation, and drug susceptibility patterns of non-fermenting bacteria, with a specific emphasis on detecting multi-drug resistance in Pseudomonas species. This detection was achieved through both phenotypic and molecular detection methods, including PCR, to identify resistant genes in the isolated samples. This may emphasize the nosocomial spread of multi-drug-resistant non-fermenters and infection control practices in ICUs.
Aim and objectives
- To isolate and speciate non-fermenting Gram-negative organisms from respiratory samples of intensive care unit patients.
- To detect antimicrobial susceptibility patterns of isolated non-fermenters.
- To detect extended-spectrum beta-lactamases among isolated non-fermenters carbapenem resistance among Pseudomonas species by phenotypic methods.
- To confirm the metallo-beta lactamase (MBL) production by phenotypic and genotypic methods among Pseudomonas species.
This cross-sectional study was conducted at the Institute of Microbiology in collaboration with Medical Intensive Care units in Rajiv Gandhi Government General Hospital. A total of 200 patients who satisfied the inclusion criteria were included in the study conducted between March 2019 and March 2020. Patients aged 18 years and above who were admitted to medical ICUs with signs and symptoms of respiratory infections, underwent intervention procedures, and had a prolonged stay were included in the study. A standard questionnaire was used to obtain the demographic and clinical profile of the patients involved in the study.
Ethical consideration
The study (Ref – EC Reg. No. ECR/270/Inst/TN/2013) was approved by the Institutional ethical committee. The study proceeded after obtaining informed consent from the patients.
Sample collection, transport and processing
Respiratory samples such as sputum, endotracheal aspirates, tracheal aspirates, and bronchoalveolar lavage were collected and immediately transported according to the standard operating procedures to the microbiology laboratory. The collected respiratory samples were homogenized mechanically by vortexing for 1 min and direct Gram’ staining was done and examined for the presence of PMN cells, squamous cells, and bacteria.
Bacterial cultures
Respiratory samples such as sputum, endotracheal aspirates, and Bronchoalveolar lavage were processed under aseptic precautions as per standard microbiological techniques.
Interpretation of cultures
The interpretation was based on colony morphology, Gram stain, motility, and biochemical reactions. The non-fermenting Gram-negative bacilli were identified based on the catalase test, oxidase test, motility, nitrate reduction test, Hugh Leifson’s oxidative fermentative test, test for indole production methyl red test, Voges Proskauer test, citrate utilization test, urease test growth at variable temperature, acetamide test, gelatin liquefication test, 1% Sugar fermentation test, lysine decarboxylase test, ornithine decarboxylase test, and arginine dihydrolase test.
Antimicrobial susceptibility
Antimicrobial susceptibility was carried on Muller Hinton Agar as per CLSI guidelines 2020, using Kirby Bauer’s disc diffusion method and interpreted.
Detection of antimicrobial-resistant pattern
Phenotypic method
ESBL screening test using cefotaxime and ceftazidime discs, and carbapenem screening test using imipenem and meropenem were performed for the non-fermenters isolated from the respiratory samples. Positive screening tests for Pseudomonas species are subjected to confirmatory testing using the appropriate method outlined in CLSI 2020.
Extended spectrum β lactamase detection9-12
All the non-fermenters that were resistant to cefotaxime, or ceftazidime (screened) were tested for Extended spectrum of β-lactamases and confirmation was done using a cephalosporin/clavulanate combination disk.9
Detection of metallo β-lactamases production in pseudomonas species using the phenotypic method
Screening for MBL detection
Production of MBL by Pseudomonas isolates was confirmed using a combined disk test (Imipenem with Imipenem-EDTA). Those isolates which were resistant to imipenem or meropenem were subjected to the confirmatory test. Enhancement of the zone of inhibition of the imipenem-EDTA combination disc of ≥ 7mm when compared to the imipenem disc was interpreted as positive for MBL production.
Modified carbapenem inactivation method (mCIM)13
Rapid Carba NP TEST 14,15 procedure done and interpreted as per kit guidelines.
