Cephalosporins are the β-lactam antibiotics for the treatment of infection diseases caused by gram-positive and gram-negative bacteria. Fungus Cephalosporium acremonium produces cephalosporin C (CPC) that is not potent for clinical use. Its molecule can be transformed to 7-amino cephalosporanic acid (7-ACA) as the intermediate compound for making semi-synthetic cephalosporin derivatives. The first method for production of 7-aminocephalosporanic acid involves the chemical reaction using toxic reagents and laborious procedures. The second method uses two-step and one-step enzymatic transformation. In two-step method, the first step involves the conversion of Ceph-C to Glutaryl-7-amino cephalosporanic acid (GL-7-ACA) by D-amino acid oxidase (DAAO) and the second step, GL-7-ACA acylase (GA) hydrolyzes GL-7- ACA to produce 7-ACA. The two-step enzymatic transformation was used widely because of its safety and environmental friendship. The one-step enzymatic transformation is developed because of process simplification and cost reduction. This method uses cephalosporin C acylase for transformation CPC to 7-ACA. Same microbes can produce cephalosporin C acylase such as Achromobacter xylosoxidans, Aeromonas sp., Arthrobacter viscosus, Bacillus laterosporus, Flavobacterium sp., Paecilomyces sp., and Pseudomonas sp. The natural of cephalosporin C acylase catalyzed the CPC to 7-ACA directly in a very low efficiency and the protein engineering of cephalosporin C acylase was used to increase activity.
Cephalosporin C, 7-aminocephalosporanic acid, D-amino acid oxidase, GL-7-ACA acylase, cephalosporin C acylase, Cephalosporium acremonium.
Cephalosporin C (CPC) was the second β-lactam antibiotics to be discovered after penicillin from the fungus Cephalosporium acremonium by Giuseppe Brotzu1,2. CPC showed a moderate antibacterial activity: minimum inhibitory concentration values were in the range of 25–100 and 12– 25 µg/mL for gram-positive and gram-negative bacteria, respectively2 and is not potent for clinical use3. Robert Morin discovered a process by which the D-a- amino adipoyl side chain of CPC could be removed, generating 7-aminocephalosporanic acid (7-ACA) in 19622. 7-ACA is the intermediate for semi-synthetic cephalosporin derivates. Some semi-synthetic cephalosporin such as ceftezole, cefotiam, cefazedone, and cefazolin were produced by 7-ACA4.
The cephalosporin antibiotics are divided into two groups; the first is derived from penicillin (G or V) and the second from CPC. The most products that have intrinsic oral activity are better made from penicillin, while the most products made from CPC are insufficiently absorbed from the human gut to be therapeutically adequate by the oral route, unless converted to pro-drugs by esterification5.The cephalosporins are clinically active against gram-positive as well as gram-negative organisms. Cephalosporins are broad-spectrum, less toxic and resistant to degradation of β-lactamase as compared to penicillins;1,3
Conversion CPC to 7-ACA
Cephalosporins derivates are semi-synthetic products that derive from the fermentative product CPC. CPC is initially converted to 7-ACA by either a chemical or an enzymatic catalysis removal of the 7-amino adipoyl side chain. The chemical catalysis was developed by Morin et al. and this process is used to 7-ACA production in an industrial scale (Scheme 1)5. Enzymatic transformation of CPC to 7-ACA is the best alternative and has industrial significance.3
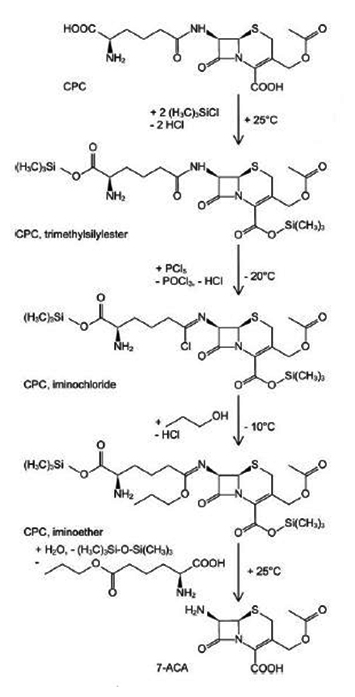 Scheme 1. Reaction scheme of chemical cleavage of CPC into 7-ACA5
Scheme 1. Reaction scheme of chemical cleavage of CPC into 7-ACA5The chemical method has been replaced by enzymatic method for producing 7-ACA because of (1) safety and environmental contamination: dangerous and toxic reagents (i.e., trimethylchlorosilane, phosphorus pentachloride, dichloromethane, dimethylaniline, etc.) are absent in the enzymatic transformation;1,2 (2) selectivity: owing to enzyme selectivity, the use of permanent or temporarily protective groups is avoided;2 (3) energy consumption: the chemical process uses low temperatures and exothermic steps, while the enzymatic approach is carried out at a fixed temperature of 20–30°C;2 (4) equipment: owing to the use of aggressive reagents, the chemical process requires glass-lined vessels and dedicated and expensive equipment to transport and charge these materials, to trap spilling or vapors, etc.;2 and (5) quality: in the enzymatic 7-ACA preparations, the levels of oligomers, 7-desacetoxycephalosporanic acid, and desacetyl- 7-ACA (or corresponding lacton) are considerably lower—the typical assay is 3–6 % higher than for the chemical process2.
