Biofilms are always a major concern in the healthcare field and food industry. The resistant properties of biofilm that allow bacteria to persist are difficult to study. Biofilms are often more resistant to antibiotics than individual planktonic cells. Thus, alternative strategies for managing biofilm formation are needed. Currently, using phages as anti-biofilm agents has been suggested. In this review, some of the diverse strategies, reported in previous studies, for preventing biofilm formation are discussed. Use of phages as anti-biofilm agents can involve phage application prior to biofilm formation, application to biofilms that are already formed, or using phages in association with other mechanisms to physically disrupt the biofilm. The development of novel methods as anti-biofilm agents will add an important dimension to the search for new potent compounds for preventing biofilm-associated infections.
Phages, Biofilm, Bacteria, Antibiotics, Resistance.
Biofilm
In general, biofilms are complex composition of bacteria that can be formed either from one or numerous different species living together inside a matrix made of extracellular polymeric substances (EPS) with the capability to attach to numerous surfaces1. EPS mainly include polysaccharides, but other biomolecules present include nucleic acids, lipids, and proteins, which form a scaffold that helps bacteria remain attached within the biofilm2, 3. This matrix displays a modified phenotype, and the regulation of specific drug resistance genes and virulence factors has been observed in bacterial biofilms. Horizontal genetic transfer can occur easily, facilitating cross-breeding of resistance genes4, 5. Biofilm formation involves five stages 6, as shown in Fig. (1)
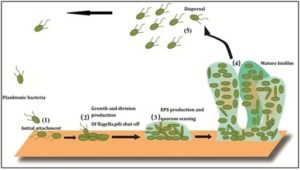
Fig. 1: Biofilm is formed in five stages, these stages are 1) initial, reversible attachment, 2) Irreversible binding and growth, 3) EPS production and inter communication through quorum sensing, 4) Mature biofilm, and 5) dispersal; essential stage for biofilm dispersion and life cycle (adapted and modified from Mizan and others 2015)7
The complex composition of the matrix increases survival ability under extreme conditions, as well as enhances the inflow of nutrients, water, and signaling molecules important for cell communication8, 9. Furthermore, the EPS matrix forms a barrier between the external environment and bacteria, which prevent antimicrobials from penetrating the biofilm10. Biofilms of Salmonella are more resistant to the triclosan antibiotic than individual planktonic Salmonella cells11. Furthermore, negative charges on the EPS can prevent antibiotics from reaching the biofilm12, 13.
Biofilms play a fundamental role in infectious diseases. Studies have shown that 60–70% of most nosocomial infections are directly linked to the clear presence of biofilms 14. Bacteria commonly associated with medical devices include Staphylococcus epidermidis and Staphylococcus aureus, followed by P. aeruginosa, as well as other bacteria that opportunistically infect weakened patients15-17. Moreover, bacteria can be present on medical implants including catheters18, 19.
Bacteria within biofilms exhibit both antibiotic and host defense resistance19, as well as a decreased growth rate, limited diffusion, and increased efflux and enzymes responsible for antimicrobial degradation20, 21. Generally, the use of antibiotics to treat biofilm-related infections is often not successful12. Many studies confirmed that for biofilms, the minimum inhibitory concentration (MIC) and minimum bactericidal concentration were generally higher compared to those for planktonic bacterial cells (approximately 10–1000-fold)22, 24. Numerous antimicrobials function against actively growing cells, and thus biofilms reduce antimicrobial function. Once bacteria are embedded within a biofilm, various factors such as altered gene expression and quorum sensing increase the resistance to antibiotics25. Treating biofilm is difficult and challenging, and thus has been given much attention26. Thus, it is extremely important to develop new antimicrobial agents or efficient methods for targeting and destroying the biofilm responsible for infection27, 28.
Studies of biofilm-phage interactions
Bacteriophages or (phages) in general are viruses that infect bacteria (Figure 2). Some viruses were created to target biofilms29. They can either reside in the bacterial host genome in a lysogenic state, or enter a lytic state to destroy bacteria, and thus can be used for therapeutic purposes. Phages show potential as alternatives to antibiotics against bacterial infections and have been widely explored to minimize pathogen loads in food products. Phages may also be safer than antibiotics. Phage isolation is rapid, simple, and inexpensive. Phages are competent against one specific host or a range of hosts, and thus are more effective than the natural microflora which are initially attacked by the biofilm. Phages are biofriendly and not associated with negative side effects30.
