ISSN: 0973-7510
E-ISSN: 2581-690X
The current study investigates the isolation identification of bioactive strain from fish collected from El-Burrllus region in Egypt. The objective of the current study is to identify and assess the biological profile of a new record strain in Egypt Bacillus maritimus. The molecular identification of the selected promising isolate and phylogenetic analysis confirmed the accurate identity of the isolate as Bacillus maritimus. The novelty of the present study was not due to only to the fact that the Bacillus maritimus isolate was the first record from brackish-water Egyptian fish gut, but also due to the fact that there is no previous study on the bioactivity of Bacillus maritimus and their efficiency as biogenic nanoparticles synthesis. The bioactivities for the Bacillus maritimus were investigated through different biochemical assays as antioxidant, anti-inflammatory and antimicrobial activity against six of the tested human pathogens namely; Staphylococcus aureus ATCC 25923, Escherichia coli ATCC 8739, Enterococcus faecalis ATCC 29212, Pseudomonas aeruginosa ATCC 15442, Klebsiella pneumoniae ATCC 13883 and Streptococcus pyogenes ATCC 19615. Bacillus maritimus bacteria were grown in nutrient broth for 24 h at 37°C and the optical density was determined at 600 nm using the UV-visible spectrophotometer. The optical density (OD) of the broth was adjusted to 0.5 with sterile phosphate-buffered saline, pH 7.4 (PBS). the bacterial isolate showed antioxidant, and anti-inflammatory activities. In addition the bacteria showed great reducing agent ability as a biogenic agent for the silver nanoparticles. The phytochemical screening of the Bacillus maritimus isolates have showed varieties of bioactive groups in addition to total phenolic compounds, flavonoids, tannic acid, carbohydrates, sulfated polysaccharides, total proteins and lipids. This study highlighted also that the Bacillus maritimus isolate could also produce novel secondary metabolites. The current study report for the first time the Bacillus maritimus in Egypt. Further studies are needed to isolate and characterize more bioactive strains from Egyptian Fishes that could possibly act as novel bioactive compounds source of medical and agricultures importance.
Bacillus maritimus, Egyptian Fishes, Biological Activities, Sliver Nanoparticles Biosynthesis
Microbial antagonism is a common phenomenon in nature (Kraemer et al. 2017), and plays an important role in reducing the opportunism of pathogens in animal aquatic gastrointestinal tract (Bahrndorff et al. 2016). Unlike other bacteria, pseudomonads develop a broad variety of exotoxic agents including full-spectrum antibiotics, bacteriocins, and other molecules of anti-competitors (Allen et al. 2010). The research on Bacillus sp. has been investigated recently as a probiotic species and as a pathogens preventive agent (Kim et al. 2019; Wosinska et al. 2019). Several researchers reported Bacillus strain isolates that has an antagonistic effect on pathogen in the intestine of fish (Kaew-on and Wanchaitanawong, 2015;.Mukherjee and Ghosh, 2019). Increasing antibiotic use leads to the formation of resistant pathogens (Aslam et al. 2018). And consequently the microbes develop resistance to current antimicrobial compounds very quickly (Cheesman et al. 2017).
Green nanotechnology proactively affects the design of nanomaterials and removes / minimizes the pollution caused by their manufacturing. It is constructed on the basis of green chemistry / engineering principles and focuses on the synthesis of nanoscale materials. Stability and impact on the environment of synthesized materials are assessed by a life cycle approach and should ensure less environmental change during product synthesis due to the use of silver nanoparticles (AgNPs) in different fields, including biomedical, biotechnological and environmental safety purposes. Researchers have recently used marine microorganisms as a possible biofactory for the specific and unique form of metallic nanomaterial development to be bio-fabricated (Manivasagan et al. 2016, et al. 2019).
Understanding the importance of developing innovative nanomaterial synthesis techniques would therefore result in less detrimental effects on human health and the ecosystem. Furthermore, a number of researchers are increasing their focus on the search for new bacteria and the development of innovative materials that can be used to develop new drugs. Recently, due to the development of newly synthesized nanomaterial’s of well-defined size and shape, the research field of nanomaterial’s has received a great deal of attention. Some natural properties of nanoparticles, such as high surface to volume ratios when their size is reduced, have been observed in previous studies (Oves et al. 2016).
Nanoparticles are typically synthesized by various physic- chemical methods, which are, however, costly and destructive to the environment. Therefore, the interest in developing cheap, clean, nontoxic and ecofriendly strategies for the synthesis of nanoparticles with wide spectrum activity is increasing every day (Oves et al. 2013). Recent eco-friendly approaches to synthetic chemistry have included some biological systems, such as yeast, fungi, etc. For nanoparticle synthesis. Some of the most widely used microorganisms for the development of silver nanoparticles (AgNPs) based on microbes include fungi (Fusarium spp. and, Aspergillus spp.)and bacteria (Enterobacteria spp. Pseudomonas spp. bacillus spp).
Nile tilapia (Oreochromis niloticus) is considered the world’s second-largest farmed fish species and for several reasons is rapid growth, high survival rate (Nakkina, 2016). In the aquaculture sector the use of probiotics is gaining more attention as an eco-friendly agent and safe products as antibiotics and disease-protective (Hoseinifar et al. 2018).
The objective of this study was to isolate a potential bioactive bacteria from Nile tilapia intestinal mucus and study their antioxidant, antimicrobial, anti-inflammatory and biogenic synthesis of sliver nanoparticles activity. The novelty of this study was not only because the Bacillus maritimus isolate was the first record of Egyptian fish from Brackish-Water Egyptian fish gut, but also due to the fact that there is no previous study on Bacillus maritimus bioactivity and its efficiency as a synthesis of biogenic nanoparticles.
Collection and preparation of Fish samples
About 10 live fish healthy tilapia were obtained from a brackish water farm in El-Burrllus lake at Kafer Elshekh, Egypt. The fish samples were then euthanized using a glove and aseptically dissected to collect weighted and homogenized intestinal contents in phosphate-buffered saline (PBS pH 7.2); Ten-fold serial dilutions of intestinal samples (TSA LOBA, India) were prepared and plated on tryptic soy agar.
Molecular Bacterial Identification
Bacterial isolates have been grown in test tubes that contain 10 ml of the autoclaved broth medium (Zimbro et al. 2009). Cultures were incubated in a cooling-heating incubator for 48 hours at 28oC. The bacterial cultures were then sent to the molecular biology research unit of the Assiut University for DNA extraction using the DNA / RRNA extraction kit Patho-gene-spin provided by Intron Biotechnology Company, Korea Before subsequent molecular analyzes .The extracted DNA was checked for purity for each bacterial isolate. DNA samples for the polymerase chain reaction (PCR) and 16S gene sequencing were shipped to SolGent Company, Daejeon, SouthKorea. PCR was carried out using 2 universal 27F primers (5’-AGAGTTTGATCCTGCTCAG-3’ and 1492R (5’-GGTTACCTTGTACGACTTT-3’), by White et al. (1990). The purified PCR products (amplicons) were reconfirmed by electrophoresis on 1 per cent agarose gel using a size nucleotide marker (100 base pairs). In the sense and antisense directions, bacterial amplicons were sequenced using 27F and 1492R primers with incorporation of dideoxynucleotides (dd NTPs) into the reaction mixture.
Phylogenetic Assessment
The sequence was carried out against the database of the National Center for Biotechnology Information (NCBI), using the Basic Local Alignment Search Tool (BLAST). The phylogenetic tree analysis was conceived using the Tamura-Nei model based on MEGA 5 version 6 Maximum Likelihood method (Tamura et al. 2013).
