ISSN: 0973-7510
E-ISSN: 2581-690X
Methicillin-resistant Staphylococcus aureus is a clinically significant pathogen that causes infections ranging from skin and soft tissue infections to life-threatening sepsis. Biofilm formation by MRSA is one of the crucial virulence factor. Determination of beta-lactamase and biofilm production among Staphylococcus aureus was obtained from various clinical specimens. Standard bacteriological procedures were used for isolation and identification and antibiotic sensitivity was determined using the Kirby Bauer disc diffusion method according to CLSI guidelines. The cloverleaf method, acidometric, iodometric and chromogenic methods were used to detect beta-lactamase while the microtiter plate method and Congo red agar method were used to detect biofilm production. Of the 288 MRSA strains isolated from various clinical specimens,198 (67.07%) were biofilm producers. Cloverleaf and chromogenic (nitrocefin) disc shows 100% results for beta-lactamase detection. Vancomycin was 100% sensitive followed by teicoplanin (92.36%) and linezolid (89.93%). Cloverleaf and nitrocefin disc methods were the most sensitive for detection of beta-lactamase in S. aureus and there was no significant relation between biofilm production and antibiotic sensitivity pattern of S. aureus.
Beta-lactamase, Biofilm, MRSA, Antibiotic susceptibility testing
MRSA (Methicillin-Resistant Staphylococcus aureus) is a dangerous bacteria that can cause minor skin infections to sepsis which can be fatal.1 The introduction of MRSA has complicated patient management by more extended hospital stays and raising costs while reducing the therapeutic efficacy of current antibacterial drugs.2 Major clinical crises have ensued from the establishment of resistance.3 Resistance to Beta-lactam is well-known mechanism of bacterial resistance which can be chromosomally or plasmid-mediated, constitutive or inductive. Beta-lactamase degrades beta-lactam antibiotics by hydrolyzing the beta-lactam ring rendering them ineffective.4,5 S. aureus in addition to its bacterial antibiotic resistance can develop a biofilm which is a complex multilayered cellular matrix and a significant virulence factor; as a result, antibiotic diffusion is inhibited.6 All of these variables contribute to a high level of antibiotic resistance among hospital-acquired bacteria. Infection-causing bacteria and their antibiotic resistance patterns differ dramatically from hospital to hospital.7 MRSA is the most commonly documented cause of biofilm-associated infections. Because these are commensals on human skin and mucosal surfaces they have a different status. As a result it is likely to be introduced as an infection during the surgical implantation of the polymeric device.8 The current investigation was carried out to look for beta-lactamase and biofilm production in S. aureus clinical isolates.
A prospective study was carried out in the Department of Microbiology, Santosh Medical College, Ghaziabad, in collaboration with Mayo Institute of Medical Sciences, Barabanki. Approval was obtained from the Institutional Ethics Committee (SU/2018/528(5)). All S. arures isolates were screened for MRSA dection by cefoxitin disc (30 µg) method. A total of 288 MRSA isolates were included in the study from various clinical specimens (blood, pus, wound swab, soft tissues, urine, sputum, body fluids, endotracheal secretions, central venous catheter tips). Bacterial isolation was done on 5% blood agar and identification was done by Gram staining and conventional methods like 3% catalase test, slide/tube coagulase, mannitol fermentation, golden yellow pigment demonstration on nutrient agar, DNase test.2,4 Antibiotic susceptibility test was done by Kirby-Bauer disc diffusion method. The following antibiotics included were penicillin (10 units), cefoxitin (30 µg), erythromycin (15 µg), azithromycin (15 µg), tetracycline (30 µg), doxycycline (30 µg), levofloxacin (5 µg), norfloxacin (10 µg), ofloxacin (5 µg), nitrofurantoin (300 µg), clindamycin (2 µg), rifampicin (5 µg), linezolid (30 µg), moxifloxacin (5 µg), chloramphenicol (30 µg), gentamicin (10 µg). Staphylococcus aureus ATCC 25923 was used as a control strain.9,10
Cefoxitin disc diffusion method
MRSA was detected by using cefoxitin (30 µg) disc. An isolate was considered to be a MRSA strain if the cefoxitin zone of inhibition was ≤ 21 mm.9,11
CHROM agar plate method