Colistin susceptibility for carbapenem-resistant isolates16,17
As per ICMR Standard Operating Procedures bacteriology,18 Colistin MIC was determined using the Micro broth dilution method. Isolates having MIC of ≤ 2μg/mL should be considered susceptible and > 2μg/mL considered resistant.
Molecular characterization
The detection of bla VIM and bla NDM genes in Pseudomonas aeruginosa which were carbapenem-resistant and MBL-producing was done using conventional polymerase chain reaction (PCR).
Among the 200 isolates, non-fermenters were 18.5% and other isolates were 21.5%. The remaining 60% were culture-negative samples. Maximum isolates were in the age groups of 39-48 years (35.1%) and 19-28 years (16.2%).
Pseudomonas aeruginosa (49%) was the predominant isolate among non-fermenters (Figure 1).
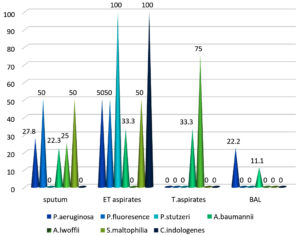
Sample distribution of isolated non-fermenters were predominantly from endotracheal aspirates (46%), followed by sputum (27%), Tracheal aspirates (5%) and Bronchoalveolar lavage (5%) (Figure 2).
ICU stay 59.4% was most common risk factor followed by mechanical ventilation 43.2%, Diabetes (27.0%), Prolonged antibiotic therapy & hypertension (21.6%), COPD (16.21%), Tracheostomy (10.8) and renal disease (5.4%) (Table 1).
Table (1):
Associated risk factors with isolated non-fermenters (n=37)
Risk Factors |
Isolates |
Percentage |
|---|---|---|
ICU stay |
22 |
59.4% |
Mechanical ventilation |
16 |
43.2% |
Diabetes |
10 |
27.0% |
Prolonged antibiotic therapy |
8 |
21.6% |
Hypertension |
8 |
21.6% |
COPD |
6 |
16.21% |
Tracheostomy |
4 |
10.8% |
Renal disease |
2 |
5.40% |
Polymyxin B showed 100% sensitivity, while meropenem also showed 100% sensitivity against P. fluorescence and stutzeri and 61.1% sensitivity to P. aeruginosa. Piperacillin tazobactam showed 66.6% sensitivity to P. aeruginosa and A. baumannii. Gentamicin (33.3%) had the lowest susceptibility among all isolates (Table 2).
Table (2):
Antimicrobial susceptibility pattern of non-fermenters
| Antibiotics | P. aeruginosa (n=18) | P. fluorescence (n=2) | P. stutzeri (n=1) | A. baumannii (n= 9) | A. lwoffii (n=4) | S. maltophilia (n=2) | C. indologenes (n=1) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S | % | S | % | S | % | S | % | S | % | S | % | S | % | |
| Gentamicin | 6 | 33.3 | 1 | 50 | 1 | 100 | 3 | 33.3 | 1 | 25 | – | – | 1 | 100 |
| Amikacin | 8 | 44.4 | 2 | 100 | 1 | 100 | 5 | 55.5 | 2 | 50 | – | – | 0 | 0 |
| Ciprofloxacin | 8 | 44.4 | 1 | 50 | 0 | 0 | 3 | 33.3 | 1 | 25 | – | – | 1 | 100 |
| Cotrimoxazole | – | – | – | – | – | – | 4 | 44.4 | 3 | 75 | 2 | 100 | 1 | 100 |
| Ceftazidime | 8 | 44.4 | 1 | 50 | 1 | 100 | 4 | 44.4 | 1 | 25 | – | – | 1 | 100 |
| Levofloxacin | 12 | 66.6 | 2 | 100 | 1 | 100 | 6 | 66.6 | 3 | 75 | 2 | 100 | – | – |
| Piperacillin tazobactam | 12 | 66.6 | 2 | 100 | 1 | 100 | 6 | 66.6 | 4 | 100 | – | – | 1 | 100 |
| Meropenem | 11 | 61.1 | 2 | 100 | 1 | 100 | 5 | 55.5 | 3 | 75 | – | – | 0 | 0 |
| Tetracycline | – | – | – | – | – | – | 5 | 55.5 | 2 | 50 | – | – | – | – |
| Polymyxin B | 18 | 100 | 2 | 100 | 1 | 100 | 9 | 100 | 4 | 100 | 2 | 100 | – | – |
Out of 21 Pseudomonas spp isolated 7(33.3%) were MBL producers. As per one proportion Z test, P value was less than 0.0001 (Table 3).