Total and extracellular biochemical and phytochemical analysis
The Microbial Culture and pathogenic isolates
The bacterium was grown for 24 hours at 37°C in the Nutrient Broth medium. A Bacillus maritimus supernatant was obtained by centrifuging the medium at 3000 rpm at 20°C for 15 minutes. The supernatant was then passed through a filter unit sterile of 0.22 μ-porous size. The filtrate was collected as a metabolite mixture and then maintained at 4°C. The “Agar Cup-Plate method” was used to investigate the antimicrobial activity of Bacillus maritimus. 20 mL of sterile Nutrient -agar medium has been poured into sterile petri-dishes and solidified. The pathogenic strains Staphylococcus aureus ATCC 25923, Escherichia coli ATCC 8739, Enterococcus faecalis ATCC 29212, Pseudomonas aeruginosa ATCC 15442, Klebsiella pneumoniae ATCC 13883 and Streptococcus pyogenes ATCC 19615. Bacillus maritimus bacteria were grown at 37°C for 24 h in nutrient broth and 600 nm of optical density was determined using the UV-visible spectrophotometer (Jenway, USA). The broth’s optical density (OD) has been adjusted to 0.5 with the addition and spread of sterile phosphate buffered saline, pH 7.4 (PBS) on the Nutrient-agar surface. Four bores were made using sterile borers on the medium in each plate. To the respective bores 100 μL of fresh supernatant have been added. The plates were kept for diffusion at 4 a.c. for 30 minutes and then incubated at 37 a.c. with appropriate positive for 24-48 hours and measured and observed zone of inhibition.
Preparation of the total bacterial and extracellular extracts
The isolate was incubated at 30°C in a medium of 500 mL brackish water nutrient broth (NB) with shaking at 220 rpm until the optical density reached 0.6. The cultivated medium was then centrifuged for 20 min at 4000 rpm at 4°C, the supernatant containing the antagonistic Bacillus maritimus substance, and the extracellular Bacillus maritimus products were extracted according to method as reported by Bordoloi et al. (2001). The supernatant was extracted 3 times, with equal ethanol and dichloromethane volumes (1:1). The extract was then evaporated under vacuum and lyophilized.
Preliminary phytochemical screening of different Bacillus maritimus extracts
Both total culture extract (TCE) and free cell extract (FCE) were screening for phytochemical screening for tannins, phlobatannis, saponins, flavonoids, alkaloids, quinine, coumarins, terpenoids, and glycosides were determined by a method reported previously (Trease and Evans, 1989; Sofowara,1993; Makkar et al. 1993).
Determination of total phenolic contents
Total phenolic compounds in the bacterial extract was determined by the method of Taga et al. (1984).
Determination of total flavonoid contents
Total flavonoid content was determined by a colorimetric method of Zhishen et al. (1999).
Determination of total carotenoid contents
Total carotenoid content was determined by the method reported by Lichtenthaler and Welburn (1983).
Determination of total protein contents
Total proteins were determined by means of the Biuret reaction as described by Gornall et al. (1949).
Determination of total lipid contents
Total lipid was determined by the method of Zollner and Kirsch (1962).
Determination of total proteins
Determination of total glucose
Total Glucose concentration in the sample was determined by Trinder (1969).
Determination of total sulfated polysaccharides
Sulfate content was determined turbidimetrically with barium chloride after acid hydrolysis as described by Dodgson et al. (1961).
Antioxidant activity by DPPH assay
The antioxidant activity of the extracts was determined based on radical scavenging activity by using 2, 2-diphenyl-1-picrylhydrazyl radical (DPPH) according to Amarowicz et al. (2000).
Anti-inflammatory activity by NO assay
Nitric oxide scavenging activity was estimated by the method described by Garrat (1964).
Biogenic synthesis of Silver nanoparticles using Bacillus maritimus
Production of biomass
The Bacillus maritimus were cultured in Nutrient broth medium to produce the cultured biomass for biogenic synthesis. The culture bacteria was incubated at 37°C and agitated at 200 rpm on shaker. The bacterial culture was harvested after 24 hours and centrifuged at 2000 rpm for 20 minutes. The supernatant was collected.
Biogenic Synthesis of silver nanoparticles
For the biogenic synthesis of silver nanoparticles, 30 ml of supernatant was mixed with 15 ml silver nitrate (AgNO3) solution (8 mM). The prepared solution was incubated at room temperature for 24 h in dark to prevent phytochemical reactions turning solution into dark brown from yellow solution after 24 was observed.
Characterization of nanoparticles
The appearance of a dark-brown color confirms the formation of silver nanoparticles and the efficient of bacterial biomass as potent reducing agent.
UV-Vis Spectroscopy analysis
Ultraviolet visible (UV_Vis) spectral analysis was done by using JENWAY 6800 Spectrophotometer in the range of 200_900 nm. The reduction and formation of pure silver ions synthesized by biological means was monitored by measuring the UV_Vis spectrum of the reaction mixture.
Scanning electron microscope
Scanning electron microscopy was performed by using a JEOL scanning electron microscope.
Transmission Electron Microscopy
Transmission electron microscopy was performed by using a JEOL instrument. TEM analysis was prepared by placing a drop of the nanoparticles suspension on a carbon-coated copper grid.
The Molecular and Phylogenetic analysis of the of Bacillus maritimus
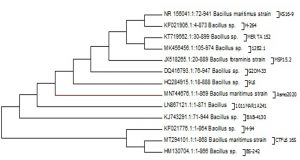
The evolutionary history was inferred by using the Maximum Likelihood method based on the Tamura-Nei model (Tamura and Nei, 1993). The tree with the highest log likelihood (-137.3418) is shown in Fig. 1. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the Maximum Composite Likelihood (MCL) approach, and then selecting the topology with superior log likelihood value. The analysis involved 12 nucleotide sequences. All positions containing gaps and missing data were eliminated. There were a total of 32 positions in the final dataset. Also the results of the BLAST for Bacillus maritimus is listed in table 1, furthermore the GC% were calculated in the whole sequence of Bacillus maritimus and shown in Table 2.
Table (1):
The taxonomical analysis of Bacillus maritimus.
Organism |
Blast Name |
Score |
No. of Hits |
Description |
|---|---|---|---|---|
Bacteria |
bacteria |
108 |
||
Terrabacteria group |
bacteria |
83 |
||
Bacillus |
firmicutes |
81 |
||
Bacillus maritimus |
firmicutes |
1581 |
5 |
Bacillus maritimus hits |
Bacillus sp. H-264 |
firmicutes |
1581 |
1 |
Bacillus sp. H-264 hits |
Bacillus sp. 9L6 |
firmicutes |
1578 |
1 |
Bacillus sp. 9L6 hits |
Bacillus foraminis |
firmicutes |
1576 |
19 |
Bacillus foraminis hits |
Bacillus sp. G2DM-33 |
firmicutes |
1572 |
1 |
Bacillus sp. G2DM-33 hitsacillu |
Bacillus sp. H-94 |
firmicutes |
1570 |
1 |
Bacillus sp. H-94 hits |
Bacillus sp. MER_TA_152 |