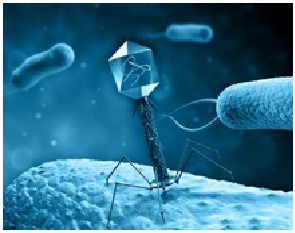
Fig. 2. Bacteriophage targeting bacteria within biofilm
Targeting of biofilms by phages has been examined in numerous studies31, 32. Phages have long been used for purposes comparable to those of antibiotics, in addition to treating bacterial infections33-35. Biofilm enucleation through phages can involve phage application to prevent biofilm formation, application to biofilms that are already formed, or in association with other mechanisms that may physically disrupt the biofilm.
Promising strategies involving phage against biofilm include the following:
Inhibition of attachment by phages
Phages are capable of affecting the initial adsorption stage of biofilms (or adhered cells). When employing lytic phages, as generally is the case for anti-biofilm phages, phage infection results in the killing and lysis of bacteria. This impacts biofilms structurally and releases new phage virions that can potentially reach and then infect adjacent bacteria36. The effect is a cyclical acquisition and killing of biofilm bacteria34. Sillankorva et al. (2008) reported that single cells on glass surfaces for 60 min were efficiently inhibited by phage jS1. Cell removal was fast and efficient and resulted in a biomass reduction of approximately 90%37.
Inhibition of EPS matrix by depolymerizing enzymes
It has been reported that some phages are effective at penetrating the EPS matrix by diffusion or through the action of phage-associated enzymes. A large range of enzymes can destroy the biofilm EPS matrix. In the case of phages, these enzymes include those mainly produced to help release phages from the host cell and tail spike proteins that allow infection of bacteria within the biofilm, but in general the activity of these enzymes and proteins are strictly localized. However, studies revealed that proteins with activity limited to the virus particle may be released from lysing cells, affecting the biofilm matrix38.
Phages are capable of producing depolymerizing enzymes that can degrade the EPS from the host genome. The genome of many phages also contains genes that specialize in producing enzymes that break down the matrix29, 39, 40. Under many conditions, these enzymes target the bacterial cell wall for release from the host cell, but these enzymes can also degrade the biofilm EPS. The T4 and HK620 phages of Escherichia coli contain enzymes that exist on the viral tail, and may play a role in degrading the matrix39, 40. Polysaccharide depolymerase is a very important part of the phage tail and many tail spike proteins have endoglycosidase activity by breaking down their polysaccharide receptors through hydrolyzation40. It has been reported that a phage-induced method of making the biofilm matrix more porous can facilitate the infection process by progeny phage, while a rapid bacterial infection reaction moves away from the focus on infection. Although the presence of polysaccharide depolymerase in phages has been reported, EPS-degrading enzymes are difficult to isolate, and thus have been reconstructed, including those from T7 phage41.
Importantly, different species of bacteria produce different EPS components. A depolymerase active that acts on polysaccharides from one species of bacteria may not digest that produced by other bacteria. However, depolymerases likely have broader activity than their parent phages among closely related bacteria, as the complexity and variability in the EPS is lower that of the host bacteria. Son et al. (2010) observed this by comparing phage of S. aureus with a specific depolymerase. However, only Staphylococci were affected, suggesting that multiple depolymerases are required to target mixed biofilms and that dynamic depolymerases are needed; haloes may be observed over the phage plaques formed on bacterial cultures, revealing the areas where bacterial polysaccharide has been broken down42. Gutiérrez et al. (2012) used this approach to detect such activity in two phages infecting S. epidermidis, both of which were then confirmed by sequencing to contain genes for pectin lyases43, while Glonti et al. (2010) identified haloes in cultures of a phage infecting P. aeruginosa and purified a depolymerase protein from the phage44. Yan et al (2013) classified phage polysaccharide depolymerases as endorhamnosidases, alginate lyases, endosialidases, and hyaluronidases40.
Pretreatment of catheter using phages
Another important challenge in medical care for reducing biofilm formation by S. epidermidis is pre-treating catheter surfaces with phages (Curtin and Donlan, 2006).
The utilization of phages for treating device-related infections has been examined since the 20th century. Pretreatment of hydrogel-coated catheters by phage was found to inhibit S. epidermidis and P. aeruginosa biofilm formation45, 46.
