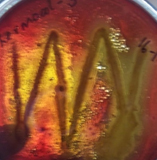ISSN: 0973-7510
E-ISSN: 2581-690X
Biosurfactants are a structurally diverse group of amphiphilic, surface-active substances produced by microorganisms which can be used as an eco-friendly approach for environmental remediation. Surface activity makes surfactants excellent emulsifiers, foaming and dispersing agents. An attempt was made to produce biosurfactant from solid waste bacteria. Organisms were isolated from solid waste. The isolated organisms were identified to be Micrococcus luteus, Serratia liquefaciens, Bacillus macquariensis, Bacillus popilliae and Corynebacterium kutscheri by using various biochemical tests. All these organisms were screened for biosurfactant production using penetration assay, microplate assay and blood haemolysis technique. The yield of biosurfactants was compared in different media with and without oil stimulant and the role of an oil stimulant in biosurfactant production was determined.
Biosurfactants, amphiphilic, emulsifiers and microplate assay
Biosurfactants are amphiphilic compounds produced on living surfaces, mostly microbial cell surfaces, or excreted extracellularly. They contain hydrophobic and hydrophilic moieties that reduce surface tension and interfacial tension between individual molecules at the surface and interface respectively. Most biosurfactants are either anionic or neutral and the hydrophobic moiety is based on long-chain fatty acids or fatty acid derivatives, whereas the hydrophilic portion can be a carbohydrate, amino acid, phosphate or cyclic peptide1
Considerable attention has been given in the past to the production of surface-active molecules of biological origin because of their potential utilization in food processing, pharmacology, and oil industry. Although the type and amount of the microbial surfactants produced depend primarily on the producer organism, factors like carbon and nitrogen, trace elements, temperature, and aeration also affect their production by the organism1
Many microbial surfactants have been purified and their structures have been elucidated. The high molecular weight microbial surfactants are generally polyanionic heteropolysaccharides containing polysaccharides and proteins and the low molecular weight microbial surfactants are often glycolipids. The productivity of microbial surfactants varies with the nutritional environment of the growing microorganism1
Screening techniques [2]Penetration assay
The penetration assay was developed by Maczek et al. in 2007. This assay relies on the contacting of two insoluble phases which leads to a colour change. For this assay, the cavities of a 96 well microplate are filled with 150 ¼l of a hydrophobic paste consisting of oil and silica gel. The paste is covered with 10 µl of oil. Then, the supernatant of the culture is coloured by adding 10 µl of a red staining solution to 90 ¼l of the supernatant. The coloured supernatant is placed on the surface of the paste. If biosurfactant is present, the hydrophilic liquid breaks through the oil film barrier into the paste. The silica enters the hydrophilic phase and the upper phase changes from clear red to cloudy white within 15 minutes.
Microplate assay
The surface activity of individual strains can be determined qualitatively with the microplate assay developed and patented by Vaux and Cottingham. This assay is based on the change in optical distortion that is caused by surfactants in an aqueous solution. Pure water in a hydrophobic well has a flat surface. The presence of surfactants causes some wetting at the edge of the well and the fluid surface becomes concave and takes the shape of a diverging lens. For this assay, a 100 ¼l sample of the supernatant of each strain is taken and put into a microwell of a 96-mircowell plate. The plate is viewed using a backing sheet of paper with a grid. If biosurfactant is present, the concave surface distorts the image of the grid below. The optical distortion of the grid provides a qualitative assay for the presence of surfactants.
Blood haemolysis technique
Biosurfactants can cause lysis of erythrocytes. This principle is used for the haemolysis assay which was developed by Mulligan et al. Cultures are inoculated on sheep blood agar plates and incubated for 2 days at 25°C. Positive strains will cause lysis of the blood cells and exhibit a colourless, transparent ring around the colonies.
Production of biosurfactants
Production of biosurfactants was tried in four different media which include nutrient broth with hydrocarbon stimulant, nutrient broth without hydrocarbon stimulant, production medium with hydrocarbon stimulant and production medium without hydrocarbon stimulant.