firmicutes |
1565 |
1 |
Bacillus sp. MER_TA_152 hits |
Bacillus persicus |
firmicutes |
1565 |
4 |
Bacillus persicus hits |
Bacillus sp. BS-242 |
firmicutes |
1565 |
1 |
Bacillus sp. BS-242 hits |
Bacillus sp. BAB-4130 |
firmicutes |
1563 |
1 |
Bacillus sp. BAB-4130 hits |
Bacillus sp. (in: Bacteria) |
firmicutes |
1559 |
12 |
Bacillus sp. (in: Bacteria) hits |
Bacillus sp. MER_TA_155 |
firmicutes |
1550 |
1 |
Bacillus sp. MER_TA_155 hits |
Bacillus sp. 281C102Y12 |
firmicutes |
1546 |
1 |
Bacillus sp. 281C102Y12 hits |
Bacillus jeotgali |
firmicutes |
1543 |
3 |
Bacillus jeotgali hits |
Bacillus caccae |
firmicutes |
1537 |
1 |
Bacillus caccae hits |
Bacillus sp. B013-7 |
firmicutes |
1537 |
1 |
Bacillus sp. B013-7 hits |
Bacillus sp. FJAT-27251 |
firmicutes |
1526 |
1 |
Bacillus sp. FJAT-27251 hits |
Bacillus sp. BJ-14 |
firmicutes |
1526 |
1 |
Bacillus sp. BJ-14 hits |
Bacillus sp. M7-8(2014) |
firmicutes |
1519 |
1 |
Bacillus sp. M7-8(2014) hits |
uncultured Bacillus sp. |
firmicutes |
1517 |
2 |
uncultured Bacillus sp. hits |
Bacillus novalis |
firmicutes |
1517 |
2 |
Bacillus novalis hits |
Bacillus sp. NRT6 |
firmicutes |
1515 |
1 |
Bacillus sp. NRT6 hits |
Bacillus horneckiae |
firmicutes |
1515 |
2 |
Bacillus horneckiae hits |
Bacillus sp. m8-51 |
firmicutes |
1513 |
1 |
Bacillus sp. m8-51 hits |
Bacillus sp. FJAT-27238 |
firmicutes |
1511 |
1 |
Bacillus sp. FJAT-27238 hits |
Bacillus sp. Bac39R |
firmicutes |
1509 |
1 |
Bacillus sp. Bac39R hits |
Bacillus sp. Bac38R |
firmicutes |
1509 |
1 |
Bacillus sp. Bac38R hits |
Bacillus sp. M71_S11 |
firmicutes |
1509 |
1 |
Bacillus sp. M71_S11 hits |
Bacillus sp. BS29 HS-2008 |
firmicutes |
1509 |
1 |
Bacillus sp. BS29 HS-2008 hits |
Bacillus sp. US7 HS-2008 |
firmicutes |
1507 |
1 |
Bacillus sp. US7 HS-2008 hits |
Bacillus sp. GB02-25 |
firmicutes |
1507 |
1 |
Bacillus sp. GB02-25 hits |
Bacillus sp. Marseille-P3608 |
firmicutes |
1506 |
1 |
Bacillus sp. Marseille-P3608 hits |
Bacillus sp. GB02-46B |
firmicutes |
1506 |
1 |
Bacillus sp. GB02-46B hits |
Bacillus sp. ADMK17 |
firmicutes |
1504 |
1 |
Bacillus sp. ADMK17 hits |
Bacillus sp. GB02-2A |
firmicutes |
1504 |
1 |
Bacillus sp. GB02-2A hits |
Bacillus sp. A46(2010) |
firmicutes |
1500 |
1 |
Bacillus sp. A46(2010) hits |
Bacillus sp. TSWCSN6 |
firmicutes |
1500 |
1 |
Bacillus sp. TSWCSN6 hits |
Bacillus drentensis |
firmicutes |
1498 |
1 |
Bacillus drentensis hits |
Bacillus sp. NSNT5 |
firmicutes |
1498 |
1 |
Bacillus sp. NSNT5 hits |
Bacillus sp. NMLR12 |
firmicutes |
1498 |
1 |
Bacillus sp. NMLR12 hits |
Nocardioides luteus |
high GC Gram+ |
1520 |
1 |
Nocardioides luteus hits |
Nocardioides sp. |
high GC Gram+ |
1520 |
1 |
Nocardioides sp. hits |
uncultured bacterium |
bacteria |
1526 |
25 |
uncultured bacterium hits |
Table (2):
The GC% comparing to other nucleotides.
Total count, all bases: |
867 |
Adenine (A) count: |
215 |
Thymine (T) count: |
162 |
Guanine (G) count: |
285 |
Cytosine (C) count: |
205 |
%G~C content: |
56.5 |
Fig. 1. Molecular Phylogenetic analysis by Maximum Likelihood method of Bacillus Maritimus strain Asmo2020 in gene bank
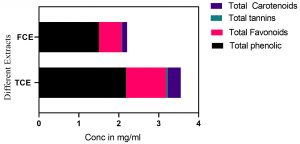
Fig. 2. The phytochemical analysis of total culture extract (TCE) and free cell extract (FCE) for total phenolics, total flavonoids, total tannic acids and total carotenoids.
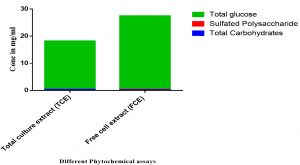
Fig. 3. The phytochemical analysis of total culture extract (TCE) and free cell extract (FCE) for total carbohydrates, sulfated polysaccharides, and total glucose.
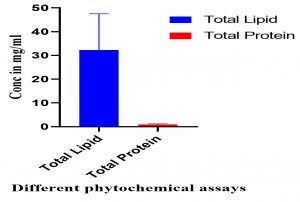
Fig. 4. The phytochemical analysis of total culture extract (TCE) and free cell extract (FCE) for total lipids and total proteins.
The phytochemical screening of different Bacillus maritimus extracts
The qualitative phytochemical screening showed that the total culture extracts have higher contents of phytochemical compounds such as phlobatannins, saponins, flavonoids,…etc as shown in Table 3.
Table (3):
Quantitive phytochemical screening of different Bacillus maritimus extracts.
Test |
Total culture extract (TCE) |
Free cell extract (FCE) |
|---|---|---|
Test for Phlobatannins |
+++ |
+ |
Test for Saponins |
+++ |
– |
Test for Flavonoids |
+++ |
+ |
Test for Alkaloids |
+++ |
+ |
Test for Quinones |
+++ |
– |
Test for Coumarin |
+++ |
– |
Test for Terpenoids |
+++ |
+ |
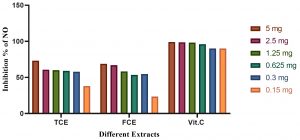
Fig. 5. The anti-inflammatory activities of vitamin C (Vit C), the total culture extract (TCE), and the free cell extract (FCE).
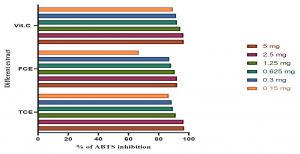
Fig. 6. The total antioxidants capacity using ABTS of vitamin C (Vit C), the total cultures extract (TCE), and the free cell extract (FCE).
The phytochemical screening for Total culture extract (TCE) and free cell extract (FCE)
The phytochemical analysis for total culture extract (TCE) and free cell extract (FCE) was presented in Figs. 4-6. The total culture extract (TCE) showed higher content of total lipid, total proteins, total carbohydrates, total phenolic compounds, total flavonoids, total tannic acids and total carotenoids. On contrast, the free cell extract (FCE) have higher concentrations of sulfated polysaccharides and total glucose. The phytochemical assays for TCE showed higher contents of total phenolic, flavonoid, total tannic acids and total carotenoids compared to free cell extract (FCE) as shown in Fig. 4.
Moreover, the screening for polysaccharides and free monosaccharide using total carbohydrates, sulfated polysaccharides and total glucose contents are shown in Fig. 5. The results indicated that the TCE have higher activity than FCE in total carbohydrates and total glucose where in sulfated polysaccharides the FCE has higher activity than TCE (Fig. 5).
The phytochemical screening for total lipids and proteins, on the other hand, are given in Fig. (6) and are revealing that the TCE have higher activity than FCE for total lipids and total proteins; respectively (Fig. 6).
The anti-inflammatory activity of different Bacillus maritimus extracts using NO assay
The anti-inflammatory results by using NO inhibition assay are shown in Fig. (5). The results showed that the vitamin C (Vit C) have higher anti-inflammatory activity as manifested by the inhibition of NO radicals. The results of the present study revealed that the total culture extract (TCE) have higher inhibition activity than Free cell extract (FCE).
The total antioxidants capacity using ABTS and DPPH assays
The total antioxidant capacity using ABTS assay is shown in Fig. 6. The present study cleared out that the total antioxidant capacity of TCE in 5 mg is higher than FCE and Vit C; while TCE in low concentration of 0.15 mg is lower than Vit C.
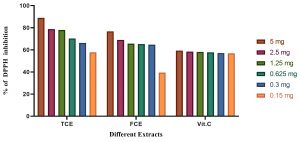
Fig. 7. The total antioxidants capacity using DPPH of vitamin C (Vit C), the total culture extract (TCE), and the free cell extract (FCE).
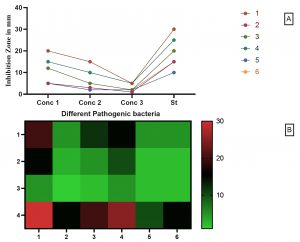
Fig. 8. The antimicrobial activity of Free Cell Culture compared to six different pathogenic strains.