Quorum sensing inhibition (QSI) by phages
One strategy that can be used against biofilm may be to inhibit quorum sensing (QS), a cell-to-cell signaling system that controls the expression of genes necessary for adding virulence factors, such as those responsible for interactions with the host bacteria and regulating biofilm development47-51. The key intent behind this strategy is not to kill the pathogens, but to disarm them by making them oversensitive to normal antimicrobial treatments. Furthermore, the QS system does not contribute any mechanisms essential for bacteria survival, but inhibiting this method does not provide firm selective pressure sufficient to cause resistance development52. Pei and Lamas-Samanamud (2014) showed that the engineered phage strain T7 which produces metalloenzymes AiiA lactonase have various actions against signaling molecules (acyl homoserine lactones) involved in bacterial quorum sensing and that these molecules are important for biofilm development53.
Phage growth within biofilms
Experimental data indicate that phages do grow well in P. aeruginosa biofilms53, at least in the primary stages of their development. In 2-day-old biofilms55, of 17 insensitive strains of P. aeruginosa phages (therefore, planktonic bacterial hosts were used), 8 strains encouraged the growth of the same phages in the biofilm. However, the effects of antibiotics could be blocked in their initial stages of formation. This agrees with the findings of another study showing that antibiotic resistance begins to appear in the first stages of biofilm formation. Thus, bacteria can be destroyed by phages in cases where antibiotics are ineffective56.
Previous studies have revealed the processes involved in regulating biofilms, showing that phages effecting P. aeruginosa can terminate bacteria in an adult biofilm and (based on their sizes) diffused through thick alginate gel. However, this activity clearly varied from that of highly-restricted tail spike proteins57. Sillankorva et al. (2004) showed that phages of both P. fluorescens and S. lentus reduced both single species and mixed biofilms with these agents. The phages of both hosts were completely sequenced, with neither found to code for a polysaccharide depolymerase (although the P. fluorescens phage encoded an endopeptidase)29. Similarly, Doolittle et al. (1996) reported that the E. coli phage T4 does not code for polysaccharide depolymerase, except for a restricted tail spike protein released from the phage tail only during host cell penetration. But could spread effectively through biofilm58.
Some studies have shown that phages can penetrate biofilms even if they cannot produce polysaccharide depolymerases, but within biofilm, effective infection has not been observed in most studies. Additionally, some researchers have proposed the existence of EPS-degrading enzymes are extremely important for biofilm-related applications38. A study by Tait et al. (2002) revealed that using a combination of three phages entirely destroyed a biofilm composed of a single species, while in the presence of other bacterial species which were insensitive, this technique had little effect59. A study by Kay et al. (2011) also demonstrated that phage efficiency is decreased in the presence of mixed biofilms60. However, Sillankorva et al. (2004) reported that efficiency remained high in model biofilms, even when individual bacterial species in the biofilm are targeted by the phage, demonstrating that phages can kill a specific type of bacterial host even when under mixed biofilm conditions. In addition, they reported that phages can effectively target an adult biofilm29.
Combining phage with other agents
Using phages as mixtures or coupled with antibiotics can completely prevent the development of phage resistance61. Verma et al. (2010) found that mature biofilms can adapt to antibiotics if lytic phages are used 62, which agrees with the results of some clinical trials of phage activity63, 64. Using phages and antibiotics in a combined or sequential manner has shown potential for therapeutic applications. In support of this, Yilmaz et al. (2013) found that using phages coupled with antibiotics to treat biofilms of S. aureus was clearly effective65. Another study suggested a polysaccharide lyase and DNase enzymes to destroy the matrix can be used in combination with phages54. Abedon et al. (2011) found similar results, although differential diffusion of phages and co-administered enzymes is difficult. The use of phages can also be combined with physical wound cleaning33. Seth et al. (2013) used a rabbit ear mold to show that individually removing damaged tissue or foreign objects from a wound and using phage treatment had no effect, while combining these methods was effective. Phages may have similar functions in biocides and sanitizers currently used, but should be applied after the primary cleaning processes to destroy bacteria on the remaining biofilms66. Similarly, Ganegama Arachchi et al. (2013) found that using a combination of three different phages could clear Listeria monocytogenes biofilms effectively from steel surfaces. Thus, when treating biofilms with phages, the biofilm cell surface should be disrupted prior to phage application67. Other combinations are also possible for use in biological systems. Liao et al. (2012) found that combining phages with commensal bacteria had synergistic effects in preventing biofilm formation on silicone catheter segments68, while Zhang and Hu (2013) observed that when using phages coupled with biocides such as chlorine, the effects on filters were increased69. However, further studies are needed to explore phage activity in a multispecies context, animal models, and in combination with other antimicrobials70. Figure 3 shows various strategies that have been used to destroy biofilms within the past 20 years.