A swab was dipped in the bacterial suspension and streaked onto a CHROM agar plate (HiCrome™ MRSA Agar, Himedia, India). The growth of any green colony was considered to be positive for MRSA detection.12
Detection of beta-lactamase using a Chromogenic Technique (Nitrocefin Disc)
As directed by the manufacturer (BD diagnostic) several colonies of the test organism were placed directly to a nitrocefin disc moistened with sterile distilled water. The hydrolysis of beta-lactam antibiotics by the induced lactamase enzyme which was identified by changing color from bright yellow to deep red within 15 seconds to 5 minutes. A negative test was considered when there was no change in color within 5 minutes.13
Cloverleaf Experiment
E. coli ATCC- 25922 was inoculated on a Muller-Hinton agar [(MHA), Himedia, India] plate. Four test isolates were streaked radially outward from a penicillin disc [(10U), Himedia, India] in the center of the plate resulting in growth around 0.25 cm wide. After an 18-hour incubation period at 37°C the plate was inspected to see if the isolate possessed beta-lactamase resulting in a cloverleaf pattern.13
Idometric Method
This test was carried out by dispensing 100 μl of penicillin solution (6000 μg/ml, Himedia, India) into each well of a microtitre plate. To make a dense suspension many colonies of the organism to be examined were emulsified into the solution. After adding two drops of starch solution 1% the plate was allowed to incubate at room temperature for 30 minutes. A drop of iodine solution (2.03g of iodine and 53.2 g of potassium iodide in 100 ml of diltilled water) was added to the solution which colored it blue. The organism was termed positive for beta-lactamase production, if the blue color faded after 10 minutes. Without any culture suspension a negative control with penicillin alone was kept.12
Acidometric Method
The test organisms were suspended in a 100 μl volume of penicillin (20 million units)-phenol red (0.5%) reagent in microtitre wells. For the generation of beta-lactamase, a change in the color from purple-pink to yellow within 15 minutes was considered positive and no change within 15 minutes was considered negative.12
Biofilm Detection by Modified Congo-Red Agar method
MRSA strains were cultured on agar containing 10g of glucose and 0.4 g of Congo-red in one litre of blood base agar and incubated at 37°C for 48 hours. Slime producers were defined as strains that appear in black colonies while non-slime producers were defined as strains in red colonies. Positive biofilm-producing strains are black-colored colonies with a dry crystalline consistency.14
Biofilm Detection by Microtiter Plate Method
The isolates were placed in BHI (Brain Heart Infusion) broth and incubated for 24 hours at 37°C. Biofilm production was detected using ninety-six well microtiter plates. First, 200 μl of brain heart infusion broth was added to each well. The wells were then filled with 20μl of each sample to obtain 105 cfu/ml as a final concentration and incubated at 37˚C for 24 hours. The contents of the wells were discarded and removed by tapping the plate after 24 hours. Then 200 μl PBS was used to wash each well four times. Then 100μl of 0.1% crystal violet was added to each well to stain it and it was left for 15 minutes. As a positive control Pseudomonas aeruginosa ATCC 27853 was used. The plates were allowed to dry before being analyzed using an ELISA plate reader at 570 nm.15
The reading values are interpreted as:
Sample OD >0.12 indicated a strong biofilm producer.
Sample OD values range from 0.06 to 0.12 indicated biofilm producers as moderate to poor
Sample OD 0.06 indicated non-biofilm producer.
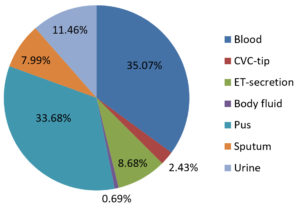
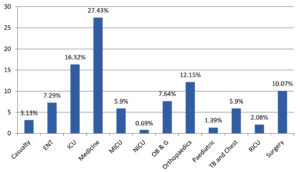
A total of 655 S. aureus were isolated from various clinical samples. Of these 288 isolates were MRSA on screening by Cefoxitin disc diffusion method and majority of these were isolated from blood (35.07%) and pus (33.68%) samples from various clinical departments. Samples received from the Medicine Department showed maximum MRSA isolates (27.43%), followed by Intensive Care Unit (16.32%), Orthopaedic (12.15%), Surgery (10.07%), Obstetrics and Gynecology (7.64%), Ear Nose Throat (7.29%), TB and Chest (5.90%), Medicine ICU (5.90%), Casualty (3.13%), Respiratory ICU (2.08%), Paediatric (1.39%) and Neonatal ICU (0.69%) as shown in Fig. 1 and 2.
