ISSN: 0973-7510
E-ISSN: 2581-690X
To study the risk factors and outcome of blood stream infection caused by non-fermenting gram negative bacilli (NFGNB) and their pattern of antibiotic susceptibility and genes. We included sepsis cases with blood culture positive for NFGNB. MIC for colistin was determined by broth microdilution method. Multiplex PCR was used to detect BlaIMP, BlaVIM, BlaKPC, BlaNDM-1 genes in cephalosporin and carbapenems resistant Acinetobacter spp. isolates. Out of 4,664 cases of sepsis, 50 (1.07%) were positive for NFGNB. Acinetobacter spp. 29 (58%) was the predominant isolate, of which 16 (55.17%) isolates were resistant to cephalosporins and carbapenems. We detected BlaKPC and BlaNDM-1 genes in two of these isolates. We did not detect BlaIMP, BlaVIM, BlaKPC and BlaNDM-1 genes in any other NFGNB isolates. Majority of the strains of Pseudomonas spp. showed sensitivity to all the antibiotics tested. NFGNB sepsis patients with respiratory illness correlated well with fatal outcome (p <0.05; OR 21). More numbers of Acinetobacter spp. sepsis cases had fatal outcome (p <0.05; OR 12.83). NFGNB sepsis patients with respiratory illness and those which yielded Acinetobacter spp. correlated positively with fatal outcome. We detected BlaKPC and BlaNDM-1 genes in two strains of drug resistant Acinetobacter spp.
Bacteremia, colistin, Drug resistance, Gram negative bacilli, Risk factors
Gram negative bacilli which are non-fermenters (NFGNB) are a heterogeneous category of microorganisms which do not have the ability to ferment sugars. NFGNB, which were considered as contaminants in the past, have emerged as important major pathogenic organisms responsible for causing a variety of infections in recent years1. NFGNB cause opportunistic healthcare-associated infections. They are commonly found on skin of healthcare-workers, instruments such as ventilator machines, humidifiers and mattresses2-4. Among NFGNB Burkholderia cepacia, Pseudomonas aeruginosa, Acinetobacter baumanni complex, Alcaligenes faecalis, Sphingomonas paucimobilis and Stenotrophomonas maltophilia are some pathogens known for causing health care associated bloodstream infections5.
Multidrug resistance (MDR) property of NFGNB is the reason that facilitated the way for the reconsideration of previously used antibiotic colistin into clinical use. For the treatment of infections caused by multi-drug resistant gram-negative bacilli (MDRGNB), colistin may have crucial and reliable role as potent antibiotic.6,7 The clinical use of colistin was restricted because of reports of serious nephrotoxicity and discovery of less toxic antibiotics6,7. So we intend to study the antibiotic sensitivity pattern of NFGNB from sepsis cases and correlate with the clinical condition of the patient. We also determine MIC of Colistin and also detect BlaIMP, BlaVIM, BlaKPC, BlaNDM-1 genes in cephalosporin and carbapenems resistant Acinetobacter spp. isolates.
It is a prospective time bound study conducted from (1st November 2018 to 30th April 2019) at the Microbiology laboratory of a tertiary care center. Sepsis cases with blood culture positive for NFGNB were included in the study. Blood culture samples which did not grow NFGNB and samples received on days other than the duration specified were excluded. Non random sampling strategy was used. Clinical data was taken from the records section of the hospital. Institutional ethics committee clearance has been taken for the project.
Bac T/Alert bottles with patient’s blood received in microbiology laboratory, for blood culture were processed as per protocol, in Bac T/Alert 3D system. From the positive bottles, the subcultures were done on MacConkey agar, Blood agar and Chocolate agar. The identification of NFGNB up to the species level was done by VITEK ®2 Compact(C) system using the Gram-negative Identification (GN-ID) 21341 card, and antibiotic sensitivity testing (AST) was done with AST 281 card. Both the Bac T/Alert 3D system and VITEK®2 Compact (C) system were from bioMerieux company (North Carolina, USA),
MIC for colistin by broth micro dilution method
MIC for colistin by broth microdilution method was carried out as per CLSI guidelines.8 A 100 mL stock solution of colistin sulfate (Sigma Aldrich, USA) of concentration 256 μg/mL was prepared according to the directions provided by the drug manufacturer. Two fold dilutions of the antibiotic were made to get a final dilution of 128, 64, 32, 16, 8, 4, 2, 1, 0.5 & 0.25 μg per ml of colistin respectively in wells 1 to 10 of microtitre plate.