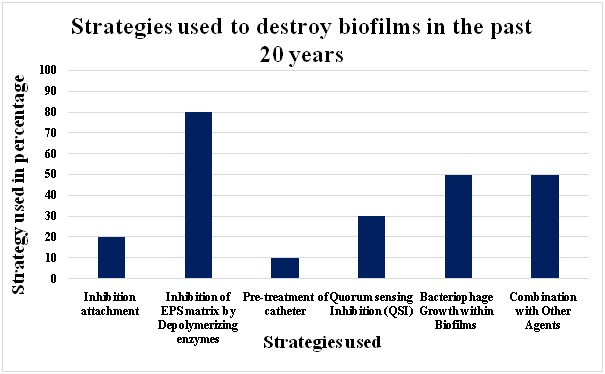 Fig. 3: Phage mediated prevention Biofilm Strategies used within the last few 20 years
Fig. 3: Phage mediated prevention Biofilm Strategies used within the last few 20 yearsStudies examining interactions between phages and biofilms indicated that phages contain some unique properties and are promising for biofilm control. Different phages have been used to infect numerous bacterial biofilms. The treatment of biofilms using phages is complex, and only strictly lytic phages should be used. Similar to phage infection of planktonic cells, numerous essential steps are required. Phage adsorption to receptors of the targeted bacteria is the initial step of infection. It is also evident that phages express enzymes that can disrupt biofilms. These enzymes are induced from the host genome. While much progress has been made in these methods, more studies are needed. Thus, strategies for destroying biofilm are currently speculative in nature. With additional studies, new and better strategies will be developed.
- Hurlow J, Couch K, Laforet K, Bolton L, Metcalf D, Bowler P. Clinical biofilms: a challenging frontier in wound care. Adv Wound Care, 2015; 4(5): 295–301.
- Cortes ME, Consuegra J, Sinisterra RD. Biofilm formation, control and novel strategies for eradication. Sci Against Microbial Pathog Commun Curr Res Technol Adv., 2011; 2:896–905.
- Flemming HC, Wingender J. The biofilm matrix. Nat Rev Microbiol., 2010; 8(9): 623–633.
- Anderson GG, O’Toole G.A. Innate and induced resistance mechanisms of bacterial biofilms. in Bacterial Biofilms (ed. Romeo T.), pp. 85–105 Springer, Heidelberg 2008.
- Donlan RM, Costerton W. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin Microb Rev., 2002; 15: 167-193.
- Stowe SD, Richards JJ, Tucker AT, Thompson R, Melander C, Cavanagh J. Anti-biofilm compounds derived from marine sponges. Mar Drugs. 2011; 9(10): 2010-35.
- Mizan MFR, Jahid IK, Ha SD. Microbial biofilms in seafood: a food-hygiene challenge. Food Microbiol., 2015; 49:41–55.
- Tarver, T. Biofilms – A threat to Food Safety. Journal of Food technology. 2009; 63(2): 47.
- Watnik, P. and Kolter, R. Biofilm, city of microbes. Journal of bacteriology, 2000; 182(10): 2675-9.
- Mah TF & O’Toole GA. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 2001; 9: 34–39.
- Tabak M, Scher K, Hartog E, Romling U, Matthews KR, Chikindas ML & Yaron S. Effect of triclosan on Salmonella typhimurium at different growth stages and in biofilms. FEMS Microbiol Lett., 2007; 267: 200–206.
- Costerton W, Veeh R, Shirtliff M, Pasmore M, Post C. The application of biofilm science to the study and control of chronic bacterial infections. Journal of Clinical Investigations., 2003; 112: 1466–77.
- Qu, Y., Daley, A. J., Istivan, T. S., Garland, S. M. & Deighton, M. A. Antibiotic susceptibility of coagulase-negative staphylococci isolated from very low birth weight babies: comprehensive comparisons of bacteria at different stages of biofilm formation. Ann Clin Microbiol Antimicrob. 2010; 9: 16.