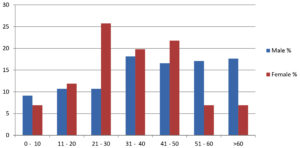
Of the 288 MRSA isolates, maximum isolates were isolated from males patients 187 (63.93%) than female patients 101 (35.06%). In addition among the male age group of 31-40 years old were more infected (18.18%) while females were more infected (25.74%) in the age group of 21–30 years as shown in Fig. 3.
The detection of beta-lactamase by various methods showed that the Cloverleaf method and Nitrocefin method showed 100% detection than the Acidometric 284 (98.61%) and Iodometric 280 (97.22%) method as shown in Table 1.
Table (1):
Beta-lactamase Detection in S. aureus.
Method |
Number (288) |
Biofilm Producer (%) |
|---|---|---|
Clover leaf |
288 |
100.0 |
Iodometric |
280 |
97.2 |
Acidometric |
284 |
98.6 |
Nitrocefin |
288 |
100.0 |
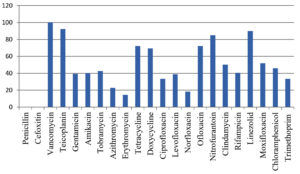
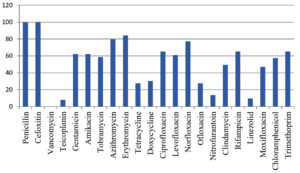
Antibiotic susceptibility patterns of isolated MRSA showed 100% susceptibility tovancomycin, followed by Linezolid (89.93%) and Teicoplanin (92.36%). Urine specimen isolates were the most susceptible to Nitrofurantoin 33 (11.46%) when compared to Norfloxacin 6 (18.18%). All isolates were resistant to penicillin and cefoxitin as they were beta lactamse producing strains as shown in Fig. 4.
Of the total 288 MRSA isolates 198 (68.75%) were biofilm producers by microtitre plate method. Strong biofilm producers were 101 (35.06%) while 97 (33.68%) isolates were weak biofilm producers where as 187 (64.93%) MRSA showed biofilm production on congored agar method. Of the 198 biofilm-producing isolates highest number were from Blood 82 (41.44%) samples, followed by pus 59 (29.80%), urine 22 (11.11%), Endotracheal-secretion 17 (5.59%) as shown in Table 2 and 3.
Table (2):
Biofilm Production in MRSA.
| Methods | Number | Results |
|---|---|---|
| Microtiter Plate methods | Strong | 101 |
| Moderate | 97 | |
| Congo Red Agar | Qualitative | 187 |
Table (3):
Distribution of Biofilm Producers According to Clinical Samples.
| Specimen | Biofilm Producer | Strongly Positive | Weakly Positive | |||
|---|---|---|---|---|---|---|
| No | % | No | % | No | % | |
| Blood | 82 | 41.41 | 37 | 36.63 | 45 | 46.39 |
| CVP-tip | 6 | 3.03 | 4 | 3.96 | 2 | 2.06 |
| ET-secretion | 17 | 8.59 | 10 | 9.90 | 7 | 7.22 |
| Pus | 59 | 29.80 | 34 | 33.66 | 25 | 25.77 |
| Sputum | 12 | 6.06 | 4 | 3.96 | 8 | 8.25 |
| Urine | 22 | 11.11 | 12 | 11.88 | 10 | 10.31 |
| Total | 198 | 68.75 | 101 | 35.05 | 97 | 33.68 |
Antibiotic resistance patterns of biofilm producers showed penicillin and cefoxitin showing 100% resistance. Azithromycin and erythromycin were tested among 176 biofilm producers excluded from urine samples in which resistance was found to be 89.77% and 84.09%. Among the urine specimens norfloxac insensitivity was 81.81%. There was no vancomycin resistance found and the least resistance was observed from teicoplanin 8.1% and linezolid 9.6%.