Inoculum was prepared from 4 to 5 isolated colonies of similar colony morphology grown over night (18- 24 hours) on Mac Conkey’s agar. The colonies were inoculated into cation adjusted Mueller Hinton broth (CAMHB), kept at 37°C for 4 to 6 h to get log phase growth and adjusted to 0.5 McFarland (1 x 108 CFU/ml). This broth was repeatedly diluted to get the final concentration of bacteria as approximately 5 x 104 CFU/ ml. Now 10μl of inoculum was pipetted into wells 1 to 11. Well 11 acted as the growth control and 12th well as broth sterility control with sterile CAMHB. The microtitre plates were incubated for 12-18 h at 37°C. E. coli ATCC 25922 and P. aeruginosa ATCC 27853 were run as controls. The lowest concentration of antibiotic at which there is no visible growth of organism was taken as MIC endpoint. According to CLSI, ≤ 2 µg/ml and ≥ 4 µg/ml were considered as the susceptible and resistant break points respectively.
Multiplex PCR9,10
The DNA was extracted according to CDC protocol using boiling method. The primer sets were purchased from sigma Aldrich Pvt. Ltd (Table 1). The master mix was prepared by adding ready mix Taq PCR reaction mix with MgCl2 (Califormia, USA), in which the respective primer sets i.e, Forward (Fˈ) as well as Reverse (Rˈ) and the testing DNA were added and this master mix was subjected to amplification. Amplification was done with following cycling conditions;10 min denaturation at 95°C, then 35 cycles of (45s denaturation at 94°C, 45 s primer annealing at 54°C, and 50 s primer extension at 72°C). The last cycle, an additional 7 min extension step at 72°C in Rotor gene –Q (Qiagen, germany) and the products were kept at 4°C. The amplified product was detected by using 2% agarose gel electrophoresis. K.pneumoniae ATCC BAA 1705 and K.pnemoniae BAA 1706 were used as positive and negative control. Clinical isolates known to be positive for BlaIMP, BlaVIM BlaNDM-1 genes were also used as controls.
Table (1):
The primer sets were in PCR reaction.
| Primers for detection of genes | Primer sequence | Product size (bp) |
|---|---|---|
| bla IMP | 5 ˈGGAATAGAGTGGCTTAAYTCTC 3ˈ | 232 |
| 5ˈ GGTTTAAYAAAACAACCACC 3ˈ | ||
| bla VIM | 5 ˈ GATGGTGTTTGGTCGCATA 3ˈ | 390 |
| 5ˈ CGAATGCGCAGCACCAG 3 ˈ | ||
| bla NDM-1 | 5ˈ CACCTCATGTTTGAATTCGCC 3ˈ | 561 |
| 5ˈ CTCTGTCACATC GAAATCGC 3ˈ | ||
| bla KPC | 5′-CATTCAAGGGCTTTCTTGCTGC-3′ | 538 |
| 5′-ACGACGGCATAGTCATTTGC-3′ |
Statistical analysis
Statistical analysis was performed using SPSS version 16.0. Multiple logistic regression analysis was done for correlation of risk factor and fatal outcome. Chi Square test was also used to correlate other variables.
Out of 4,664 cases of sepsis 50(1.07%) patient’s blood samples yielded non fermenting gram negative bacilli. Of these 3(6%) were Acinetobacter lwoffi; 23(46%) A. baumannii; 1(2%) A. juni; 2(4%) Acinetobacter spp; 14(28%) Pseudomonas aeruginosa; 2(4%) P. putida; 3(6%) were P.stutzeri; 1(2%) Burkholderia cepacia; 1(2%) Brevundomonas vescularis. Of the 50 cases of NFGNB, 28(56%) were male and 22(44%) were females; 22(44%) were more than 50 years of age, 16(32%) were 31 to 50 years of age, 7(14%) were 11-30 years of age and 5(10%) were less than 10 years of age.