- Percival S. L., Hill K. E., Williams D. W., Hooper S. J., Thomas D. W., Costerton J. W. A review of the scientific evidence for biofilms in wounds. Wound Repair Regen. 2012; 20: 647–657.
- Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science., 1999; 284(5418):1318–1322.
- Hall-Stoodley L, Costerton JW & Stoodley P. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol., 2004; 2: 95–108.
- Romling, U., and Balsalobre, C. Biofilm infections, their resilience to therapy and innovative treatment strategies. J. Intern. Med., 2012; 272: 541–561.
- Maric, S., Vranes, J. Characteristics and significance of microbial biofilm formation. PeriodicumBiologorum., 2007; 109: 1-7.
- Vasudevan R. Biofilms: Microbial Cities of Scientific Significance. Journal of Microbiology & Experimentation., 2014; 1(3).
- Hall-Stoodley L, Stoodley P. Evolving concepts in biofilm infections. Cell Microbiol., 2009; 11(7): 1034-43.
- Kumar, A. & Schweizer, H.P. Bacterial resistance to antibiotics: active efflux and reduced uptake. Advanced Drug Delivery Reviews., 2005; 57: 1486-1513.
- Hoiby N, Bjarnsholt T, Givskov M, Molin S, Ciofu O. Antibiotic resistance of bacterial biofilms. Int J Antimicrob Agents, 2010; 35: 322–332.
- Hengzhuang W, Wu H, Ciofu O, Song Z & Hoiby N. Pharmacokinetics/pharmacodynamics of colistin and imipenem on mucoid and nonmucoid Pseudomonas aeruginosabiofilms. Antimicrob Agents Chemother, 2011; 55: 4469–4474.
- Hengzhuang W., Wu H., Ciofu O., Song Z., Høiby N. In vivopharmacokinetics/pharmacodynamics of colistin and imipenem in P. aeruginosa biofilm infection. Antimicrob. Agents Chemother., 2012; 56: 2683–2690.
- Clutterbuck AL, Woods EJ, Knottenbelt DC, Clegg PD, Cochrane CA, Percival SL. Biofilms and their relevance to veterinary medicine. Vet Microbiol., 2007; 121:1-17.
- Wu, K., Fang, Z., Guo, R., Pan, B., Shi, W., Yuan, S., et al. Pectin enhances bio-control efficacy by inducing colonization and secretion of secondary metabolites by Bacillus amyloliquefaciens SQY 162 in the rhizosphere of tobacco. PLoS ONE, 2015; 10: e0127418.
- Yarwood JM, Paquette KM, Tikh IB, Volper EM, Greenberg EP. Generation of Virulence Factor Variants in Staphylococcus aureus Biofilms. J. Bacterol., 2007; 189: 7961-7967.
- Issam, I., Xiang, F., Xavier, M., Ying, J., Robert, S. and Ray, H. The role of chelators in preventing biofilm formation and catheter-related bloodstream infections. Curr Opin Infect Dis, 2008; 21: 385–392.
- Sillankorva S, Oliveira R, Vieira MJ, Sutherland IW, Azeredo J. Bacteriophage phiS1 infection of Pseudomonas fluorescens planktonic cells versus biofilms. Biofouling., 2004; 20: 133-38.
- Olszowska-Zaremba, N.; Borysowski, J.; Dabrowska, J.; Górski, A. Phage translocation, safety,and immunomodulation. In Bacteriophages in Health and Disease; Hyman, P., Abedon, S.T.,Eds.; CABI Press: Wallingford, UK,; 2012; pp. 168–184.
- Brussow H. Bacteriophage-host interaction: from splendid isolation into a messy reality. Curr Opin Microbiol., 2013; 16:500–6.
- Harper DR, Parracho HM, Walker J, Sharp R, Hughes G, Werthe´n M, Lehman S, Morales S. Bacteriophages bioûlms. Antibiotics, 2014; 3(3): 270–284.
- Kutter E, De Vos D, Gvasalia G, Alavidze Z, Gogokhia L, Kuhl S, Abedon ST. Phage therapy in clinical practice: treatment of human infections. Curr Pharm Biotechnol., 2010; 11(1): 69–86.
- Abedon ST, Kuhl SJ, Blasdel BG, Kutter EM. Phage treatment of human infections. Bacteriophage., 2011; 1: 66-85.