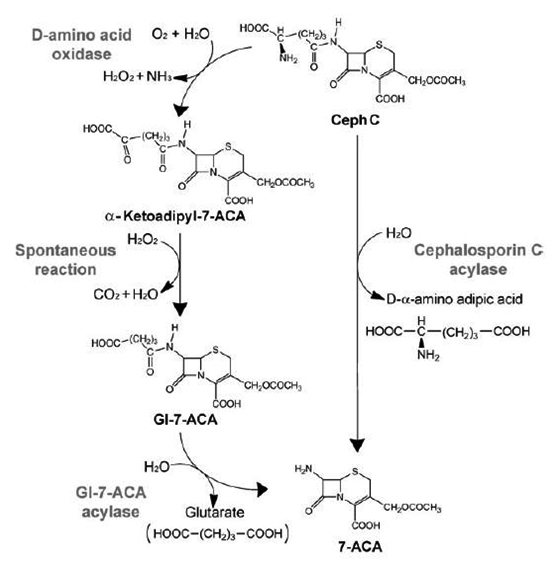
The biocatalytic conversion for CPC into 7-ACA, the key intermediate to cephalosporin derivates was developed5. CPC can be transformed by two-step enzymatic method by D-amino acid oxidase (DAAO) and glutaryl acylase (GAC) or One-step enzymatic method by a CPC acylase (CA).1,2,5,7,8
The most widely used enzymatic approach is represented by a two-step enzymatic method that employs DAAO (D-amino acid oxidase) and Gl-7-ACA acylase (Scheme 2)5. The flavoenzyme D-amino acid oxidase (DAAO; EC 1.4.3.3) catalyzes the oxidative deamination of neutral and basic D-amino acids but not acid D-amino acid, which are subtrates of D-aspartate oxidase9. D-Amino acid oxidases are rather frequent in nature; they are present in mammalian organs, mainly in the kidneys. Different microbes too are known to produce DAAO, such as the yeasts Candida tropicalis5, Rhodotorula gracillis,5 and Trigonopsis variabilis,5 the fungi Fusarum oxysporum5, Fusarium solani5, Neurospora crassa,5 Penicillium chrysogenum5 and Rhodosporidium sp.;5 prokaryotic microbial Arthrobacter protophormiae,5 Rubrobacter xylanophilus9, and Streptomyces coelicolor5. Only two enzymes, namely DAAO from Rhodotorula gracilis and from Trigonopsis variabilis, have been developed into an industrial biocatalyst.5
GAs (glutaryl acylases; EC 3.5.1.93) have been classified into five classes on the basis of their sequence, molecular mass and enzymatic properties. Members of each class are very similar on the basis of substrate specificity and sequence conservation (they share >90%of nucleic acid or amino acid sequence identity). Members of class I (e.g. P130 from Pseudomonas sp. 130) and class III (e.g. N176 from Pseudomonas sp. N176) show the highest activity to CPC (4%relative to Gl-7ACA)10.