Table (2):
The antibiotic sensitivity pattern of Acinetobacter spp. isolated from blood of patients with symptoms of sepsis.
Antibiotics |
Sensitive (%) |
Intermediate (%) |
Resistant (%) |
|---|---|---|---|
Amikacin |
26 (89.7) |
0 |
3(10.34) |
Ciprofloxacin |
15(51.72) |
0 |
14(48.28 |
Ceftazidime |
17(58.62) |
0 |
10(34.48) |
Cefepime |
11(37.93) |
0 |
18(62.07) |
Cefaperazone_Sulbactam |
16(55.17) |
5(17.24) |
8(27.59) |
Ceftriaxone |
10(34.48) |
2(6.7) |
17(58.62) |
Gentamicin |
13(44.83) |
0 |
16(55.17) |
Imipenem |
12(41.38) |
0 |
17(58.62) |
Meropenem |
13(44.83) |
0 |
16(55.17) |
Netillin |
16(55.17) |
0 |
13(44.82) |
Ofloxacin |
18(62.07) |
0 |
10(34.48) |
Piperacillin |
17(58.62) |
1(3.45) |
11(37.93) |
Piperacillin_Tazobactam |
11(37.93) |
1(3.45) |
17(58.62) |
Ticarcillin_Clavulinic acid |
15(51.72) |
1(3.45) |
13(44.83) |
Trimethoprim _Sulphamethoxazole |
16(55.17) |
0 |
13(44.83) |
Tigecycline |
25(86.21) |
3(10.34) |
1(3.45) |
Of the 29 isolates of Acinetobacter spp. most of them were sensitive to amikacin 26(89.7%) followed by tigecycline 25(86.21%) and 16(55.17%) isolates were resistant to carbapenems and cephalosporins (Table 2). All the isolates were sensitive to colistin with MIC values from 0.25 to 2 µg/ml. Majority of Pseudomonas spp. strains showed sensitivity to all the antibiotics tested (Table 3). Only one of these isolates was resistant to cephalosporins and carbapenems. MIC values for colistin among these isolates were between 0.25 and 8 µg/ml of which two isolates were resistant. The single isolate of Brevundomonas vescularis was sensitive to cephalosporin, carbapenems and resistant to aminoglycosides, fluoroquinolones, Trimethoprim/sulfamethoxazole. The single isolate of Burkholderia cepacia was resistant to most of the antibiotics tested, but sensitive to carbapenems, fluoroquinolones and tigecycline.
Table (3):
The antibiotic sensitivity pattern of Pseudomonas spp. isolated from blood of patients with symptoms of sepsis.
