ISSN: 0973-7510
E-ISSN: 2581-690X
Bioversity of fungal antagonist, Trichoderma from different locations of Nilgiri district of Tamil Nadu, India, were characterized through molecular methods. Thirty four isolates were tentatively identified as Trichoderma and confirmed upto species level through molecular tools. PCR amplification of the 18s-28s rRNA gene region revealed that all thirty four isolates produced amplicon size of 600bp and were further confirmed through sequencing. The sequences of various Trichoderma spp. were compared with both NCBI and TrichOKEY database to validate their molecular identity. Among 34 isolates, 31 isolates were identified as T. asperellum (KT462693, KU361372, KX533978 to KX533999, KX523262 to KX523264, KX555650, KX147092 to KX147094, KX5334000), 2 isolates as T. harzianum (KX533989, KX533990) and 1 isolate as T. virens (KU666466) through NCBI data base. However, all the T. asperellum isolates identified through NCBI database were identified as T. koningiopsis using TricHOKEY data base. Identity of T. harzianum isolates (TRI 35 and TRI 36) and T. virens isolate (TRI 37) were same in both NCBI and TricHOKEY database. Antagonistic assay with diverse species of Trichoderma revealed that T. virens (TRI 37) was effective in inhibiting the radial growth of Pythium aphanidermatum (87.78%) followed by T. harzianum (TRI 35), (TRI 36) and T. asperellum (TRI 9) in vitro. The effective isolates T. virens (TRI 37), T. harzianum (TRI 35, TRI 36) and T. asperellum (TRI 9) were compatible with each other. Biopriming of cucumber seeds with talc based formulation of the consortia comprising of T. virens isolate ( TRI 37), T. harzianum isolates (TRI 35 and TRI 36) and T. asperellum TRI 9 @ 108 cfu/g and soil application suppressed damping off to an extent of 76.82% over untreated control.
Chlamydospore, Conidiophore, ITS, Phylogenetic analysis, Trichoderma.
Trichoderma is a filamentous, soil-borne, mycoparasitic fungus found in plant root ecosystem32 with antifungal activity against several soil-borne plant pathogens.15 Antagonistic action is attributed through competition for space and nutrients,23 production of siderophores,3,29 synthesis of inhibitory compounds (pyrone antibiotics)5 and the release of cell wall lytic enzymes including cellulytic, chitinolytic, pectinolytic, proteolytic and lipolytic enzymes.6 Besides the antagonistic activity, Trichoderma is an opportunistic, avirulent plant symbiont. Rhizosphere colonization by Trichoderma stimulate plant defense, plant growth and reproductive capacity.4,12 Meyer21 demonstrated the diversity in conidial ornamentation, mitochondrial DNA and plasmids among strains having warted conidia and warranted for taxonomic revision. Recently Polymerase Chain Reaction (PCR) has been used for identification of fungal species. Genotypic techniques involving the amplification of a phylogenetically informative target, such as the small-subunit (18S) rRNA gene are increasingly gaining importance.35 rRNA gene is essential for the survival of all cells and the genes encoding the rRNA are highly conserved in the fungal kingdom. The rRNA genes are universally conserved, while the ITS region and intergenic spacer (IGS) are highly variable.19 The ITS and IGS region are the fastest evolving regions and varies among the species within a genus. Thus, the sequences of these regions were used for identification of closely related species.34 The diversity of Trichoderma has been used for the management of soil-borne diseases. Among the soil-borne diseases, damping off caused by the genus Pythium is a common problem in fields and greenhouse grown crops which kills the seedlings. This disease complex usually involves other pathogens such as Fusarium, Phytophthora and Rhizoctonia. Pre – and post-emergence damping-off caused by Pythium spp. in vegetable crops are economically important worldwide33. Rapid germination of sporangia of Pythium in 1.5–2.5 h after exposure to exudates or volatiles from seeds or roots22 followed by immediate infection makes management of the pathogen very difficult.33 Pythium spp. tends to be generalistic and non-specific in their host range, which causes extensive and devastating root rot is often very difficult to prevent or control.17 With this background the present study was undertaken for characterization of bio geographical diversity of Trichoderma by morphological and molecular means to explore the antagonistic potential against damping off pathogen in cucumber under protected cultivation.