- Abedon, S.T. Ecology of anti-biofilm agents ii: bacteriophage exploitation and biocontrol of Biofilm Bacteria. Pharm. Basel Switz., 2015; 8: 559–589.
- Abedon ST. Spatial vulnerability: bacterial arrangements, microcolonies, and biofilms as responses to low rather than high phage densities. Viruses., 2012; 4: 663–87.
- Sillankorva S, Neubauer P, Azeredo J. Pseudomonas fluorescensbiofilms subjected to phage philBB-PF7A. BMC Biotechnol., 2008; 8: 79.
- Cornelissen A., Ceyssens P. J., T’Syen J., Van Praet H., Noben J. P., Shaburova O. V., et al. The T7-related Pseudomonas putidaphage phi15 displays virion associated biofilm degradation properties. PLoS ONE, 2011; 6: e18597 10.1371/ journal.pone.0018597.
- Leiman PG, Chipman PR, Kostyuchenko VA, Mesyanzhinov VV, Rossmann MG. Three-dimensional rearrangement of proteins in the tail of bacteriophage T4 on infection of its host. Cell., 2004; 118: 419–429.
- Yan Y., Su S., Meng X., Ji X., Qu Y., Liu Z., et al. Determination of sRNA expressions by RNA-seq in Yersinia pestisgrown in vitro and during infection. PLoS ONE, 2013; 8: e74495. 10.1371/journal.pone.0074495.
- Lu TK & Collins JJ. Dispersing biofilms with engineered enzymatic bacteriophage. P Natl Acad Sci USA., 2007; 104: 11197–11202.
- Son JS, Lee SJ, Jun SY, Yoon SJ, Kang SH, Paik HR, Kang JO & Choi YJ. Antibacterial and biofilm removal activity of a podoviridae S. aureusbacteriophage SAP-2 and a derived recombinant cell-wall-degrading enzyme. Appl Microbiol Biotechnol. 2010; 86: 1439–1449.
- Gutiérrez D., Martínez B., Rodríguez A., García P. Genomic characterization of two Staphylococcus epidermidisbacteriophages with anti-biofilm potential. BMC Genomics, 2012; 13: 22810.1186/1471-2164-13-228.
- Glonti T., Chanishvili N., Taylor P. W. Bacteriophage-derived enzyme that depolymerizes the alginic acid capsule associated with cystic fibrosis isolates of Pseudomonas aeruginosa. J. Appl. Microbiol., 2010; 108: 695–702.
- Curtin, J.J. and Donlan, R.M. Using bacteriophages to reduce formation of catheter-associated biofilms by Staphylococcus epidermidis. Antimicrob Agents Chemother., 2006; 50: 1268–1275.
- Fu W, Forster T, Mayer O, Curtin JJ, Lehman SM & Donlan RM. Bacteriophage cocktail for the prevention of biofilm formation by Pseudomonas aeruginosa on catheters in an in vitro model system. Antimicrob Agents Ch., 2010; 54: 397–404.
- Cotar AI, Chifiriuc MC, Banu O, Lazar V. Molecular characterization of virulence patterns in Pseudomonas aeruginosa strains isolated from respiratory and wound samples. Biointerface Res Appl Chem., 1213; 3: 551-55.
- Mellbye B., Schuster M. “More than just a quorum: integration of stress and other environmental cues in acyl-homoserine lactone signaling,” in Bacterial Stress Responses, eds Storz G., Hengge R., editors. (Washington, DC: ASM Press), 2011; 349–363.
- Atkinson S, Williams P. Quorum sensing and social networking in the microbial world. J.R.Soc. Interface., 2009; 6(40): 959-978.
- Rutherford ST, Bassler BL. Bacterial quorum sensing: Its role in virulence and possibilities for its control. Cold Spring Harbor Perspect Med., 2012; 2(11): a012427.
- Waters CM & Bassler BL. Quorum sensing: cell-to-cell communication in bacteria. Annu Rev Cell Dev Bi., 2005; 21: 319–346.
- Defoirdt T, Boon N, Bossier P. Can bacteria evolve resistance to quorum sensing disruption? PLoS Pathog., 2010; 6: e1000989.