MRSA is a common cause of nosocomial infection that causes high morbidity and mortality in inpatients.16 Infected or colonized patients are key reservoirs of MRSA in hospitals and transient hand carriage on the hands of health care personnel is the most common mechanism of patient-to-patient transmission.17 In this investigation of 288 methicillin-resistant S. aureus isolates from different clinical specimens majority were isolated from blood 101 (35.07%) followed by pus 97 (33.68 %) and body fluid 2 (0.69 %) whereas Rajaduraipandi et al. reported 35.7% of MRSA strains were obtained from throat swabs while 33.6% were collected from pus.18 A similar observation was found by Mehta et al. who reported a 33% isolation rate from pus and wound swabs.19 However, Qureshi et al. from Pakistan reported an 83% MRSA isolation rate from pus.20 Other investigations have found in blood isolates 29 %,21 while in urine isolates 76%.22 Majority of MRSA isolates were found in Department of Medicine (27.43 %) and ICU (16.32 %) while the least was found in NICU (0.69%). On the other hand Wilfred Gitau et al. reported 20% isolates from the Medicine Department.23 This maybe because of the prolonged duration of stay as most isolates in our investigation were from intensive care units. MRSA was shown to be more prevalent in males (63.93 %) than females (35.06%). The male age group of >60 years old were more infected 17.65% than the female age group with more isolates discovered in the 21–30-year-old group. Males were shown to have a higher MRSA infection rate (19.9%) than females in Tebelaydilnessa et al. (17% ) study. The age group 35–44 years24 had the highest frequency of MRSA found 8.30%, demonstrating that gender and age are not risk factors for MRSA acquisition or colonization. MRSA was present in 17.5% of the population which is lower than that reported in Addis Ababa.25,26
Cloverleaf and Nitrocefin methods detected 100 percent of beta-lactamase while Acidometric and Iodometric methods detected 98.61% and 97.22%, respectively. In the current investigation the rate of Nitrocefin tests was 100% similar to the findings of Odonkor and Addo27 and Meeaadand Kadhim Ali Al-Kudheiri.28 Other investigations on the other hand, revealed lower rates than the other investigation: 85.7%,29 88%,30 and 75%.31 Depending on the manufacturer of the Nitrocefin assay the rates may vary from one trial to the next.32 The Cloverleaf technique like the Chromogenic approach had high rates. High sensitivity and specificity (100 percent ) for these approaches have recently been reported,33 supporting this finding.
Vancomycin showed 100% sensitivity followed by Linezolid (89.93%) and Teicoplanin in the antibiotic susceptibility patterns of isolated MRSA. The total number of isolates found in the urine sample was 11.46% and isolates were more vulnerable to Nitrofurantoin (18.18%). Other researchers from Iran and other countries have reported similar findings.34 In this investigation 68.75% produced biofilms with the biggest number of biofilm producers coming from blood samples (82.41%), pus (59.79%), urine (22.11%) and Endotracheal-secretion (8.59%).