The total antioxidant capacity using DPPH assay is illustrated in Fig. 7. The results revealed that the total antioxidant capacity of TCE in 5 mg is higher than FCE and Vit C.
The antimicrobial activity of Free Cell Culture
The antimicrobial activity of Free Cell Culture is shown in Fig. 8 where the conc 1, 2, 3, are 100, 50, 25 µL; respectively. The pathogenic strain from 1 up to 6 are: (1) Staphylococcus aureus ATCC 25923, (2) Escherichia coli ATCC 8739,(3) Enterococcus faecalis ATCC 29212,(4) Pseudomonas aeruginosa ATCC 15442, (5) Streptococcus pyogenes ATCC 19615, (6) Klebsiella pneumoniae ATCC 13883 compared to Ampicillin trihydrate. The highest antimicrobial activity of free cell culture (FCC) was against Staphylococcus aureus and Pseudomonas aeruginosa with 20 and 15 mm; respectively. On contrast, the Escherichia coli, Streptococcus pyogenes and Klebsiella pneumoniae showed the lowest activity with 5 mm inhibition zone for each strain Figs. 8-9.
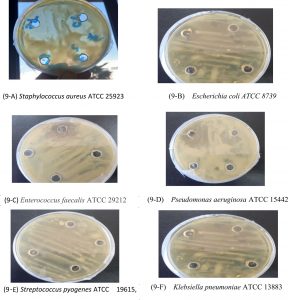
Fig. 9. Inhibition zone produced by the B. Maritimus . The Plates A.B.C.D.E.and F in Fig. 1 represent antibacterial activity six different pathogenic bacteria Staphylococcus aureus ATCC 25923, Escherichia coli ATCC 8739, Enterococcus faecalis ATCC 29212, Pseudomonas aeruginosa ATCC 15442, Klebsiella pneumoniae ATCC 13883 and Streptococcus pyogenes ATCC 19615), respectively.
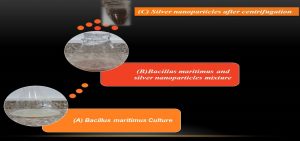
Fig. 10. The formation of Nano silver using Biogenic synthesis of Bacillus maritimus where (A) is the Bacillus maritimus , (B) is the Bacillus maritimus and silver nanoparticles mixture and (C) is the formation of Nano silver after centrifugation.
Fig. 11. UV Vis absorption spectra for both chemically and biologically synthesized silver nanoparticle.
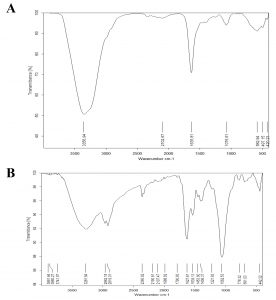
Fig. 12. (A ) FTIR spectra of Bacillus maritimus Culture and ( B ) : FTIR spectra of nano silver and Bacillus maritimus mixture.
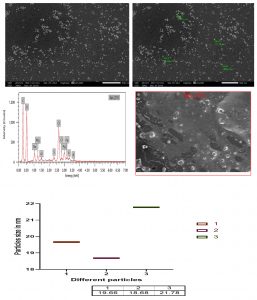
Fig. 13. SEM and EDX micrograph of the Ag-NPs produced with the Bacillus maritimus using nutrient broth media.
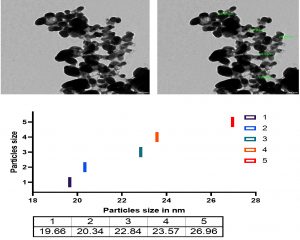
Fig. 14. TEM micrograph and diameter distribution graphic of the Ag-NPs obtained with the Bacillus maritimus using nutrient broth media.
Nanoparticles characterization
UV Vis spectroscopy analysis
The biogenic synthesis of silver nanoparticles by using bacillus maritimus was observed in the beginning by the change in color from Fig. 10A to dark brown in Fig. 10B followed by centrifugation and confirmed by optical measurements using UV Vis spectrophotometer. An absorbance peak maximum was observed at 420 nm (Fig. 11).
FT-IR analysis
The results of the FT-IR analysis for Bacillus maritimus culture and Bacillus maritimus-silver nanoparticles mixture are shown in Fig. 12 A & 12B and are listed in Table 4.
Table (4):
FT-IR analysis of function group in Bacillus maritimus culture and Bacillus maritimus culture silver nanoparticles mixture .
| FTIR peak for Bacterial culture for Bacillus maritimus | FTIR peak for Bacillus maritimus Silver nanoparticles mixture | ||||
|---|---|---|---|---|---|
| Peak | Function group | Peak | Function group | ||
| 3356.94 | Stretching vibration of O-H groups; | related to OH of phenolic compounds | 3891.64 | NH2 | |
| 2102.67 | 3860.27 | C-H stretching | |||
| 1635.81 | C=C N-H C=O |
vibration of aromatic rings and to the vibration of N-H of amines, C=O of amides and carboxylic groups |
3741.97 | O-H stretching | Alcohol |
| 1076.61 | stretching vibration of (C-O-C) | Ester group | 3281.64 | C-H tretching | alkyne |
| 582.94 | 2962.15 | N-H stretching | Amine salt | ||
| 497.15 | 2919.01 | O-H stretching | Alcohol Intermolecular | ||
| 420.23 | Silver | Ag metal | 2360.62 | O=C=O stretching | carbon dioxide |
| 2190.18 | CΞC stretching | Alkyne disubstituted | |||
| 2107.47 | N=C=S stretching | Isothiocyanate | |||
| 1988.82 | C=C=C stretching | Allene | |||
| 1790.83 | C=O stretching | Acid halide | |||
| 1627.81 | N-H bending | Amine | |||
| 1539.13 | N-O stretching/ C=C).. |
Nitro compound/ related to aromatic ring deformations and to flavonoids and aromatic rings (stretching of aromatic C=C) |
|||
| 1453.60 | C-H bending alkane | methyl group | |||
| 1406.37 | C-F stretching | Fluoro compound | |||
| 1221.83 | C-O stretching | vinyl ether OR C-O group of polyols, such as hydroxyflavonoids |
|||
| 1062.53 | S=O stretching | Sulfoxide OR secondary alcohols |
|||
| 778.32 | C-Cl stretching | halo compound | |||
| 691.03 | C-Br stretching | halo compound | |||
| 442.32 | Ag metal | Ag metal | |||
Scanning electron microscope
The scanning electron microscope micrograph confirm that the silver in Nano scale the range of nanoparticles from 18.68 up to 21.78 as in Fig. 13. The EDX result also confirm the presence of carbon, oxygen, chlorine, sulfur and sodium beside the silver.
Transmission electron microscope
The Transmission electron microscope micrograph confirmed that the Sliver in Nano scale 19.66 up to 26.96 nm.
The genus Bacillus consists of endospore forming rod-shaped bacteria that are members of the phylum firmicutes. Because species within the genus are largely common inhabitants of soil and aquatic sediments, they are widespread in nature and are found in virtually any environment. (Rooney et al. 2009). The reason Bacillus exploited for various biotechnological applications, because of their genetic variation lead to many of which were used for biotechnological and industrial applications (Price et al. 2007). The importance of Bacillus species in medicine, environment and industrial applications is reflected in the large number of Genome sequences deposited in the GenBank database 2017 (Anbu, et al. 2015).
Hernandez-Gonzalez et al, (2018) reported that by November 2017, more than 2000 complete and draft genome sequences of this genus had been deposited in the GenBank database. Most of these Bacillus sequences are either medically relevant strains, or are isolated from terrestrial environments. The number of Bacillus genome sequences publicly available isolated from aquatic environments has recently increased.
The change is a preliminary tool for the confirmation that the particles in nano size as well as the ability of bacterial extract in nanoparticle biogenic synthesis. Turn the culture to brown color in the reaction mixture (silver nitrate and bacterial culture) confirm the formation of silver nanoparticles (Fig.13 and 14). The reaction mixture turned into brown color after 48 hr of incubation. The occurrences and turning to brown color indicate the formation of silver nanoparticles in the culture of reaction mixture and the efficacy of the broth media as reducing agent the Ag ions as in Fig. 10 and that with agreement with previous studies (Skiba and Vorobyova, 2019).