- Pei, R. and Lamas-Samanamud, G.R. Inhibition of Biofilm Formation by T7 Bacteriophages Producing Quorum-Quenching Enzymes. Applied and Environmental Microbiology., 2014; 80(17): 5340 –5348.
- Sharp, R.; Hughes, G.; Hart, A.; Walker, J.T. Bacteriophage for the treatment of bacterial biofilms. 2006; U.S. Patent 7758856 B2.
- Olson JC, Fraylick JE, McGuffie EM, Dolan KM, Yahr TL, Frank DW, Vincent TS. Interruption of multiple cellular processes in HT-29 epithelial cells by Pseudomonas aeruginosa exoenzype S. Infect Immun., 1999; 67: 2847-2854.
- Gupta, K.; Marques, C.N.H.; Petrova, O.E.; Sauer, K. Antimicrobial tolerance of Pseudomonas aeruginosa biofilms is activated during an early developmental stage and requires the two-component hybrid SagS. J. Bacteriol. 2013; 195: 4975–4981.
- Hanlon, G.W.; Denyer, S.P.; Olliff, C.J.; Ibrahim, L.J. Reduction in exopolysaccharide viscosity as an aid to bacteriophage penetration through Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol., 2001; 67: 2746–2753.
- Doolittle, M. M.; Cooney, J. J.; Caldwell, D. E. Tracing the interaction of bacteriophage with bacterial biofilms using fluorescent and chromogenic probes. Journal of Industrial Microbiology., 1996; 16(6): 331-341.
- Tait, K., Skillman, L.C., Sutherland, I.W. The efficacy of bacteriophage as a method of biofilm eradication. Biofouling, 2002; 18: 305e311.
- Kay M.K., T.C. Erwin, R.J. McLean and G.M. Aron. Bacteriophage ecology in Escherichia coli and Pseudomonas aeruginosa mixed-biofilm communities. Appl. Environ. Microbiol. 2011; 77: 821–829.
- Ho K. Bacteriophage therapy for bacterial infections: rekindling a memory. Perspect Biol Med., 2001; 44: 1-16.
- Verma, V., K. Harjai, and S. Chhibber. Structural changes induced by a lytic bacteriophage make ciprofloxacin effective against older biofilm of Klebsiella pneumoniae. Biofouling. 2010; 26:729-737.
- Soothill, J.S.; Hawkins, C.; Harper, D.R. Bacteriophage-containing therapeutic agents. U.S. Patent 8105579 B2; 2011.
- Harper, D.R. Beneficial effects of bacteriophage treatment.U.S. Patent 8475787 B2; 2010.
- Yilmaz C, Colak M, Yilmaz BC, Ersoz G, Kutateladze M, Gozlugol M. Bacteriophage therapy in implant-related infections: an experimental study. J Bone Joint Surg Am., 2013; 95(2): 117–25.
- Seth, A.K.; Geringer, M.R.; Nguyen, K.T.; Agnew, S.P.; Dumanian, Z.; Galiano, R.D.; Leung, K.P.; Mustoe, T.A.; Hong, S.J. Bacteriophage therapy for Staphylococcus aureus biofilm-infected wounds: A new approach to chronic wound care. Plast. Reconstr. Surg. 2013; 131: 225–234.
- Ganegama Arachchi, G.J.; Cridge, A.G.; Dias-Wanigasekera, B.M.; Cruz, C.D.; McIntyre, L.; Liu, R.; Flint S.H.; Mutukumira, A.N. Effectiveness of phages in the decontamination of Listeria monocytogenes adhered to clean stainless steel, stainless steel coated with fish protein, and as a biofilm. J. Ind. Microbiol. Biotechnol., 2013; 40: 1105–1116.
- Liao, K.S.; Lehman, S.M.; Tweardy, D.J.; Donlan, R.M.; Trautner, B.W. Bacteriophages are synergistic with bacterial interference for the prevention of Pseudomonas aeruginosa biofilm formation on urinary catheters. J. Appl. Microbiol., 2012; 113: 1530–1539.
- Zhang, Y. Hu, Z. Combined treatment of Pseudomonas aeruginosa biofilms with bacteriophages and chlorine. Biotechnol. Bioeng. 2013; 110: 286–295.
- Szafranski, S.P., Winkel, A., Stiesch, M. The use of bacteriophages to biocontrol oral biofilms. Journal of Biotechnology., 2017; 7763: 16 pages.
© The Author(s) 2017. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.