Antibiotics |
Sensitive (%) |
Intermediate (%) |
Resistant (%) |
|---|---|---|---|
Amikacin |
19(100) |
0 |
0 |
Ciprofloxacin |
19(100) |
0 |
0 |
Ceftazidime |
18(94.74) |
1(5.26) |
0 |
Cefepime |
19(100) |
0 |
0 |
Cefaperazone_Sulbactam |
18(94.74) |
0 |
1(5.26) |
Ceftriaxone |
17(89.47) |
1(5.26) |
1(5.26) |
Gentamicin |
18(94.74) |
1(5.26) |
0 |
Imipenem |
19(100) |
0 |
0 |
Meropenem |
17(89.47) |
1(5.26) |
1(5.26) |
Netillin |
19(100) |
0 |
0 |
Ofloxacin |
19(100) |
0 |
0 |
Piperacillin |
18(94.74) |
1(5.26) |
0 |
Piperacillin_Tazobactam |
17(89.47) |
1(5.26) |
1(5.26) |
Ticarcillin_Clavulinic acid |
13(68.42) |
5(26.32) |
1(5.26) |
Trimethoprim _Sulphamethoxazole |
17(89.47) |
0 |
2(10.53) |
Tigecycline |
18(94.74) |
0 |
1(5.26) |
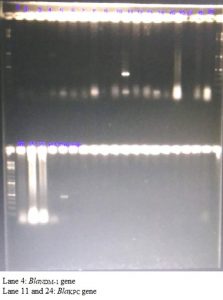
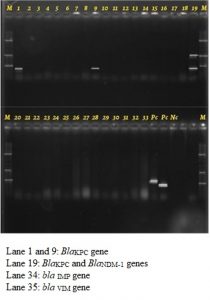
Of the 16 isolates of the Acinetobacter spp. resistant to cephalosporins and carbapenems BlaKPC and BlaNDM-1 genes were detected in two of these isolates. We did not detect BlaIMP, BlaVIM, BlaKPC and BlaNDM-1 genes in any other NFGNB isolates. (Fig. 1 and 2)
Fig. 1. Detection of BlaIMP, BlaVIM, BlaKPC and BlaNDM-1 genes by multiplex PCR in the drug resistant Acinetobacter species isolated from cases of blood stream infection
Fig. 2. Multiplex PCR conducted on Klebsiella clinical isolates run as positive and negative controls
Of the 50 cases of NFGNB we got detailed clinical history for 23 cases. Of these 23 cases, 14(60.9%) expired due to multiple underlying conditions. Most of the fatal outcome was due to a combination of chronic obstructive pulmonary disease (COPD) respiratory failure, cardiac arrest, renal failure or in other words multi-organ failure. Most of the cases of NFGNB, the patients had multiple underlying clinical conditions like diabetes mellitus 9(39.13%), cardiac anomalies 8(34.8%), malignancy 6(26%), respiratory illness 14(60.9%), malaria 4(17.4%), HIV 3(13%), GIT anomalies 3(13%), renal anomalies 4(17.4%), hepatic anomalies 7(30.4%), CNS infection 3(13%), wound infection 4(17.4%). Cardiac anomalies included cases of cardiac arrest, cardiac valve dysfunction, tachycardia, atrial fibrillation; malignancy included lymphoma, leukemia, nasopharyngeal carcinoma, multiple myeloma, osteocarcinoma; respiratory illness included COPD, ventilator associated pneumonia (VAP), lower lobe pneumonia, aspiration pneumonia, bronchial asthma, respiratory failure; GIT anomalies included esophageal candidiasis, progressive abdominal fistulation, GIT bleeding; renal anomalies included acute renal failure, decreased urine output, renal insufficiency; hepatic anomalies included portal hypertension, hepatomegaly, jaundice; CNS anomalies included myasthenia gravis, PCOM aneurysm; wound infection included cellulitis, surgical wound infection.
When underlying clinical conditions were correlated with fatal outcome, patients with respiratory illness (p <0.05) correlated well with fatal outcome (OR 21, Cl = 2.397-183.99) (Table 4). When the organisms were correlated with fatal outcome, more numbers of patients infected with Acinetobacter spp. had fatal outcome (p <0.05; OR 12.83, Cl = 1.695-97.193) (Table 4).
Table (4):
Correlation of underlying clinical conditions with fatal outcome.