The information about the formation of silver nanoparticles using Bacillus species is confirmed in many previous studies. Reduction of silver ions and formation of silver may be due to the existence of bioactive molecules for examples polyphenolic compounds such as phenolic and flavonoids (Kaurinovic and Vastag, 2019). Other bioactive molecules were also confirmed to be excellent reducing agent in nanoparticles formation such as protein molecules, enzyme for examples nitrate reeducates which reported as good regulating agent in the nanoparticles biosynthesis and particularly for silver nitrate (Lee and Jun 2019).
The present study revealed a GC ratio of the bacillus martiums of 56.5% (Table 2). This finding is in line with previous studies confirming that firmicutes have GC content of up to 55 percent (e.g. Geobacillus thermocatenulatus). Some firmicutes make resistant progeny called endospores, while others can reproduce by binary fission only. It’s obvious that firmicutes are as varied as important. Different strains were isolated from Nile Tilapia in the present study, among which one strain named Asmo2020 (Fig. 1) showed excellent antimicrobial activity. The percentage identity is to estimate how closely the strain profile matches the taxon relative to the Bacillus Maritimus. The strain’s 16S rRNA sequence showed that the alignment similarity to Bacillus maritimus strain KS16-9 and Bacillus sp. was > 99 per cent. H-264 16S (Fig. 1).These species are considered to be members of the Bacillus group, and their similarity has been improved based on the 16S rRNA sequence. Chun and Bae (2000) reported that if the identity of the sequence showed similarity above 99 %. It could definitely conclude that the two strains belong to the same species in the current study it was identified as Bacillus maritimus. In addition, they reported that strains are classified as the same genus or the same family if the identity is higher than 97 %. Therefore the isolated strain was confirmed as Bacillus maritimus based on the morphological, physiological, biochemical characteristics, phylogenetic position, and 16S rRNA gene sequences.. The antioxidant capacity for the Bacillus Maritimus culture was investigated using two antioxidant model DDPH and ABTS; respectively (Figs 6 & 7). In the present study, Bacillus maritimus culture showed high antioxidant activity compared to vitamin C in agreement with other previous study (Elshaghabee et al. 2017). This is may be due to the presence of different bioactive compounds such as phenolic, flavonoids, carotenoids, fatty acids and lipids, carbohydrates and sulfated polysaccharide, amino acids and proteins (Figs 2 & 4) (Abd El Moneam et al. 2017a-e & 2018; Nabil-Adam et al. 2019 & 2020; Shreadah et al. 2017 & 2018a-c). Moreover, the current study revealed high anti-inflammatory activity of Bacillus maritimus compared to vitamin C (Fig. 5). This can be attributed to the presence also of over mentioned bioactive secondary metabolites (Abd El Moneam et al. 2017a-e & 2018; Nabil-Adam et al. 2019 &2020; Shreadah et al. 2017 & 2018a-c). It has been reported also that polysaccharides and sulfated polysaccharides particularly derived from Bacillus induce anti-inflammatory cytokine leading to suppressing T cell-dependent immune responses by inhibiting T cell activation (Xia et al. 2019). One reason behind the anti-inflammatory effect of Bacillus species is the production of the bioflocculant QZ-7 (Abu Tawila et al. 2018).
Class Bacilli organisms are capable of producing larger amounts of polysscharide compared to lactic acid bacteria (Ruiz Rodriguez, et al. 2019). In an earlier study, a bioflocculant-producing thermotolerant species Bacillus subtilis F9 was isolated from wastewater sludge (Giri et al. 2015). NO is a signaling molecule which has important functions in inflammatory pathogenesis. It appears to be an anti-inflammatory in normal condition, but NO functions as a pro-inflammatory mediator thus promoting inflammation in abnormal physiological states through its overproduction (Chen, et al. 2017). Thus it has been shown that NO inhibitors play a vital role in controlling inflammatory diseases. The present study showed that, in agreement with that of Paynich et al. (2017), different concentrations of Bacillus maritimus (mg / ml) led to maximum inhibition of NO production. Bacillus’ anti-inflammatory effect was investigated against pretreated Bacillus Caco-2 cells, which resulted in the release of IL-8 to various pro-inflammatory signals IL-1β, deoxynivalenol, or flagellin. In all stressed conditions tested, Bacillus was able to decrease IL-8 production considerably. Mechanistically, during IL-1β exposure Bacillus maritimus appeared to limit the nuclear translocation of NF-kB by pre-exposure by preventing degradation of IκB (Rhayat et al. 2019). The effects of Bacillus 29784 were observed independently with supernatant and cells, but to a lesser extent than with the combination, indicating that both secreted metabolites and cell-associated compounds are thus likely to be attributed to them. In addition, Bacillus 29784 significantly reduced the upregulation of the iNOS protein levels under inflammatory conditions further underlining its anti-inflammatory intestinal potential. Previous studies have reported an immunomodulatory effect of soluble secondary metabolites of Bacillus, inhibiting the signaling pathway of compounds associated with NF-yB, or / and bacterial membranes (Valdivieso-Ugarte et al. 2019). As antibiotics have been known for decades, the potential of Bacillus species to produce a broad diversity of bioactive secondary metabolites acts. It is well known that at least 4–5 per cent of the genome of Bacillus acts as antimicrobial compounds. The nature of the mainly antimicrobial peptides in these compounds. Their structures are hydrophobic cyclic, and for example intermolecular thioether bonds and/or D-amino acids (AA) properly contain peculiar moieties. In addition, volatile compounds also have a large family of antimicrobials that exhibit variety of functional and metabolic roles in antimicrobial compounds. the main biosynthesis pathway and the secondary metabolites bioactivity clades of antimicrobial compounds within the Bacillus group: i.e., the polyketides (PKs), (Enzymes and bacteriocins),the ribosomal peptides (RPs) the non-ribosomal peptides (NRPs) and the volatiles (Caulier, et al. 2019).
The growing antimicrobial resistances in modern clinical pathology and medicine interest and focus on bioactive compounds from natural products in order to generate new and novel antimicrobials in general and particular antibiotics (Fiedler et al. 2019). Bacilli have capability able to synthesis more than two dozen of antibiotics with variety in function and structures as they well known for the production of a vast array of structurally unrelated antimicrobial compounds. The formation of these antibiotics is depending on the environmental characteristics, the metabolic pathways including lipopeptides such as surfactin, iturin, bacteriocins, bacteriocin and fengycins (Ramachandran, et al. 2014). one of known antimicrobial mechanisms against pathogen is that lipopeptides allows certain Bacillus to change their outer surface which form biofilm through allow them to regroup together to proliferate and distributed in the area.
Formation of silver nanoparticles is easily detected by spectroscopy because the colored nanoparticle solution shows a peak ~400 nm. In the current study, the optical density of the solution was measured by using a Jenway spectrophotometer and a transmission electron microscopy was applied. The result of micrograph of TEM analyzed of biogenic silver nanoparticles using Bacillus maritimus. as reducing agent showed that nanoparticles is essentially spherical furthermore they are well-disperse and size ranging from 18.68-26.9nm. The Ag nanoparticles have particle size in the range of 18.68- 26.96 nm. However, in the current study we investigate the possible mechanism of metal and microbes interaction which may be due to structural specificity of the cell of microbes and how metal availability influences microbia. In this paper we report the culture form bacteria Bacillus for synthesis of silver nanoparticles using the culture filtrate figure. the Bacillus maritimus able to biogenic synthesis of silver nanoparticles which confirmed using UV, FT-IR, SEM and TEM (Fig. s 11-14 & table 4). Image showed that the nano-silver particles shape and crystal structure of silver nanoparticles as well as size of the particles. Furthermore, the EDX analysis confirm this finding with presence of other element such as sodium, chlorine, magnesium Sulfur and potassium in addition to carbon and oxygen and that due to culture of bacillus that contain all that also the metabolite of the Bacillus maritimus. as in Fig. 13 The UV-Vis absorption spectra (Fig. 11). The control samples were subjected to the solution of silver nitrate have been shown the wide spectrum range around 400 nm where in case of nanoparticles formation of silver nitrate a broad resonance confirm the aggregation of the silver nanoparticles in the solution (Adebayo-Tayo et al.2019). Many studies have confirmed the advantages of biogenic synthesis over physical, chemical, since these methods attach to toxic substances on the surface of nanoparticles, in addition to the instability of nanoparticles, in addition to producing hazardous as by-products (Singh et al. 2018). In addition, Igiri et al. (2018) investigated the microorganism mechanism that reduces hazard and toxic substance by accumulating metallic ions to reduce its toxicity. Reduction of nanoparticles occurs intracellularly with different reduction of bioactive compounds that have been extracellularly produced by different metabolites, inside the cell and/or on the cell wall. The biogenic synthesis of bioactive compounds such as polyphenolic (flavonoids, phenolic and tannic) carotenoids and water-soluble biomolecules produces stable nanoparticles, additional active surface area without formation for interaction in the biological environment, hazardous by-product (Singh, et al. 2018; Siddiqi et al. 2018).