Table (3):
MBL producers among Pseudomonas spp.(N=21)
Organisms |
MBL Producers |
One Proportion Z Test |
|---|---|---|
Pseudomonas aeruginosa |
7(33.3%) |
5.950 |
Pseudomonas fluorescence |
– |
– |
Pseudomonas stutzeri |
– |
– |
Total |
7(33.3%) |
Presence of resistant genes VIM and NDM (Metallo beta-lactamase genes) for resistant isolated was tested by PCR. 3 were positive for VIM (42.8%) and 2 were positive for NDM (28.5%) production among 7 tested resistant isolates (Table 4).
Table (4):
Resistance gene distribution in pseudomonas aeruginosa (n=7)
Gene Tested |
Positive |
Percentage |
|---|---|---|
bla VIM |
3 |
42.8% |
bla NDM |
2 |
28.5% |
TOTAL |
5 |
71.1% |
Among different methods of MBL detection, Combined disc test was 100% sensitive and modified Carbapenemases inhibitor method was 71.5% sensitive and RAPID Carba NP was 85.7% sensitive and PCR was positive in 71.5% isolates (Table 5).
Table (5):
Comparison of different methods for MBL detection
| Organism | No of isolates | CDT | mCIM | Rapid Carba NP | PCR | ||||
|---|---|---|---|---|---|---|---|---|---|
| +ve | -ve | +ve | -ve | +ve | -ve | +ve | -ve | ||
| Pseudomonas aeruginosa | 7 | 7 | – | 5 | 2 | 6 | 1 | 5 | 2 |
| Percentage | 100% | – | 71.5% | 28.5% | 85.7% | 14.3% | 71.5% | 28.5% | |
Non-fermenting respiratory pathogens have exhibited a dramatic increase in antimicrobial resistance due to prophylactic antibacterial therapy. Saad et al.19 also identified prolonged ICU stay and mechanical ventilation as significant risk factors.
Predominant isolates among the non-fermenters were Pseudomonas aeruginosa (49%) followed by Acinetobacter baumannii (24.3%), Acinetobacter lwoffii (10.8%), Pseudomonas fluorescence (5.4%), Stenotrophomonas maltophilia (5.4%), Pseudomonas stutzeri (2.7%) and Cryseobacterium indologenes (2.7%). Pseudomonas aeruginosa (56.7%), followed by Acinetobacter species (39.3%) were the predominant isolates (Figure 1) reported in the study by Chawla et al., which correlates with the current study.20
The present study showed that the sample distribution of isolated non-fermenters was predominantly from endotracheal aspirates (46%), which correlates with the study done by Vasundhra Devi P et al., wherein non-fermenters were predominantly isolated from endotracheal aspirates (55%). Higher resistance was observed in ICU patients, mainly due to device-associated infections in the hospital environment. These pathogens are often highly resistant to hospital disinfectants and can easily spread from patient to patient.1
Pseudomonas stutzeri exhibited 100% sensitivity to gentamicin, amikacin, ciprofloxacin, ceftazidime, piperacillin tazobactam, meropenem, and polymyxin B. Pseudomonas fluorescence exhibited 100% sensitivity to amikacin, piperacillin tazobactam, and meropenem, whereas gentamicin, ciprofloxacin, and ceftazidime exhibited 50% sensitivity pattern. This is indifferent to the study done by Malini et al.21 where it was 95% sensitive to imipenem, only 9 % sensitive to piperacillin, 47% sensitive to amikacin, and 14% to ceftazidime.