| Variables | Non-fatal (n=9) | Fatal (n=14) | p value | Odds ratio |
|---|---|---|---|---|
| Age group (11-30) | 2(22.2%) | 1(7.1%) | 0.295 | 0.26 |
| 31-50 | 5(55.5%) | 6(42.9%) | 0.552 | 0.6 |
| More than 50 | 2(22.2%) | 7(50%) | 0.183 | 3.5 |
| Male | 5(55.5%) | 11(78.6%) | 0.242 | 2.933 |
| Female | 4(44.4%) | 3(21.43%) | 0.242 | 0.341 |
| Under lying disease conditions | ||||
| Diabetes Mellitus | 2(22.2%) | 7(50%) | 0.311 | 2.625 |
| Malignancy | 3(33.3%) | 3(21.4%) | 0.526 | 0.545 |
| Cardiac anomalies | 2(22.2% ) | 6(42.9%) | 0.311 | 2.625 |
| Respiratory illness* | 2(22.2%) | 12(85.71%) | 0.002* | 21* |
| Hepatic anomalies | 2(22.2%) | 5(35.7%) | 0.493 | 1.944 |
| Renal anomalies | 1(11.1%) | 3(21.4%) | 0.524 | 2.182 |
| GIT anomalies | 1(11.1%) | 2(14.3%) | 0.825 | 1.33 |
| HIV | 0 | 3(21.4%) | 0.136 | 1.273 |
| Malaria | 1(11.1%) | 3(21.4%) | 0.524 | 2.182 |
| Wound infection | 11(84.6) | 3(21.4%) | 0.524 | 2.182 |
| CNS complaints | 2(22.2%) | 1(7.1%) | 0.295 | 0.269 |
| Organisms | ||||
| Pseudomonas spp. | 6(66.7%) | 2(14.3%) | 0.01 | 0.083 |
| Acinetobacter spp.* | 2(22.2% ) | 11(78.6) | 0.008* | 12.83* |
| Burkholderia cepacia | 1(11.1%) | 0 | – | – |
| Brevundomonas vesiculosis | 0 | 1(7.1%) | – | – |
* Statistically significant
Among NFGNB Burkholderia cepacia, Pseudomonas aeruginosa, Acinetobacter baumannii complex, Alcaligenes faecalis and Sphingomonas paucimobilis are some pathogens known for causing health care associated bloodstream infections5-7. In the present study, Acinetobacter spp. 29(58%), Pseudomonas spp. 19(38%), followed by one isolate each of Burkholderia cepacia and Brevundomonas vescularis were isolated.
In a study on resistance of NFGNB isolated from blood cultures from an emergency hospital in Brazil found that from 87(100%) isolated and analyzed strains, 11(13%) were NFGNB. Acinetobacter spp. was most often occurring organism (55%) succeeded by Pseudomonas spp. (18%), Stenotrophomonas spp. (18%) and Burkholderia spp. (9%). Acinetobacter spp. were resistant to gentamycin, meropenem, imipenem, amikacin, ciprofloxacin, ceftazidime and ceftriaxone. Pseudomonas spp. was sensitive to gentamycin, meropenem, imipenem, amikacin, levofloxacin, norfloxacin, ceftazidime and cefepime. All Stenotrophomonas spp. were resistant to levofloxacin and chloramphenicol. Burholderia spp. were sensitive to the antibiotics tested11. A study showed A.baumannii isolated from sepsis cases was highly resistant to aminoglycosides in comparison to other Acinetobacter spp. P. aeruginosa strains were sensitive to all the aminoglycosides tested.12 We have similar results.
Rate of isolation of NFGNB from blood vary in studies conducted from various places from 0.7 to 20.813,14. In a study by Grewal US et al isolation rate of NFGNB was 11.6%. Commonest NFGNB isolated in the study was P. aeruginosa (87.96%), followed by A. baumannii (7.87%). 24.2 % of P. aeruginosa strains were identified as multi drug resistant (MDRPA). About 100% of the MDRPA isolates were found to be susceptible to polymyxin B. However, 60.9% of the MDRPA isolates in their study showed resistance to imipenem, which is usually the first therapeutic choice for treating the infections caused by them. Isolates of A. baumannii in their study showed maximum susceptibility to imipenem (88.2%), followed by cefoperazone + sulbactam (76.4%). Maximum resistance was shown to aztreonam (11.7%). 11 isolates of A. baumannii showed multidrug resistance (MDRAB) in the study. Isolation of multi drug resistant P. aeruginosa and multi drug resistant A. baumannii in the study raised the concern of emerging antibiotic resistance in this group14.
In an another study on predominance of NFGNB and sensitivity pattern in Tamil Nadu, found that out of 5052 clinical samples, 517 (10.2%) samples were NFGNB. Acinetobacter and Pseudomonas spp. were the most frequently found organisms. Colistin and imipenem showed maximum sensitivity for all clinical samples. Highest resistance was seen with gentamycin ceftazidime and ciprofloxacin15.