One possible mechanism behind the stability of nanoparticles is the bioactive compounds that make capping layers that usually affect the biological activity of nanoparticles with particular and highly stable long-term stability during biosynthesis. Previous study confirmed that both nucleation and surface growth undergoes the biosynthesis of nanoparticles from microorganisms. In both positive gram negatives such as Streptomyces and gram negative such as Bacillus, antimicrobial metabolites are widely produced among genus bacteria in many bacterial genera (Chen et al. 2019).
Bacillus strains in particular are widely distributed in the environment, and have also been isolated from various environmental sources. Most species of Bacillus members consider good sources of biologically active compounds, in particular enzyme inhibitors, therapeutic proteins, antibiotics, and bioactive pharmacological agents (Perez et al. 2017). They produce bioactive metabolites with vast numbers of secondary antimicrobial metabolites, as well as bio-peptides with varied diversity and chemical structures (Andryukov et al. 2019).). Although most of these compounds are bioactive against Gram-positive bacteria, some of them have a wide range of bioactivity towards Gram-negative bacteria and filamentous fungi, such as bacteriocins, bacteriocin-like substances and lipopeptides (Silva et al. 2018).
Although several studies have been conducted on the isolation and characterization of antimicrobial metabolite producing Bacillus from different origins, few or no studies were evaluations of their anti-microbial performance from Bacillus maritimus. Seven peaks were shown in FT-IR profile (Fig. 5A & 5B). The position were at 3356.94, 2102.67, 1635.81, 1076.61, 582.94, 497.15 and 420.23 cm−1 ; respectively for Bacillus maritimus culture whereas the Bacterial culture AgNPs reaction mixture at the end of 30 hours showed 23 spectral bands (Fig. 10 B) at the following wave numbers 3891.64, 3860.27, 3741.97, 3281.64, 2962.15, 2919.01, 2360.62, 219018, 2107.47, 1988.82, 1790.83, 1627.81, 1539.13, 1453.60, 1406.37, 1221.83, 1062.53, 778.32, 691.03 and 442.32 cm−1. With respect to the AgNPs reaction mixture FT-IR analysis after 30 hours (Fig. 10B), the absorbance bands located between 3000–3500 cm−1 were assigned to the stretching vibrations of amine groups and hydroxyl (Geana et al. 2019). Illustrated distinctive broad spectral bands around 3356.94 -3891.64 cm−1 are characteristic of the O-H stretching vibration type of the hydroxyl functional group in polyphenols and N-H stretching vibrations of amino acids , peptides, and proteins in primary and secondary amines. The peak change in the spectral profile of AgNPs could be attributed to the interactions between these chemical functional groups and AgNPs. Protein molecules have been documented collaborating with AgNPs via free amide groups (Hamouda et al. 2019). FT-IR data showed that the amide protein linkage possessed a higher potential for joining silver and thus forming protein covering around AgNPs to prevent agglomeration and thus stabilize the medium (Elamawi et al. 2018).The involvement of hydroxyl groups in the bioreduction process could be confirmed in this study by shifting the deformation vibration of O-H positioned at about wave number 1100 cm−1 in the AgNP preparations as well as the observed shift from 1100 to 1132 cm−1 which did not appear in the crude extract of Bacillus maritimus. The FTIR showed spectral peaks at 2962.15, 2919.01, 2360.62, 2190.18, and 2107.47 cm−1 may refer to N-H bending vibration and hydrocarbon chains stretching C-H vibration (Coates, 2000).The spectral vibration of the aldehyde group (C = O) was illustrated by a wave length of 1740 cm−1, whereas the peak at a range of 1648–1621 cm−1 could be attributed to amides (N-H) stretching in addition to peptide bond and C = C stretching which may involve the stabilization of nanoparticles by proteins as explained by Castro et al. (2013), who investigated the biosynthesis of AgNPs using aqueous extract of either t. The spectral band at 1384 cm−1 could be designated to the residual amount of AgNO3, which appeared only in the three AgNPs preparations. The data revealed the presence of 1221,83cm−1 absorption bands that may have been attributed to vibration of proteins as a stabilizing agent is attributed to free amine groups or cysteine groups (Coates, 2000). The peaks in the Bacillus maritimus extract IR profile were positioned at wave numbers 1551 and 1415 cm−1 and disappeared in the AgNPs preparations. The bending region of the aliphatic chain was indicated by spectral peaks of 616–672, 522–575 & 441–493 cm−1 In addition, bands assigned at 1381, 1269, 1132 & 1034 cm−1 can be attributed to either sulfur or phosphorus function groups that may attach silver and perform both nanoparticles capping and stabilisation processes (Castro et al. 2013).
The current study investigated the potential bioactivity of new recorded strain in Egypt from Bacillus species (Bacillus maritimus). isolated from Nile tilapia and their ability to produce variety of secondary metabolites and potent activity them further investigation needed to explore the new strain in-vitro and in-vivo studies.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
ANA performed the experiments, involved in planning and supervision, processed the experimental data, performed the analysis, drafted the manuscript and designed the Figures. MAS interpret the results and worked on the manuscript. Both the authors read and apporved the manuscript for publication.
FUNDING
None.
ETHICS STATEMENT
Not applicable.
AVAILABILITY OF DATA
All datasets generated or analyzed during this study are included in the manuscript and/or the Supplementary Files.
- Abd El Moneam NM, Al-Assar SA, Shreadah MA, Nabil-Adam A. Isolation, Identification and Molecular Screening of Psudomance Sp. Metabolic pathways NRPs and PKS associated with the Red Sea sponge, Hyrtios aff. Erectus, Egypt. J pure Appl Microbiol. 2017a;11(3):1299-1311.
Crossref - Abd El Moneam NM, Assar SA,, Shreadah MA, Nabil-Adam A. The hepatoprotective effect of Hyrtios aff. Erectus sponge isolated from the Red sea extract against the toxicity of Persistent organic pollutants (POPs) from Sediments of Lake Mariout. Journal Biotechnology and Biotechnological Equipment. 2018b;32(3):734-743.
Crossref - Abd El Moneam NM, Shreadah MA, Assar SA, Nabil-Adam A. Protective role of antioxidants capacity of Hyrtios aff. Erectus sponge extract against mixture of persistent organic pollutants (POPs)-induced hepatic toxicity in mice liver: biomarkers and ultrastructural study. Environmental Science and Pollution Research. 2017c.
Crossref - Abd El Moneam NM, Yacout GA, Aboul-Ela HM, Shreadah MA. Hepatoprotective Activity of Chitosan Nanocarriers Loaded With the Ethyl Acetate Extract of Astenotrophomonas sp. Bacteria Associated with the Red Sea Sponge AmphimedonOchracea In CCl4 Induced Hepatotoxicity in Rats. Advances in Bioscience and Biotechnology (ABB). 2017d;8(1):27-50.
Crossref - Nabil-Adam A, Shreadah MA. Red Algae Natural Products for Prevention of Lipopolysaccharides (LPS)- induced liver and kidney inflammation and Injuries. Biosci Rep. 2020 Nov 27:BSR20202022.
Epub ahead of print. PMID: 33245092.