Glutaryl acylase (GA) is a metal free heterodimer of 16 kDa+54 kDa subunits without any prosthetic group. This enzyme has been isolated from many organisms, but only the one from Pseudomonas diminuta was cloned and expressed in a recombinant E. coli and has been developed into an industrial biocatalyst.5 GA activity has been reported in bacteria from Aeromonas2, Arthrobacter2, Bacillus2,and Flavobacterium2, Pseudomonas,2,5 and from the fungus Paceilomyces.2,3
The alternative enzymatic approach is a one-step enzymatic method process in which CPC is converted directly into 7-ACA by a CPC acylase (CA; Scheme 2). As shown in Scheme 2, acylases can cleave the amide bond through which the cephem nucleus is linked to the acyl side chain2. Cephalosporin acylase (CA, EC 3.5.1.11) is a member of the N-terminal nucleophile (Ntn) hydrolase superfamily, in which the precursor gene is translated into a single polypeptide chain and then folded into a self-activating pre-protein activated by intra-molecular double cleavages. The first cleavage is most important for generating the active center catalysis residue (Ser) of the enzyme at the N terminus of the b-subunit. The second cleavage is also essential for activating the enzyme, forming a mature heterodimer with the a- and b- subunits and releasing a spacer peptide of different lengths (8–11 aa) in varying CAs. According to the diversity in substrate specificity, CAs can be divided into two classes, the cephalosporin C (CPC) acylase and the glutaryl-7-aminocephalosporanic acid (GL-7-ACA) acylase classes. Both enzymes classes form 7-aminocephalosporanic acid (7-ACA).11
Sonawane (2006) classified CA base on their substrate (CPC, GL-7-ACA, and GL-7-ADCA) specificity. The types of cephalosporin acylase are: type 1 is true cephalosporin C acylase. CPC acylase can hydrplyze only CPC and is not active for GL-7-ACA; type 2 GL-7-ACA acylase. This enzymes can hydrolyze GL-7-ACA to 7-ACA in high yield but have less activity against CPC; type 3 GL-7-ADCA acylase. Their enzymes have activity against GL-7-ADCA but their activity against GL-7-ACA is not known.12
Recently, the CPC acylase is very attractive at industrial level because it possesses very simple with reducing cost, although its catalytic performance, including its activity and product inhibition resistance, is not yet as satisfactory as that of the GL-7-ACA acylase2,11. Some studies focused on identifying more active enzymes from different microbial sources, such as Achromobacter xylosoxidans,12 Aeromonas sp.,1,12,13 Alcaligenes xylosoxidans MTCC*491,1,13 Arthrobacter viscosus,1,12 Bacillus megaterium,1 Bacillus laterosporus12, Bacillus sp.1, Flavobacterium sp.,12,13 Micrococcus luteus NCIM 21708, Paecilomyces sp.12, Pseudomonas diminuta N176,1,2,7,11 Pseudomonas cepacia12, Pseudomonas nitroreducens12, Pseudomonas paucimobilis12, Pseudomonas putida12, Pseudomonas sp. 13011, Pseudomonas sp. SE831,7, Pseudomonas sp. V222.
The natural of cephalosporin C acylase catalyzed the CPC to 7-ACA directly in a very low efficiency14, 15, 16, significant substrate inhibition14, and product inhibition14. In recent years, protein engineering of cephalosporin C acylase was developed to overcome the above limitations14, 15. Protein engineering approach based on combined use of error-prone PCR mutagenesis, molecular modeling, site saturation and site-directed mutagenesis17 were used to enhance the catalytic efficiency of CPC acylase. Wang et al. found a mutant of CPC acylase activity from Pseudomonas diminuta N176, Ala675Gly showed 1.12-fold compared with its wild type7; Golden et al. found two mutants of cephalosporin acylase from Pseudomonas N176, H57aS/H70bS and M165áS/H57bS/H70bS had Vmax 4.2- and 6.0-fold compared with wild type, respectively10; Zhang et al. found two mutants of CPC acylase from Pseudomonas sp. SE83, mutant protein D2 (227-AM-228 deletion) and D4 (212-ADLA-215 deletion) had Kcat/Km values 1.46- and 2.02-fold compared with the original control, respectively11; Xiao et al. found two mutants of CPC acylase form Pseudomonas SE83, mutant H57bA/H70bY and mutant H57bA/H70bY/I176bN had Kcat/Km values 1.2- and 5.6-fold compared with the original control, respectively14; Pollegiani et al. found two mutants of the CPC acylase from Pseudomonas N176, M31bF/H57bS/H70âS and A215aY/M31bF/H70bS showed 3.3- and 4.3 fold improvements in Vmax on CPC compared with its starting template (M31bF), respectively with eliminated substrate inhibition and reduced product inhibition14; A mutant of the CPC acylase acyII from Pseudomonas SE83, V122aA/G140aS/F58bN/I75bT/I176bV/S471bC (S12) showed a 7.5- and 3.7-fold improvement in the specific activity and Kip, respectively, compared with the wild type14; Ishii et al. mutated Met269 and Ala271 of cephalosporin C acylase can increase activity 1.6-fold and 1.7-fold respectively15; Ren et al. found a mutant of the cephalosporin acylase from gene acyII in Pseudomonas sp., G139aS/F58bN/I176bV/S471bC showed 2.86-fold improvement in Kcat/Km compared with wild type14; Oh et al. found that the deacylation activity of the mutation Q50bM-Y149aK-F177aG toward CPC improved by 7.9-fold16.