An antibiotic trend analysis over a period of seven years in patients suffering from nosocomial infections showed elevated levels of resistance to frequently used antibiotics and hence a significant rise in colistin use especially in patients who were critical16.
In a retrospective study factors deciding the existence of patients with Acinetobacter baumannii bacteremia were evaluated. Relevant details of hospitalized patients who developed Acinetobacter baumannii bacteremia were collected in Beirut between 2010 and 2015. Presence of diabetes mellitus, large amount of steroids, ventilator use, early treatment with tigecycline and colistin, critical care unit and septic shock correlated with worse result by univariate analysis. Multivariate analysis showed significant correlation with large doses of steroids and septic shock. Approximate fatality was 63.5%. Bacteremia showed 70.3% of the deaths. An extended hospital admission is usually correlated with failure of performance, prior use of steroid and early exposure of antibiotic17.
In our study there were no positive correlation among death and diabetes mellitus. But as in previous studies we got positive correlation between death and underlying respiratory illness like, VAP, COPD, bronchial asthma and pneumonia. We also found that mortality was associated with sepsis in Acinetobacter spp.
In a study on carbapenem resistant strains of coliforms isolated from cases of blood stream infection blaNDM-1 & blaKPC genes were detected. They found that 16 isolates harbored blaNDM-1, and blaKPC gene was not detected in any of the isolates9. In a study period of 2008 and 2012, 166 uropathogens isolated from cases with complicated UTI, 34(43.6%) isolates had blaVIM gene, 5(6.4%) had blaIMP, blaNDM-1 and blaKPC genes were absent. Among the isolates from 2012, 47(53.4%) had blaNDM-1 gene 19(24.4%) had blaVIM, one (1.1%) had blaIMP and blaKPC gene was absent10. In the present study one each of BlaKPC and BlaNDM-1 genes were detected in two strains of drug resistant Acinetobacter spp. Further studies with a larger sample size may give more conclusive results.
Rate of isolation of NFGNB from blood from sepsis cases was 1.07%. Most commonly isolated bacteria were A. baumannii and P.aeruginosa. NFGNB sepsis patients with respiratory illness and those which yielded Acinetobacter spp. correlated positively with fatal outcome. Most isolates of Acinetobacter spp. were sensitive to amikacin and tigecycline. Sixteen isolates of Acinetobacter spp. were resistant to cephalosporin and carbapenems. Most strains of Pseudomonas spp. were sensitive to all antibiotics tested. Only one isolate was resistant to carbapenems and cephalosporin. All strains of Acinetobacter spp. were sensitive to colistin and Two isolates of Pseudomonas species were resistant to colistin. One each of BlaKPC and BlaNDM-1 genes were detected in two strains of drug resistant Acinetobacter spp. Further studies with a larger sample size may give more conclusive results.
ACKNOWLEDGMENTS
We are extremely grateful to our institution Kasturba Medical College, Mangalore, Manipal Academy of Higher Education, Manipal, Karnataka, India, for providing us with required infrastructure and all the materials for the conduct of this research project.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
Ms. AV: Review of literature, conduct of the research and data collection, result analysis, writing the project report, Dr. JU: Planning of the topic, review of literature, research protocol, guiding the first author in conduct of research project, result analysis, writing project report, Dr. PR: Guidance regarding the gene detection, result analysis and writing of the project report, Dr. ES: Guidance regarding the topic selection, result analysis and writing of the project report. All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
All financial support was provided by the institution, Kasturba Medical College, Mangalore, Manipal Academy of Higher Education, Manipal, Karnataka, India
ETHICS STATEMENT
Institutional ethics committee clearance has been taken for the project. IEC KMC MLR 10-18/355.
AVAILABILITY OF DATA
All datasets generated or analyzed during this study are included in the manuscript.
- Viswanathan R, Singh A, Chatterjee S, Basu S, Isaacs D, Roy S. Non-fermenting, Multi drug resistant, gram negative bacilli in neonatal sepsis in Kolkata, India, a 4-year study. Paediatr. Int Child Health. 2014;34(1):56-59.