Acinetobacter lwoffii showed various sensitivities, including 100% sensitivity to piperacillin tazobactam and polymyxin B, 75% to meropenem and cotrimoxazole, and 50% to amikacin and tetracycline. Its sensitivity was lower at 25% for gentamicin, ciprofloxacin, and ceftazidime, consistent with the findings by Malini et al.21 In their study, amikacin, cotrimoxazole, and ciprofloxacin had 60% sensitivity, imipenem exhibited 100% sensitivity. Stenotrophomonas maltophilia exhibited 100% sensitivity to cotrimoxazole, levofloxacin, and minocycline, consistent with the findings of Malini et al.
Knowledge about the prevalence of ESBL is essential in initiating appropriate therapy. In this study, among the ESBL-producing non-fermenters (n=37), Pseudomonas aeruginosa is the highest ESBL producer (44.4%) followed by baumannii (33.3%), Acinetobacter lwoffii (50%), and Pseudomonas fluorescence (50%). There is no ESBL production by Stenotrophomonas maltophilia, Pseudomonas stutzeri, and Cryseobacterium indologenes. Amirth Koirala et al.,22 also reported that out of 158 (52%) confirmed ESBL producers, 46% were Pseudomonas aeruginosa and 63.3% were Acinetobacter spp.
In this study, the detection of meropenem-resistant Pseudomonas spp (n=21) showed sensitivity to 14 (66.7%) isolates and resistance to 7 (33.3%) isolates, which is analogous to the analysis done by Kali et al.,23 wherein imipenem-resistant isolates were 11 (22.4%) according to their study. Choudry et al.24 reported multi-drug resistant and MBL-producing Pseudomonas aeruginosa to be 44.4% which was higher compared to the present study.
Additionally, carbapenem resistance in Pseudomonas aeruginosa among respiratory isolates of ICU patients in our hospital was determined. We phenotypically characterized the resistance mechanisms and also evaluated the in vitro activity of colistin against the isolates. A susceptibility of 100% to colistin was observed among MBL-producing Pseudomonas aeruginosa (n=7). This did not correlate with the study done by Mohanty et al.25 in India, who reported an 8% colistin resistance in Pseudomonas aeruginosa.
Colistin resistance variation may be due to different antibiotic policies among different hospitals and geographic variations. One isolate of MBL-producing Pseudomonas aeruginosa showed MIC of 2, which indicates that colistin should be mandatorily tested for MIC before administration to patients which will prevent the emergence of colistin resistance in a community. This is of utmost importance because the treatment of carbapenem-resistant infections relies on colistin, which is the only available effective antibiotic.
Molecular detection of MBL-producing Pseudomonas aeruginosa revealed that among the 7 isolates, 3 tested positive for bla VIM, and 2 tested positive for bla NDM. This finding correlates with the study conducted by Mohanam & Menon,26 where, out of 20 isolates, 7 carried the bla VIM gene, and 6 carried the bla NDM gene. The primary mechanism of resistance among Pseudomonas aeruginosa27 is due to the loss of OprD porin expression, without the expression of a carbapenemase.
Metallo β-lactamase-producing non-fermenters were phenotypically detected using a combined disc test (CDT), modified carbapenem inactivation method (mCIM), and Rapid Carba NP test. Among different methods, the combined disc test was 100% sensitive, modified Carbapenem inactivation method (mCIM) was 71.5% sensitive, RAPID Carba NP was 85.7% sensitive, and PCR was positive in 71.5% of isolates. These findings are consistent with Behra et al.,28 where CDT showed 100% sensitivity. However, in a study conducted by Huang et al.29 Rapid carba NP showed 100% sensitivity, which contrasts with the results of this study.
Pseudomonas species showed maximum sensitivity to polymyxin B and colistin. The use of these antibiotics must be strictly restricted to critically ill ICU patients with severe infections, to avoid rapid emergence of resistance.
Rapid Carba NP test proves to be an effective screening test in addition to the phenotypic test with 85% sensitivity and a turnaround time of 30 minutes to two hours. Although the gold standard for carbapenamase detection is the genotypic method, the costs associated with these tests in a resource-poor laboratory limit its use. Rapid Carba NP, mCIM, and combined disc test were the most satisfactory simple, easy, rapid, and reliable methods among all the phenotypic methods for detecting carbapenemase-producing non-fermenting GNB.