Crossref - Abd El Moneam NM, Shreadah MA, Assar SA, N. J. De Voogd, Nabil-Adam A. Hepatoprotective effect of Red Sea sponge extract against the toxicity of a real-life mixture of persistent organic pollutants.Biotechnol & Biotechnol Eq. 2018;32(3):734-743.
Crossref - Abu Tawila ZM, Ismail S, Dadrasnia A, Usman MM. Production and Characterization of a Bioflocculant Produced by Bacillus salmalaya 139SI-7 and Its Applications in Wastewater Treatment. Molecules (Basel, Switzerland). 2018;23(10):2689.
Crossref - Adebayo-Tayo B, Salaam A, Ajibade A. Green synthesis of silver nanoparticle using Oscillatoria sp. extract, its antibacterial, antibiofilm potential and cytotoxicity activity. Heliyon. 2019;5(10):e02502.
Crossref - Allen HK, Donato J, Wang HH, Cloud-Hansen KA, Davies J, Handelsman J. Call of the wild: antibiotic resistance genes in natural environments. Nat Rev Microbiol. 2010;8:251-259.
Crossref - Amarowicz R, Naczk M, Zadernowski R, Shahidi F. Antioxidant activity of condensed tannins of beach pea, Canola hulls, evening primrose, and faba bean. J FOOD LIPIDS. 2000;7:195-205.
Crossref - Anbu P, Gopinath SCB, Chaulagain BP, Tang TH, Citartan MM. Microbial Enzymes and Their Applications in Industries and Medicine 2014. BioMed Research International. 2015;2015:816419.
Crossref - Andryukov B, Mikhailov V, Besednova N. The Biotechnological Potential of Secondary Metabolites from Marine Bacteria. J Mar Sci Eng. 2019;7(6):176;
Crossref - Aslam B, Wang W, Arshad MI, et al. Antibiotic resistance: a rundown of a global crisis. Infect Drug Resist. 2018;11:1645-1658.
Crossref - Bahrndorff S, Alemu T, Alemneh T, Nielsen JL. The Microbiome of Animals: Implications for Conservation Biology. Int J Genomics. 2016;2016:5304028.
Crossref - Bordoloi G, Kumari B, Guha A, et al. Isolation and structure elucidation of a new antifungal antibiotic produced by Streptomyces Sp. 201. Biosci Biotechnol Biochem. 2001;65:1856-1858.
Crossref - Castro L, Blazquez ML, Munoz JA, Gonzalez F, Ballester A. Biological synthesis of metallic nanoparticles using algae. IET Nanobiotechnol. 2013;7(3):109-116.
Crossref - Caulier S, Nannan C, Gillis A, Licciardi F, Bragard C, Mahillon J. Overview of the Antimicrobial Compounds Produced by Members of the Bacillus subtilis Group. Front Microbiol. 2019;10:302.
Crossref - Cheesman MJ, Ilanko A, Blonk B, Ian E, Cock IE. Developing New Antimicrobial Therapies: Are Synergistic Combinations of Plant Extracts/Compounds with Conventional Antibiotics the Solution? Pharmacogn Rev. 2017;11(22):57-72.
Crossref - Chen L, Deng H, Cui H, et al. Inflammatory responses and inflammation-associated diseases in organs. Oncotarget. 2017;9(6):7204-7218.
Crossref - Chun J, Bae KS. Phylogenetic analysis of Bacillus subtilis and related taxa based on partial gyr A sequences. Antonie van Leeuwenhoek. 2000;78(2):123-127.
Crossref - Coates J. Interpretation of infrared spectra, a practical approach. Encyclopedia of Analytical Chemistry. 2000;12:10815-10837.
- Dodgson KS. Determination of Inorganic Sulphate in Studies on the Enzymic and Non-Enzymic Hydrolysis of Carbohydrate and Other Sulphate Esters. Biochem J. 1961;78:312-319.
Crossref - Elshaghabee FMF, Rokana N, Gulhane RD, Sharma C, Panwar H. Bacillus As Potential Probiotics: Status, Concerns, and Future Perspectives. Front Microbiol.2017;8:1490.
Crossref - Fiedler G, Schneider C, Igbinosa EO, et al. Antibiotics resistance and toxin profiles of Bacillus cereus-group isolates from fresh vegetables from German retail markets. BMC Microbiol. 2019;19:250
Crossref - Garrat DC. The Quantitative analysis of Drugs. Chapman and Hall Ltd., Japan. 1964;3:456-458.
Crossref - Geana EI, Ciucure CT, Apetrei C, Artem V. Application of Spectroscopic UV-Vis and FT-IR Screening Techniques Coupled with Multivariate Statistical Analysis for Red Wine Authentication: Varietal and Vintage Year Discrimination, Molecules. 2019;24(22):4166.
Crossref - Giri SS, Harshiny M, Sen SS, Sukumaran V, Park SC. Production and characterization of a thermostable bioflocculant from Bacillus subtilis F9, isolated from wastewater sludge. Ecotoxicol Environ Saf. 2015;121:45-50.
Crossref - Gornall AG, Bardawil CJ, David MM. Determination of serum proteins by means of the biuret reaction. J Biol Chem. 1949;177(2):751-766.
- Hamouda RA, Hussein MH, Abo-elmagd RA, Bawazir SS. Synthesis and biological characterization of silver nanoparticles derived from the cyanobacterium Oscillatoria limnetica. Scientific Reports. 2019;9:13071.
Crossref - Hernandez-Gonzalez IL, Moreno-Hagelsieb G, Olmedo-Alvarez G. Environmentally-driven gene content convergence and the Bacillus phylogeny. BMC Evol Biol. 2018;18:148.
Crossref - Hoseinifar SH, Sun YZ, Wang A, Zhou Z. Probiotics as Means of Diseases Control in Aquaculture, a Review of Current Knowledge and Future Perspectives. Front Microbiol. 2018;9:2429.
Crossref - Igiri BE, Okoduwa SIR, Idoko GO, Akabuogu EP, Adeyi AO, Ibe K, Ejiogu IK. Toxicity and Bioremediation of Heavy Metals Contaminated Ecosystem from Tannery Wastewater: A Review. Journal of Toxicology. 2018;2018:2568038.
Crossref - Kaew-on S, Wanchaitanawong P. Antagonistic activity against Fish pathogens and in vitro probiotic properties of lactic acid bacteria. Proceeding of International Conference On Research, Implementation and Education of Mathematics And Sciences, Yogyakarta State University. 2015.
- Kaurinovic B, Vastag D. Flavonoids and Phenolic Acids as Potential Natural Antioxidants, Antioxidants, Emad Shalaby. Intech Open. 2019. doi: 10.5772/intechopen.83731. Available from: https://www.intechopen.com/books/antioxidants/flavonoids-and-phenolic-acids-as-potential-natural-antioxidants
- Kim JA, Bayo J, Cha J, et al.. Investigating the probiotic characteristics of four microbial strains with potential application in feed industry. PLoS ONE. 2019;14(6):e0218922.
Crossref - Kraemer SA, Jean-Paul R, Soucy JR, Kassen R. Antagonistic interactions of soil pseudomonads are structured in time. FEMS Microbiology Ecology. 2017;93(5).
Crossref - Lee SH, Jun BH. Silver Nanoparticles: Synthesis and Application for Nanomedicine. Int J Mol Sci. 2019;20(4):865.
Crossref - Lichtenthaler HK, Wellburn AR. Determinations of total carotenoids and chlorophylls a and b of leaf extracts in different solvents. Biochemical Society Transactions. 1983;11:591-592.
Crossref - Makkar HPS, Blummel M, Borowy NK, Becker K. Gravimetric determination of tannins and their correlations with chemical and protein precipitation methods. Journal of the Science of Food and Agriculture. 1993;61:161-165.
Crossref - Manivasagan P, Nam SY, Oh J. Marine microorganisms as potential biofactories for synthesis of metallic nanoparticles, Crit Rev Microbiol. 2016; 42(6): 1007-1019.
Crossref - Mukherjee A and Ghosh K. Antagonism against fish pathogens by cellular components and verification of probiotic properties in autochthonous bacteria isolated from the gut of an Indian major carp, Catla catla (Hamilton). Aquac Res. 2019; 47(7):2243–2255.