Crossref - Devi VP, Reddy S, John MS. Incidence of carbapenem resistant nonfermenting gram negative bacilli from patients with respiratory tract infections among intensive care units. Int J Res Med Sci. 2015;3(6):1368-1371.
Crossref - Bhuwaneshwari G. Multiple antibiotic resistance indexing of non-fermenting gram-negative bacilli. Asian J Pharm Clin Res. 2017;10 (6):78-80.
Crossref - Ritu B, Kumar S, Bansal G, Mehra SK. Identification and antimicrobial susceptibility pattern of clinical isolates of non-fermentative gram negative bacilli. Int J Pharma Res Health Sci. 2014;2(4):347-351.
- Baruah FK, Hussain A, Kausalya N, Grover RK. Antibiotic resistance profile of non-fermenting Gram-negative bacilli isolated from the blood cultures of cancer patients. J Global Infect Dis. 2015;7(1):46-47.
Crossref - Loho T, Dharmayanti. A. Colistin: an antibiotic and its role in multiresistant Gram-negative infections. Acta Med Indones. 2015;47(2):157-168.
- Dhariwal AK, Tullu MS. Colistin: Re-Emergence of The ′Forgotten′ Antimicrobial Agent. J Postgrad Med. 2013;59(3): 208-215.
Crossref - CLSI. Performance Standards for Antimicrobial Susceptibility Testing: Twenty eighth Informational Supplement. CLSI document M100-S28.Wayne PA: Clinical and Laboratory Standards Institute. 2018.
- Ankit P, Sevitha B, Ashwini H, Pooja R, Shalini S. Clinico-microbiologicalstudy of bacteremia caused by coliforms in adults. J Clin Diag Res. 2018;12(7): DC15-DC19.
- Balvinder M, Vinay kumar H, Gagandeep S, Harkiran Kaur S, Suma BA, Neelam T. Occurrence of blaNDM-1 & absence of blaKPC genes encoding carbapenem resistance in uropathogens from a tertiary care centre from North India. Indian J Med Res. 2015;142(3): 336-343.
Crossref - Oliveira ME, Araujo DG, Oliveira SR. Resistance of nonfermenting Gramnegative bacilli isolated from blood cultures from an emergency hospital. J Bras Pathol Med Lab. 2017;53(2):87-91.
Crossref - Liu JY, Wang FD, Ho M W, el al. In vitro activity of aminoglycosides against clinical isolates of Acinetobacter baumannii complex and other non-fermentative gram-negative bacilli causing healthcare-associated bloodstream infections in Taiwan. J Microbiol Immol Infect. 2016;49 (6):918- 923.
Crossref - Ashish J, Radhika R, Bhargavi L, Geetha B, Ramani B. Current Trend of Nonfermenting gramnegative bacilli in a Tertiary Care Hospital in Trivandrum. J Pure Appl Microbial. 2016;10(1):425-442.
- Grewal US, Bakshi R, Walia G, Shah PR. Antibiotic susceptibility profiles of nonfermenting gramnegative bacilli at a tertiary care hospital in Patiala, India. Niger Postgrad Med J. 2017;24(2):121-125.
Crossref - Krishnan S, Santharam P, Shanmugavadivoo N, Usha B. Prevalence of nonfermenting gramnegative bacilli and their antibiotic sensitivity pattern at a tertiary care hospital in Tamil Nadu, India. Int J Curr Microbiol Appl Sci. 2018;7(2):2751-2758.
Crossref - Zorana MD, Marko MF, Slobodan MJ. Previous antibiotic exposure and antimicrobial resistance patterns of Acinetobacter spp. and P.aeruginosa isolated from patients with nosocomial infections. Balkan Med J. 2017;34:527-533.
Crossref - Ballouz T, Aridi J, Afif C, et al. Risk factors clinical presentation and outcome of A. baumannii bacteraemia. Front Cell Infect Microbiol. 2017;156: 1-7.
Crossref
© The Author(s) 2020. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.