There were 101 (51.01%) strong biofilm producers and 97 (48.98%) weak biofilm producers among the 198 biofilm producers. According to Cha et al.35 86 (68.3%) of the 126 MRSA isolates determined biofilm-forming capacity with five strong levels (OD570 1.0) and 81 weak levels (0.2 OD570 1.0) biofilm producers. Rezaei et al.36 looked at how common biofilm development was among MRSA isolates from nasal carriers. They discovered that all MRSA isolates generate biofilms with 15.4%, 19.2%, and 65.4% of them being strong, medium and weak biofilm makers respectively.37 Biofilm development was reported in 182 (78.78%) isolates in another investigation by Dardicharankaur et al.38 Strong biofilm development was found in 121 isolates (52.38%), while mild biofilm formation was found in 26.40%. Biofilm formation was found to be negative among 21.21% isolates.39 According to Singh,40 the isolates determined to be high biofilm formers accounted for 85.72% of the isolates. Miscellaneous samples had the highest prevalence of biofilm development (86.11%), followed by urine 81.81%, sputum 81.25% and pus 81.25%. Hassan A et al.41 found a lower incidence of biofilm formation from MRSA isolates from blood (64.28 percent); Fatima Khan et al.42 Microtiter plate technique 64.89 percent, tube method 63.74%, and Congo red Agar method 47.79%. Biofilm-producing S. aureus was more resistant to antimicrobials than non-producing S. aureus.43 Antibiotic resistance trends in biofilm producers and non-biofilm producers on the other hand showed no correlation. Another study also found no link between biofilm production and antibiotic resistance a conclusion that has been previously reported by other researchers.44
Beta-lactamase detection is very crucial for the management of infections caused by S. aureus. In the present study, Cloverleaf and Nitrocefin disc methods were most sensitive for detection of beta-lactamase in S. aureus. The study also revealed that good number of MRSA strains from clinical samples producing biofilm. Microtiter plate method showed good detection over Congo red agar method for biofilm detection. There were no significant corelation between biofilm production and antibiotic resistance in MRSA therefore vancomycin and linezolid remains drug of choice for treatment of MRSA infection.
ACKNOWLEDGMENTS
The authors acknowledged Santosh Deemed to be University, India for providing the research facilities.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
ETHICS STATEMENT
This article does not contain any study with human participants or animals performed by any of the authors.
AVAILABILITY OF DATA
All datasets analysed during this study are included in the manuscript.
- Vaishampayan A, de Jong A, Wight DJ, Kok J, Grohmann E. A Novel Antimicrobial Coating Represses Biofilm and Virulence-Related Genes in Methicillin-Resistant Staphylococcus aureus. Front Microbiol. 2018;9:221.
Crossref - Kumar D, Bisht D, Faujdar SS. Incidence of Mupirocin Resistance in Staphylococcus Aureus Isolated from Rural Population: A New Emerging Challenge. Int J Cur Res Rev. 2020;12(22):82-85.
Crossref - Ali AM, Rafi S, Qureshi AH. Frequency of extended-spectrum beta-lactamase-producing Gram-negative bacilli among clinical isolates at clinical laboratories of Army Medical College, Rawalpindi. J Ayub Med Coll Abottabad. 2004;16:35-37.
- Deepa K, Faujdar SS, Azmi W, Mehrishi P, Solanki S. Screening and optimization of staphylokinase from Staphylococcus aureus isolated from nasal swab of healthy students in Himachal Pradesh University, India. Biomed Biotechnol Res J. 2019;3(4):228-232.
Crossref - Devapriya F, Ramesh R, Sajit Khan AK, Shanmugam J. β-lactamase production of Staphylococcus aureus: a comparison study of different iodometric methods. Gulf Medical Journal. 2013;2(1):16-21.
- Gupta R, Malik A, Rizvi M, Ahmed M, Hashmi A. Multidrug Resistant Gram-Positive Pathogens with Special Reference to MRSA and Biofilm Productionin ICU Patients: Recurrent Challenge for Clinicians. Int J Curr Microbiol App Sci. 2015;1:207-212.
- Otto M. Staphylococcal biofilms. Curr Top Microbiol Immunol. 2008;322:207-228.
Crossref - Barid D. Staphylococcus: Cluster-forming Gram-positive cocci, Chapter 11. In: Mackie & McCartney Practical Medical Microbiology, 14th Edn. Collee JG, Fraser AG, Marmion BP, Simmons A, Editors. (Churchill Livingstone: New York); 1996:247.
- Kumar D, Mehrishi P, Faujdar SS, Kumar S, Sharma A. Osteomyelitis in Children from Rural Population of Uttar Pradesh. J Pure Appl Microbiol. 2021;15(3):1320-1324.
Crossref - Datta P, Gulati N, Singla N, et al. Evaluation of various methods for the detection of meticillin-resistant Staphylococcus aureus strains and susceptibility patterns. J Med Microbiol. 2011;60(11):1613-1616.