Procedure
The bacteria are inoculated in 50 ml of the four different media. Incubate the media at 30° C for 3 days with 2 min of light shaking each day
Measurement of dry weight [4]
Instead of obtaining the dry weight of the bacterial pellet obtained by centrifugation, the media was filtered and the dry weight of the bacterial residues on the filter paper was measured.
Procedure
The initial weight of the Whatmann filter paper is taken.The media is filtered using Whatmann filter paper (Fig 2.1) and the weight of filter is taken at 0 min after filtration.The weights are taken for every 5 min until constant weight is obtained (Fig 2.2).This gives the dry weight of bacteria
Extraction of biosurfactants [3]
Extraction of biosurfactants is done using solvent extraction where the solvent system is in 2:1 ratio (Fig 3).
Procedure
50 ml of the culture filtrate is taken and its pH is adjusted to 2 using 1N HCl.It is then taken in a separating funnel.To this, Chloroform and Methanol is added in 2:1 ratio.The bottom solvent layer will contain biosurfactants and this is carefully collected in a initially weighed beaker.The upper aqueous layer is reextracted with the solvent mixture.The biosurfactant is then collected by evaporating the solvent.The increase in weight of the beaker indicates the amount of biosurfactants obtained.The biosurfactant is stored by diluting it as 1 g/l of water.
Confirmation of biosurfactants
Oil spreading technique [5]
The oil spreading assay was developed by Morikawa et al. 30 ml of distilled water is taken in petri plate. 1 ml of coconut oil and gingely oil is added to the centre of these plates. 20 µl of cell free supernatant is then added to centre. The biosurfactant producing organism can displace the oil and spread in water.
Thin layer chromatography [6]
10 cm long and 1 cm wide TLC strips are cut. The test solution is spotted on the strips using capillary tubes. The strips were dipped in a solvent mixture of chloroform: methanol: water (70:10:0.5). The solvent front is marked. Then the TLC plate is exposed to iodine vapour, biosurfactant if present will appear as bright yellow coloured spots.
Emulsification capacity assay [7]
Emulsification capacity assay of biosurfactants was developed by Cooper and Goldenberg. Kerosene or any other hydrophobic compound is added to an equal volume of aqueous sample. The mixture is vortexed at high speed for 2 minutes. After 24 hours, the height of the stable emulsion layer is measured. The emulsification index E24 is calculated as the ratio of the height of the emulsion layer and the total height of liquid. E24 correlates to the surfactant concentration.
E24=hemulsion / htotal ×100 %
Screening of organisms for biosurfactant production
Penetration assay
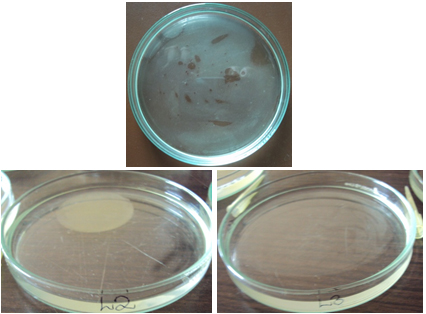
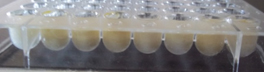
The culture supernatants of all the organisms showed colour change from clear red (Fig 3.70) to cloudy white within 15 minutes (Fig 1) indicating that they are biosurfactant producing organisms.
Microplate assay
The culture supernatants of all the organisms showed optical distortion of the grid indicating that they are biosurfactant producing organisms. (Fig 2)
Blood Haemolysis technique
All the organisms showed haemolysis of sheep blood and a colourless, transparent zone (Fig 3) and greenish zone (Fig 4) was formed around the colonies indicating that they are biosurfactant producing organisms.
Production of biosurfactants
Confirmation of the obtained biosurfactants
Oil spreading technique
All the extracted samples showed displacement of the oil (Fig 5) and spread in water confirming that they are biosurfactants.