Carbapenem resistance is considered an emerging drug-resistant mechanism in the NFGNB although not highly reported. Rapid intra-institutional spread of such resistant strains is common and hence must be notified to infection control teams. Antibiotic therapy, whether empirical or documented, should be based on a combination of antibiotics, taking into consideration the local epidemiology of sensitivity patterns to choose an appropriate combination.
The study revealed that infections spread by non-fermenters represent a considerable health problem in ICUs in tertiary care hospitals. Their capacity to survive in a hospital environment underscores the necessity for accurate species-level identification and the detection of resistant strains to control their spread.
The most common organisms isolated were Pseudomonas and Acinetobacter. The infections caused by multi-drug-resistant β-lactamases producing Pseudomonas are capable of causing significant morbidity and mortality.
The present study showed increased isolation of MDR non-fermenters from ICUs causing ventilator-associated pneumonia. VAP caused by these MDR pathogens can be prevented by using invasive devices for a short-term, judicious use of antibiotics, and by following strict infection control practices.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
This study was approved by the Institutional Ethical Committee, Madras Medical College, Chennai, India, with reference number ECR/270/Inst/ TN/2013).
INFORMED CONSENT
Written informed consent was obtained from the participants before enrolling in the study.
- Wadhwa R, Sharma Y, Upadhyay RP, Bala K. Nosocomial infection by non-fermenting gram negative bacilli in tertiary care hospital: screening and cure. Int J Pharm Pharm Sci. 2016;8(3):274-277.
- Connie R. Mahon, Donald C. Lehman Text book of diagnostic Microbiology 6th edition.
- Pachori P, Gothalwal R, Gandhi P. Emergence of antibiotic resistance Pseudomonas aeruginosa in intensive care unit; a critical review. Genes Dis. 2019;6(2):109-119.
Crossref - Qu TT, Zhang JL, Wang J, et al. Evaluation of phenotypic tests for detection of metallo-beta-lactamase-producing Pseudomonas aeruginosa strains in China. J Clin Microbiol. 2009;47(4):1136-1142.
Crossref - Ciocan O-A, Carare MC, Cozma A-P, et al. Isolation and identification of Pseudomonas aeruginosa strains producing b-lactamases (ESBL) and carbapenemases (MBL) of human origin. 2015.
Crossref - Nwadike VU, Ojide CK, Kalu EI. Multidrug resistant acinetobacter infection and their antimicrobial susceptibility pattern in a nigerian tertiary hospital ICU. Afr J Infect Dis. 2014;8(1):14-8.
Crossref - Parandekar PK, Peerapur BV. Non-Fermenters in Human Infections with Special Reference to Acinetobacter Species in a Tertiary Care Hospital from North Karnataka, India. JKIMSU. 2012;1(1):84-88.
- Pragasam AK, Vijayakumar S, Bakthavatchalam YD, et al. Molecular characterisation of antimicrobial resistance in Pseudomonas aeruginosa and Acinetobacter baumannii during 2014 and 2015 collected across India. Indian J Med Microbiol. 2016;34(4):433-441.
Crossref - Agatha D, Subitha B. Detection of Extended Spectrum â Lactamase and Amp C b-Lactamase Resistance in the Gram Negative Bacterial Isolates of Ventilator Associated Pneumonia. Int J Curr Microbiol App Sci. 2019;8(2):1139-1145.
Crossref - Gupta R, Malik A, Rizvi M, Ahmed M. Presence of metallo-beta-lactamases (MBL), extended-spectrum beta-lactamase (ESBL) &AmpC positive non-fermenting Gram-negative bacilli among Intensive Care Unit patients with special reference to molecular detection of blaCTX-M & blaAmpC genes. Indian J Med Res. 2016;144(2):271-275.
Crossref - Garg R, Gupta V, Chander J, Kaur M. Report of carbapenem resistant Ps. aeruginosa, isolates carrying ESBLs, AmpC and MBL enzymes based on phenotypic methodology and susceptibility to Fosfomycin. Indian J Med Microbiol. 2015;33(Suppl 1):160-161.