- Nabil-Adam A, Assar SA, Shreadah MA, Abd El Moneam NM. Pesudomance sp. Bacteria Associated with Marine Sponge as a Promising and Sustainable Source of Bioactive Molecules. Curr Pharm Biotechnol. 2019;20(1).
Crossref - Nabil-Adam A, Shreadah MA, El Moneam NMA, El-Assar SA. Various In Vitro Bioactivities of Secondary Metabolites Isolated from the Sponge Hyrtios aff. Erectus from the Red Sea Coast of Egypt. Turk J Pharm Sci. 2020;17(2):127-135.
Crossref - Nakkina M. Study of Growth Rate in Nile Tilapia (Oreochromis niloticus). J Aquac Res Development. 2016;7:8.
Crossref - Oves, M., Rauf, M. A., Hussain, A., Qari, H. A., Khan, A., Muhammad, P., Rehman, M. T., Alajmi, M. F., & Ismail, I. (2019). Antibacterial Silver Nanomaterial Synthesis From Mesoflavibacter zeaxanthinifaciens and Targeting Biofilm Formation. Frontiers in pharmacology, 10, 801.
Crossref - Oves, M., Qari, H. A., Felemban, N. M., Khan, M. Z., Rehan, Z. A., Ismail, I. M. I. (2016). Marinobacter lipolyticus from Red Sea for lipase production and modulation of silver nanomaterials for anti-candidal activities. IET Nanobiotechnol. 11 (4), 403–410.
Crossref - Oves, M., Khan, M. S., Zaidi, A., Ahmed, A. S., Ahmed, F., Ahmad, E., Sherwani, A., Owais, M., & Azam, A. (2013). Antibacterial and cytotoxic efficacy of extracellular silver nanoparticles biofabricated from chromium reducing novel OS4 strain of Stenotrophomonas maltophilia. PloS one, 8(3), e59140.
Crossref - Paynich ML, Jones-Burrage SE, Knight KL. Exopolysaccharide from Bacillus subtilis Induces Anti-Inflammatory M2 Macrophages That Prevent T Cell-Mediated Disease. J immunol. 2017;198(7):2689- 2698.
Crossref - Perez KJ, Viana JdS, Lopes FC, et al. Bacillus spp.Isolated from Puba as a Source of Biosurfactants and Antimicrobial Lipopeptides. Front Microbiol. 2017;8:61.
Crossref - Price NP, Rooney AP, Swezey JL, Perry E, Cohan FM. Mass spectrometric analysis of lipopeptides from Bacillus strains isolated from diverse geographical locations. FEMS Microbiol Lett. 2007;271:83-89.
Crossref - Rhayat L, Maresca M, Nicoletti C, et al. Effect of Bacillus subtilis Strains on ntestinal Barrier Function and Inflammatory Response. Front Immunol. 2019;10:564.
Crossref - Ramachandran R, Chalasani AG, La R, Roy U. A BroadSpectrum Antimicrobial Activity of Bacillus subtilis RLID 12.1. The Scientific World Journal. 2014;2014:968487.
Crossref - Rooney AP, Price NPJ, Ehrhardt C, Swezey JL, BannanJD. Phylogeny and molecular taxonomy of the Bacillus subtilis species complex and description of Bacillus subtilis subsp. inaquosorum subsp. nov. Int J Syst Evol Microbiol. 2009;59:2429-2436.
Crossref - Rodriguez LGR, Mohamed F, Bleckwedel J, Medina R, De Vuyst L, Hebert EM, Mozzi F. Diversity and Functional Properties of Lactic Acid Bacteria Isolated From Wild Fruits and Flowers Present in Northern Argentina. Front Microbiology. 2019;10:1091.
Crossref - Shreadah MA, Abd El Moneam NM, Al-Assar SA, Nabil-Adam A. The Ameliorative Role of a Marine Sponge Extract against Mixture of Persistent Organic Pollutants induced Changes in Hematological Parameters in Mice. Expert Opinion environmental Biology. 2017;6(2).
Crossref - Shreadah MA, Abd El Moneam NM, Assar SA, Nabil Adam A. Isolation, Identification and Screening of Metabolic Pathways NRPs and PKS of Psudomance Sp Associated with the Marine Sponge, Hyrtios aff. Erectus, from the Red sea; Egypt. J Pure Appl Microbiol. 2018a;11(30):1299-1311.
Crossref - Shreadah MA, Abd El Moneam NM, Assar SA, Nabil-Adam A. Phytochemical and pharmacological screening of Sargassium vulgare from Suez Canal, Egypt. Food Sci Biotechnol. 2018b;27(4):963-979.
Crossref - Shreadah MA, Abd El Moneam NM, Yakou G, Abu Ella H. Bacteria from Marine Sponges: A Source of Biologically Active Compounds. Biomed J Sci &Tech Res. 2018c; 10(5):1-20.
Crossref - Siddiqi KS, Azamal Husen A, Rao RAK. A review on biosynthesis of silver nanoparticles and their biocidal properties. J Nanobiotechnology. 2018;16:14.
Crossref - Silva CCG, Silva SPM, Ribeiro SC. Application of Bacteriocins and Protective Cultures in Dairy Food Preservation. Front Microbiol. 2018;9:594.
Crossref - Singh P, Garg A, Pandit S, Mokkapati VRSS, Mijakovic I. Antimicrobial Effects of Biogenic Nanoparticles. Nanomaterials (Basel). 2018;8(12):1009.
Crossref - Skiba MI, Vorobyova VI. Synthesis of Silver Nanoparticles Using Orange Peel Extract Prepared by Plasmochemical Extraction Method and Degradation of Methylene Blue under Solar Irradiation. Adv Mater Sci Eng. 2019;2019:8306015.
Crossref - Sofowora A. Recent trends in research into African medicinal plants. J Ethnopharmacol. 1993;38(2-3):197- 208.
Crossref - Taga MS, Miller EE, Pratt DE. Chia seeds as a source of natural lipid antioxidants. JAOCS. 1984;61(5): 928-931.
Crossref - Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10(3):512-26
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol Biol Evol. 2011;28(10):2731-2739.
Crossref - Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30(12):2725-9.
Crossref - Trease GE, Evans MD. A text book of Pharmacognosy. 13th Ed. Builler Trindall and Canssel London. 1989:176- 180
- Trinder P. Determination of blood glucose using glucose oxidase. Ann Clin Biochem. 1969;6:24-25.
Crossref - Valdivieso-Ugarte M, Gomez-Llorente C, PlazaDiaz J, Gil A. Antimicrobial, Antioxidant, and Immunomodulatory Properties of Essential Oils: ASystematic Review. Nutrients. 2019;11:2786-2815.
Crossref - White TJ, Bruns TD, Lee SB, Taylor JW. Amplification and direct sequencing of fungal ribosomal RNA Genes for phylogenetics, In book: PCR – Protocolsand Applications – A Laboratory Manual, Publisher: Academic Press. 1990.
Crossref - Wosinska L, Cotter PD, O’Sullivan O, Caitriona Guinane C. The Potential Impact of Probiotics on the Gut Microbiome of Athletes. Nutrients. 2019;11(10):2270.
Crossref - Xia A, Zhang Y, Xu J, Yin T, Lu X-J. T Cell Dysfunction in Cancer Immunity and Immunotherapy. Front Immunol. 2019;10:1719.
Crossref - Zhishen J, Mengcheng T, Jianming W. The determination of flavonoid contents in mulberry and their scavenging effects on superoxide radicals. Food Chem. 1999;64(4):555-559.
Crossref - Zimbro MJ, Power DA, Miller SM, Wilson GE, Johnson JA. Difco & BBL Manual Manual of Microbiological Culture Media, Second Edition. BD Diagnostics Diagnostic Systems, 7 Loveton Circle Sparks, Becton, Dickinson and Company Sparks, Maryland 21152 USA. 2009
- Zollner N, Kirsch K. Colorimetric Method for Determination of Total Lipids. Journal of Experimental Medicine. 1962;135:545-550.
Crossref
© The Author(s) 2020. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.