Crossref - Isenberg HD. Clinical Microbiology Procedure Handbook. Vol. 2, American Society for Microbiology, ASM Press. 2004
- Samant SA, PAI CG. Comparative Evaluation of β-lactamase detection methods in Staphylococci. Int J Res Pharm Biomed Sci. 2012;3(4):1580-1588.
- Pitkala A, Salmikivi L, Bredbacka P, Myllyniemi AL, Koskinen MT. Comparison of tests for detection of beta-lactamase-producing staphylococci. J Clin Microbiol. 2007;45(6):2031-2033.
Crossref - Jayachandran AL, Sarasa S, Doris TS, et al. Biofilm formation and Antibiotic susceptibility pattern among Staphylococcus aureus in a tertiary care hospital in Kanchipuram: An Evaluation of screening methods for biofilm formation. International Journal of Bioassays. 2016;5(4):4991-4995.
Crossref - Sachdev D, Amladi S, Nataraj G, et al. An Outbreak of Methicillin-resistant Staphylococus aureus (MRSA) infection in dermatology indoor patients. Indian J Dermatol Venereol Leprol. 2003;69(6):377-380. PMID: 17642945
- McDonald M. The epidemiology of methicillin-resistant Staphylococcus aureus: Surgical relevance 20 years on. Aust N Z J Surg. 1997;67(10):682-685.
Crossref - Rajaduraipandi K, Mani KR, Panneerselvam K, Mani M, Bhaskar M, Manikandan P. Prevalence and antimicrobial susceptibility pattern of methicillin resistant Staphylococcus aureus: a multicentre study. Indian J Med Microbiol. 2006;24(1):34-38.
Crossref - Mehta AP, Rodrigues C, Sheth K, Jani S, Hakimiyan A, Fazalbhoy N. Control of methicillin-resistant Staphylococcus aureus in a tertiary care Centre-A five-year study. Indian J Med Microbiol. 1998;16:31-34.
- Qureshi AH, Rafi S, Qureshi SM, Ali AM. The current susceptibility patterns of methicillin-resistant Staphylococcus aureus to conventional anti Staphylococcus antimicrobials at Rawalpindi. Pak J Med Sci. 2004;20:361-364.
- Soltani R, Khalili H, Rasoolinejad M, Abdollahi A. Antimicrobial susceptibility pattern of Staphylococcus aureus strains isolated from hospitalized patients in Tehran, Iran. Iran J Pharm Sci. 2010;6:125-132.
- Obiazi H, Nmorsi O, Ekundayo A, Ukwandu Ncd. Prevalence and antibiotic susceptibility pattern of Staphylococcus aureus from clinical isolates grown at 37 and 44°C from Irrua, Nigeria. African J Microbiol Res. 2007;1(5):57-60.
- Gitau W, Masika M, Musyoki M, Museve B, Mutwiri T. Antimicrobial susceptibility pattern of Staphylococcus aureus isolates from clinical specimens at Kenyatta National Hospital. BMC Res Notes. 2018;11(1):226.
Crossref - Kumar M. Multidrug-resistant Staphylococcus aureus, India, 2013-2015. Emerg Infect Dis. 2016;22:1666-1667.
Crossref - Geyid A, Lemeneh Y. The incidence of methicillin-resistant strains of Staphylococcus aureus strains in clinical specimens in relation to their beta-lactamase-producing and multiple-drug resistance properties in Addis Ababa. Ethiop Med J. 1991;29:149-161.
- Gedebou M. Staphylococcus aureus strains from a teaching hospital: clinical sources and antibiograms. E Afr Med J. 1982;59(12):810-815.
- Sindhu CP, Shanmugam J. Beta-lactamase production by Staphylococcus aureus and coagulase-negative staphylococci: detection by different iodometric and acidometric techniques. Biomedicine 1998;18:74-80.
- Odonkor ST, Addo KK. Evaluation of three methods for detection of methicillin-resistant Staphylococcus aureus. Int J Biol Med Res. 2011;2(4):1031-1034.
- Al-Kudheiri MK, Hussein AN, Al-Mohana AM. Detection of β-lactamases producing Methicillin-Resistant Staphylococcus aureus (MRSA). AL-Qadisiyah Journal of pure Science. 2018;23(2):229-238.