Crossref - Deshmukh DG, Damle AS, Bajaj JK, Bhakre JB, Patwardhan NS. Metallo-b-lactamase-producing clinical isolates from patients of a tertiary care hospital. J Lab Physicians. 2011;3(2):93-97.
Crossref - Poirel L, Nordmann P. Rapidec Carba NP Test for Rapid Detection of Carbapenemase Producers. J Clin Microbiol. 2015;53(9):3003-3008.
Crossref - Tijet N, Boyd D, Patel SN, Mulvey MR, Melano RG. Evaluation of the Carba NP test for rapid detection of carbapenemase-producing Enterobacteriaceae and Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2013;57(9):4578-4580.
Crossref - Campana EH, Chuster SG, da Silva IR, et al. Modified Carba NP test for the detection of carbapenemase production in gram-negative rods: optimized handling of multiple samples. Braz J Microbiol. 2017;48(2):242-245.
Crossref - Carretto E, Brovarone F, Russello G, et al. Clinical Validation of Sensi Test Colistin, a Broth Microdilution-Based Method To Evaluate Colistin MICs. J Clin Microbiol. 2018;56(4):e01523-17.
Crossref - Manohar P, Shanthini T, Ayyanar R, et al. The distribution of carbapenem- and colistin-resistance in Gram-negative bacteria from the Tamil Nadu region in India. J Med Microbiol. 2017;66(7):874-883.
Crossref - Indian Council of Medical Research, Standard operating procedure Bacteriology-Antimicrobial resistance surveillance and research network 2nd Edition 2019.
- Nseir S, Deplanque X, Di Pompeo C, Diarra M, Roussel-Delvallez M, Durocher A. Risk factors for relapse of ventilator-associated pneumonia related to nonfermenting Gram negative bacilli: a case-control study. J Infect. 2008;56(5):319-325.
Crossref - Chawla K, Vishwanath S, Munim FC. Nonfermenting Gram-negative Bacilli other than Pseudomonas aeruginosa and Acinetobacter Spp. Causing Respiratory Tract Infections in a Tertiary Care Center. J Glob Infect Dis. 2013;5(4):144-8.
Crossref - Bougle A, Foucrier A, Dupont H, et al. Impact of the duration of antibiotics on clinical events in patients with Pseudomonas aeruginosa ventilator-associated pneumonia: study protocol for a randomized controlled study. Trials. 2017 23;18(1):37.
Crossref - Amrit K, Agrahari G, Dahal N, Ghimire P, Rijal K. ESBL and MBL mediated resistance in clinical isolates of non-fermenting gram-negative bacilli (NFGNB) in Nepal. 2017:18-24.
- Monicacheesbrough. District Laboratory Practice in Tropical Countries, Part-1 Second Edition:76- 85
- Choudhary V, Pal N, Hooja S. Prevalence and antibiotic resistance pattern of Metallo-b-lactamase-producing Pseudomonas aeruginosa isolates from clinical specimens in a tertiary care hospital. J Mahatma Gandhi Inst Med Sci. 2019;24:19-22.
Crossref - Mohanty S, Maurya V, Gaind R, Deb M. Phenotypic characterization and colistin susceptibilities of carbapenem-resistant of Pseudomonas aeruginosa and Acinetobacter spp. J Infect Dev Ctries. 2013;7(11):880-887.
Crossref - Mohanam L, Menon T. Coexistence of metallo-beta-lactamase-encoding genes in Pseudomonas aeruginosa. Indian J Med Res. 2017;146(Supplement):S46-S52.
Crossref - Queenan AM, Bush K. Carbapenemases: the versatile beta-lactamases. Clin Microbiol Rev. 2007;20(3):440-58.
Crossref - Sachdeva R, Sharma B, Sharma R. Evaluation of different phenotypic tests for detection of metallo-b-lactamases in imipenem-resistant Pseudomonas aeruginosa. J Lab Physicians. 2017;9(4):249-253.
Crossref - Morris AJ, Tanner DC, Reller LB. Rejection criteria for endotracheal aspirates from adults. J Clin Microbiol. 1993;31(5):1027-9.
Crossref
© The Author(s) 2023. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.