Thin layer chromatography
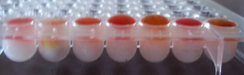
Yellow spots developed on the silica plates on exposure to iodine vapour indicating that all the extracted samples are biosurfactants. (Fig 6)
Emulsification capacity assay
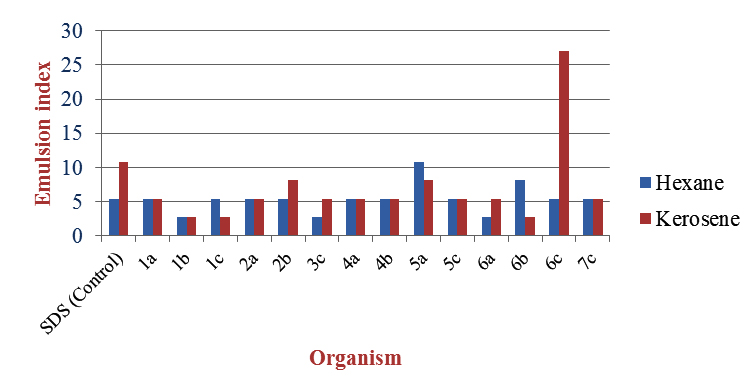
The emulsion index E24 (Fig 7) was calculated for all the extracted samples taking SDS as control.
Total height = 37 mm
The bacteria screened using Penetration assay, Microplate assay and Blood haemolysis test proved to be positive for biosurfactant production.
For the production of biosurfactants from different bacteria, production medium and nutrient broth with and without hydrocarbon stimulant (hexane) were used. For most of the bacteria, yield of the biosurfactants was found to be higher in the production medium in the presence of hexane and in the nutrient broth the yield was higher in the absence of hexane.
Micrococcus luteus (1a), Serratia liquefaciens (1b), Bacillus popilliae (1c), Aeromonas spp. (2a), Bacillus macquariensis (2b), Bacillus azotoformans (3c), Staphylococcus spp. (4a, 4b), Corynebacterium xerosis (5a), Corynebacterium kutsceri (6a), Streptococcus mitis (6b) and Staphylococcus saprophyticus (7c) showed higher yield in production medium with hexane. Hexane being an insoluble substrate and the sole carbon source, bacteria produce biosurfactants to facilitate their diffusion into the cell. [1] (Fig 1)
Serratia liquefaciens (1b), Bacillus popilliae (1c), Aeromonas spp. (2a), Bacillus azotoformans (3c), Staphylococcus spp. (4a, 4b), Corynebacterium xerosis (5a), Corynebacterium kutsceri (6a), Streptococcus mitis (6b), Bacillus pantothenticus (6c) and Staphylococcus saprophyticus (7c) showed higher yield in nutrient broth without hexane. This medium consists of complex, soluble carbon and nitrogen sources that can be easily used by the bacteria for biosurfactant production. [25] When hexane is added to the medium, there is secondary metabolite production causing alteration of pH, thereby limiting the biosurfactant production. [9]. (Fig 1)
Table (1):
Yield of biosurfactants in production medium without hexane.
Organism |
Dry weight of organism (g) |
Weight of biosufactant (g) |
Yield |
|---|---|---|---|
1a |
0.098 |
0.017 |
0.173 |
1b |
0.105 |
0.005 |
0.048 |
1c |
0.0489 |
0.005 |
0.102 |
2a |
0.0718 |
0.003 |
0.042 |
2b |
0.0681 |
0.010 |
0.147 |
3c |
0.0441 |
0.009 |
0.204 |
4a |
0.0517 |
0.004 |
0.077 |
4b |
0.0561 |
0.005 |
0.089 |
5a |
0.076 |
0.014 |
0.184 |
5c |
0.073 |
0.024 |
0.329 |
6a |
0.067 |
0.005 |
0.075 |
6b |
0.075 |
0.002 |
0.027 |
6c |
0.076 |
0.024 |
0.316 |
7c |
0.059 |
0.007 |
0.119 |
Table (2):
Yield of biosurfactants in production medium with hexane.