Crossref - Broekema NM, Van TT, Monson TA, Marshall SA, Warshauer DM. Comparison of cefoxitin and oxacillin disk diffusion methods for detection of mecA mediated resistance in Staphylococcus aureus in a large-scale study. J Clin Microbiol. 2009;47(1):217-219.
Crossref - Oliveira DC, de Lencastre H. Methicillin-resistance in Staphylococcus aureus is not affected by the over expression in trans of the mecAgene repressor: a surprising observation. PLoS ONE. 2011;6(8):e23287.
Crossref - Ba X, Harrison EM, Edwards GF, et al. Novel mutations in penicillin-binding protein genes in clinical Staphylococcus aureus isolates that are methicillin resistant on susceptibility testing, but lack the mec gene. J Antimicrob Chemother. 2014;69(3):594-597.
Crossref - Kaase M, Lenga S, Friedrich S, et al. Comparison of phenotypic methods for penicillinase detection in Staphylococcus aureus. Clin Microbiol Infect. 2008;14(6):614-616.
Crossref - Robles BF, Nobrega DB, Guimaraes FF, Wanderley GG, Langoni H. b-lactamase detection in Staphylococcus aureus and coagulase-negative Staphylococcus isolated from bovine mastitis. Pesq Vet Bras. 2014;34(4):325-328.
Crossref - El Aila NA, AlLaham NA, Ayesh BM. Nasal carriage of methicillin-resistant Staphylococcus aureus among health care workers at Al Shifa hospital in Gaza Strip. BMC Infect Dis. 2017;17(1):28.
Crossref - Cha JO, Yoo JI, Yoo JS, et al. investigation of biofilm formation and its association with the molecular and clinical characteristics of methicillin-resistant Staphylococcus aureus. Osong Public Health Res Perspect. 2013;4(5):225-232.
Crossref - Rezaei M, Moniri R, Mousavi SG, JabariShiade M. Prevalence of biofilm formation among methicillin resistance Staphylococcus aureus isolated from nasal carriers. Jundishapur J Microbiol. 2013;6(6):e9601.
Crossref - Wu WS, Chen CC, Chuang YC, et al. efficacy of combination oral antimicrobial agents against biofilm-embedded methicillin-resistant Staphylococcus aureus. J Microbiol Immunol Infect. 2013;46(2):89-95.
Crossref - Kaur DC, Chate SS. Study of Antibiotic Resistance Pattern in Methicillin Resistant Staphylococcus Aureus with Special Reference to Newer Antibiotic. J Glob Infect Dis. 2015;7(2):78-84.
Crossref - Mathur T, Singhal S, Khan S, Upadhyay D, Fatma T, Rattan A. Detection of biofilm formation among the clinical isolates of Staphylococci: an evaluation of three different screening methods. Indian J Med Microbiol. 2006;24(1):25-29.
Crossref - Singh S, Katiyar R, Kaistha SD. High oxacillin, vancomycin and fluoroquinolone resistance amongst biofilm forming Staphylococcus aureus isolates from ulcerative keratitis infections. Indian J Med Microbiol. 2011;29(3):312-313.
Crossref - Hassan A, Usman J, Kaleem F, Omair M, Khalid A, Iqbal M. Detection and antibiotic susceptibility pattern of biofilm producing Gram positive and Gram negative bacteria isolated from a tertiary care hospital of Pakistan. Malays J Microbiol. 2011;7(1):57-60.
Crossref - Khan F, Shukla I, Rizvi M, Mansoor T, Sharma SC. Detection of Biofilm Formation in Staphylococcus aureus. Does it have a role in Treatment of MRSA Infections? Trends in Medical Research. 2011;6(2):116-123.
Crossref - Pal J, Bisht D. Detection of biofilm production in Candida species isolates recovered from bloodstream patients. Int J Biomed Adv Res. 2016;7(5):226-229.
Crossref - Stickler DJ. Bacterial biofilms and the encrustation of urethral catheters. Biofouling. 1996;9(4):293-305.
Crossref
© The Author(s) 2021. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.