- The biotransformation of cephalosporin C to 7-ACA in one step enzymatic with cephalosporin acylase is a simple process with low cost and potential to applied in industrial production.
- The protein engineering of cephalosporin C acylase was developed to increase its activity and stability of CPC acylase.
ACKNOWLEDGMENTS
The author sincerely thanks Ministry of Research, Technology and Higher Education of the Republic Indonesia for financial support to this work through research grant.
- Jobanputra, A.P., Rajendrabhai D.V. Cephalosporin C acylase from Pseudomonas species: Production and enhancement of its activity by optimization of process parameters. Biocat. Agric.Biotechnol., 2015;465-470.
- Pollegioni, L., Rosini, E., Molla, G. Cephalosporin C acylase: dream and (or) reality. Appl. Microbiol. Biotechnol., 2013; 97:2341-2355.
- Vasait, R.D., Jobanputra, A.H. Single step bioconversion of cephalosporin C by strain of Achromobacter species isolated from rhizosphere soil, Adv. Biores., 2015; 6(3): 124-127.
- Egorov, A.M., Kurochkina B., Sklyarenko A.V., Nys P.S. Enzymatic transformation of betalactam antibiotics. Trend of development and approaches to practical implementation, Biocat. Fundament. Appl., 2000; 41(6): 43-46.
- Barber, M.S., Giesecke, U., Reichert, A., Minas, W: Industrial enzymatic production of cephalosporin-based â-lactams in Advances Biochemistry Engineering/Biotechnology (Scheper T, ed.). New York, Spinger-Verlag, 2004; pp179-215.
- Li, Y., Jiang, W., Yang, Y., Zhao, G., Wang, E. Overproduction and purification of glutaryl 7-aminocephalosporanic acid acylase, Prot. Exp. Purif., 1998; 12:233-238.
- Wang, Y., Yu, H., Song, W., An, M., Zhang, J., Lui, H., Shen, Z. Overexpression of synthesized cephalosporin C acylase containing mutations in the substrate transport tunnel, J. Biosci. Bioeng., 2012; 113(1):36-41.
- Gaurav, K., Kundu, K., Kundu, S. Biosynthesis of cephalosporin-C acylase enzyme: optimal media design, purification, and characterization, Artif. C.B. Subst. Biotechnol., 2010; 38:277-283.
- Takahashi, S., Makoto F., Keishi O., Namiho T., Yayoi S., Katsumas A., Yoshio K. A highly stable d-amino acid oxidase of the thermophilic bacterium Rubrobacter xylanophilus, Appl. Environ. Microbiol. 2014; 80(23):7219-729.
- Golden, E., Paterson, R., Tie, W.J., Anandan, A., Flematti, G., Molla, G., Rosini, E., Pollegioni, L., Vrielink, A. Structure of a class III engineered cephalosporin acylase: comparisons with class I acylase and implications for differences in substrate specificity and catalytic activity, Biochem. J., 2013; x: 217-226.
- Zhang, J., Yu, H., Luo, H., Shen, Z. Determination of the second autoproteolytic cleavage site of cephalosporin C acylase and the effect of deleting its flanking residues in the á-C-terminal region, J. Biotechnol., 2014; 184: 138-145.
- Sonawane, V.C. Enzymatic modifications of cephalosporin by cephalosporin acylase and other enzymes, C. Rev. Biotechnol., 2006; 26(95): 95-120.
- Nupura, H., Asmita, T., Sharath, B., Asmita, P., Media optimization for the production of cephalosporin C acylase from a novel bacterial source: Alcaligenes xylosoxidans MTCC+491, Res. J., Biotechnol., 2008; 3(1): 16-21.
- Xiao, Y., Xiangdong, H., Yu, Q., Yan, Z., Guoqiang, C., Pingkai, O., Zhanglin, L. Engineering of a CPC acyalse using a facile pH indicator assay, J Ind. Microbiol.Biotechnol., 2014; 41(11): 1617-1625.
- Ren, Y., Lei, Y., Zhu, Y. Site-directed mutagenesis of cephalosporin c acylase and enzymatic conversion of cephalosporin C to 7-aminocephalosporanic acid, Turk. J. Biochem., 2014; 39(1): 51-56.
- Ren, Y., Modeling and experimental analysis of cephalosporin C acylase and its mutant, Op. Biotechnol. J., 2013; 7: 30-37.
- Conti, G., Pollegioni, L., Molla, G., Rosini, E. Strategic manipulation of an industrial biocatalyst-evolution of a cephalosporin C acylase, FEBS J., 2014; 281: 2443-2455.
© The Author(s) 2016. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.