Organism |
Dry weight of organism (g) |
Weight of biosufactant (g) |
Yield |
|---|---|---|---|
1a |
0.050 |
0.011 |
0.22 |
1b |
0.051 |
0.004 |
0.078 |
1c |
0.055 |
0.010 |
0.182 |
2a |
0.050 |
0.005 |
0.100 |
2b |
0.0152 |
0.003 |
0.197 |
3c |
0.013 |
0.020 |
1.538 |
4a |
0.004 |
0.004 |
1.000 |
4b |
0.059 |
0.008 |
0.136 |
5a |
0.015 |
0.003 |
0.200 |
5c |
0.0466 |
0.004 |
0.086 |
6a |
0.0426 |
0.005 |
0.117 |
6b |
0.0404 |
0.003 |
0.074 |
6c |
0.0422 |
0.009 |
0.213 |
7c |
0.034 |
0.011 |
0.324 |
Table (3):
Yield of biosurfactants in nutrient broth without hexane.
Organism |
Dry weight of organism (g) |
Weight of biosufactant (g) |
Yield |
|---|---|---|---|
1a |
0.071 |
0.019 |
0.268 |
1b |
0.0595 |
0.042 |
0.706 |
1c |
0.0144 |
0.070 |
4.861 |
2a |
0.069 |
0.061 |
0.884 |
2b |
0.079 |
0.093 |
0.177 |
3c |
0.027 |
0.021 |
0.777 |
4a |
0.051 |
0.025 |
0.490 |
4b |
0.032 |
0.044 |
1.375 |
5a |
0.0137 |
0.024 |
1.752 |
5c |
0.061 |
0.010 |
0.164 |
6a |
0.063 |
0.097 |
1.539 |
6b |
0.1249 |
0.138 |
1.105 |
6c |
0.0273 |
0.133 |
4.872 |
7c |
0.077 |
0.032 |
0.416 |
Table (4):
Yield of biosurfactants in nutrient broth with hexane.
Organism |
Dry weight of organism (g) |
Weight of biosufactant (g) |
Yield |
|---|---|---|---|
1a |
0.0921 |
0.028 |
0.304 |
1b |
0.0155 |
0.007 |
0.452 |
1c |
0.042 |
0.014 |
0.333 |
2a |
0.0548 |
0.039 |
0.712 |
2b |
0.0396 |
0.018 |
0.455 |
3c |
0.1394 |
0.059 |
0.423 |
4a |
0.1125 |
0.055 |
0.489 |
4b |
0.1037 |
0.062 |
0.598 |
5a |
0.091 |
0.013 |
0.143 |
5c |
0.071 |
0.024 |
0.338 |
6a |
0.100 |
0.038 |
0.380 |
6b |
0.087 |
0.023 |
0.264 |
6c |
0.065 |
0.032 |
0.492 |
7c |
0.085 |
0.010 |
0.118 |
Exceptions were Bacillus pantothenticus (6c) and Staphylococcus spp. (5c) which showed higher yield in production medium without hexane and Micrococcus luteus (1a), Bacillus macquariensis (2b) and Staphylococcus spp. (5c) which showed higher yield in nutrient broth with hexane. This may be because yield of biosurfactants is also dependent on the organism, type of biosurfactant and the environmental conditions. [10].The extracted samples were confirmed to be biosurfactants by using oil spreading assay, thin layer chromatography and emulsification capacity assay.The effectiveness of the extracted biosurfactants was estimated using oil spreading assay and emulsification capacity assay. In oil spreading assay, all the biosurfactants showed oil displacement zones. Out of these, the ones from Corynebacterium kutsceri (6a), Streptococcus mitis (6b) gave displacements of 30 and 60 mm which are considerable with 35 mm displacement given by the SDS control. (Fig 2) Based on emulsification capacity assay, Corynebacterium xerosis (5a) and Bacillus pantothenticus (6c) showed higher emulsification indices of 10.811 with hexane and 27.027 with kerosene respectively when compared to 5.405 with hexane and 10.811 with kerosene given by the SDS control. (Fig 3)
Table (5):
Oil displacement zones.
Sample |
Diameter of Oil displacement zone (mm) |
|---|---|
SDS (Control) |
35 |
1a |
2 |
1b |
4 |
1c |
7 |
2a |
4 |
2b |
4 |
3c |
5 |
4a |
2 |
4b |
8 |
5a |
4 |
5c |
2 |
6a |
30 |
6b |
60 |
6c |
4 |
7c |
5 |
Table (6):
Emulsification index of the extracted samples.
| Sample | Height of the emulsion(mm) | |||
|---|---|---|---|---|
| Hexane | Kerosene | Hexane | Kerosene | |
| SDS (Control) | 2 | 4 | 5.405 | 10.811 |
| 1a | 2 | 2 | 5.405 | 5.405 |
| 1b | 1 | 1 | 2.703 | 2.703 |
| 1c | 2 | 1 | 5.405 | 2.703 |
| 2a | 2 | 2 | 5.405 | 5.405 |
| 2b | 2 | 3 | 5.405 | 8.108 |
| 3c | 1 | 2 | 2.703 | 5.405 |
| 4a | 2 | 2 | 5.405 | 5.405 |
| 4b | 2 | 2 | 5.405 | 5.405 |
| 5a | 4 | 3 | 10.811 | 8.108 |
| 5c | 2 | 2 | 5.405 | 5.405 |
| 6a | 1 | 2 | 2.703 | 5.405 |
| 6b | 3 | 1 | 8.108 | 2.703 |
| 6c | 2 | 10 | 5.405 | 27.027 |
| 7c | 2 | 2 | 5.405 | 5.405 |
Isolated organisms proved positive for biosurfactant production by using penetration assay, microplate assay and blood haemolysis technique. The biosurfactant yield was compared in production medium and nutrient broth with and without hexane. The yield was found to be higher in production medium with hexane and nutrient broth without hexane for most of the bacteria. The samples extracted were confirmed to be biosurfactants by using thin layer chromatography and measuring their effectiveness in terms of oil displacement zones formed and emulsification indices obtained with respect to that of SDS control.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
- N. Karanth, P. Deo, and N. Veenanadig, “Microbial production of biosurfactants and their importance,” Current Science, 1999; 77(1), pp. 116–126.
- R. Sen, “Biosurfactants,” in Advances in Experimental Medicine and Biology, 2010, pp. 3-11.
- K. B. Cheng, Z. Jian, and D. Z. Wang, “Emulsification properties of bacterial biosurfactants native to the Yellow River Delta on Hexadecane and Diesel Oil,” Biotechnology, 2008; 7(2): pp. 360-370.
- G. Bratbak and I. Dundas, “Bacterial dry matter content and biomass estimations.,” Applied and environmental microbiology, 1984; 48(4), pp. 755-757.
- R. Thavasi, S. Sharma, and S. Jayalakshmi, “Evaluation of Screening Methods for the Isolation of Biosurfactant Producing Marine Bacteria,” Petroleum & Environmental Biotechnology, 2011; pp. 1-6.
- R. A. Bayourni, B. M. Haroun, E. A. Ghazal, and Y. A. Maher, “Structural Analysis and Characteristics of Biosurfactants Produced by Some Crude Oil Utilizing Bacterial Strains,” Australian Journal of Basic and Applied Sciences, 2010; 4(8): pp. 3484-3498.
- N. Mathiyazhagan, K. Danashekar, and D. Natarajan, “Amplification of Biosurfactant Producing Gene ( rhlB ) from Pseudomonas aeruginosa isolated from oil contaminated soil,” International Journal of Pharma and Bio Sciences, 2(1): pp. 497-504.
- F. Nunizawati Daud, A. Yahya, M. Md Salleh, N. Suhaimi, and C. Hui Xuan, “Production of Biosurfactant by Locally Isolated Thermophilic Facultatively Anaerobic Bacteria strain B160,” Journal of Kustem, 2010; pp. 1-5.
- M. Abouseoud, R. Maachi, A. Amrane, and S. Boudergua, “Evaluation of different carbon and nitrogen sources in production of biosurfactant by Pseudomonas fluorescens,” Desalination, 2008; 223; pp. 143-151.
- L. Al-araji, R. N. Z. R. A. Rahman, M. Basri, and A. B. Salleh, “Microbial Surfactant,” Asia Pacific Journal of Molecular Biology and Biotechnology, 2007; 15(3), pp. 99-105.
© The Author(s) 2018. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.