Sampling and Isolation of Trichoderma
Rhizospheric soil samples were collected from different crop fields of Nilgiri district, Tamil Nadu, India. Trichoderma were isolated from the rhizospheric soil samples on Trichoderma selective medium10 using serial dilution technique.26 The plates were incubated at 28±2°C for 4 to 7 days. Visible fungal colonies were transferred to Potato dextrose agar (PDA) plates and incubated at 28±2°C for 5 days and maintained on PDA medium for subsequent studies.
Molecular characterization of Trichoderma spp.
Genomic DNA extraction from Trichoderma isolates
Extraction of genomic DNA of all the isolates of Trichoderma spp. were extracted by harvesting the mycelium grown in potato dextrose broth for 3-4 days at 28±2°C. Mycelial mat was collected on filter paper, washed with distilled water for 2-3 times, frozen and used for DNA extraction. Genomic DNA was extracted as per the protocol described by Raeder and Broda.24 DNA was suspended in 50 µl of TE buffer and quantified with ethidium bromide fluorescence.
PCR amplification and sequencing
Primers ITS1 (5’-TCCGTAGGTGAACCT GCGG-3’) and ITS4 (5’-TCCTCCGCTTAT TGATATGC-3’) described by White et al34 were used to amplify a fragment of rDNA including ITS1 and ITS2 and the 5.8S rDNA gene. The PCR amplification reactions were performed in a 50 ¼l mixture containing 50 mM KCl, 20 mM Tris HCl (pH 8.4), 2.0 mM MgCl2, 200 ¼M of each of the four deoxynucleotide triphosphates (dNTPs), 0.2 ¼M of each primer, 40 ·g/¼l of template and 2.5 U of Taq polymerase. The cycle parameters included an initial denaturation of 1 min at 95°C, followed by 35 cycles of 1 min at 95°C, 30 s at 60°C and 1.5 min at 72°C, with a final extension of 10 min at 72°C. The PCR products were resolved in 1% agarose gel, purified PCR product was sequenced in SciGenome Labs Pvt Ltd , Kerala.
Phylogenetic analysis
The rDNA homology searches were performed using ITS gene sequences by BLAST program (http://www.ncbi.nlm.nih.gov). Sequeces were compared with Trichoderma spp. isolates retrieved from the Genbank database. Newly obtained sequences were submitted in
Genbank database (NCBI). Sequences were analyzed in pairwise and multiple sequence alignment and the identity was scored with the Bio-Edit V 7.0.511. Phylogenetic tree was constructed by the neighbor joining method and tree topologies were evaluated by performing bootstrap analysis of 1000 data sets performed with MEGA 6 (Molecular Evolutionary Genetic Analysis) software31. The rDNA homology searches were performed using ITS gene sequences by TrichOKEY program (http://www.isth.com). Sequences were compared with Trichoderma spp. isolates retrieved from the TrichOKEY database.
Isolation of Pythium from infected cucumber plants
The pathogen Pythium was isolated from damping off affected cucumber plants collected from major cucumber growing areas of Coimbatore, Erode and Madurai districts of Tamil Nadu. The infected plant tissue was washed with sterile water and cut into small pieces from the leading edges of lesions. Then surface sterilized with 0.1% mercuric chloride, washed with sterile distilled water thrice and shade dried on sterile filter paper. The dried pieces were plated on PDA and incubated at 28°C ± 2°C for 5 days.
Molecular Characterization of Pythium isolate
Isolation of genomic DNA of Pythium sp.
Genomic DNA was extracted from the suspension cultures of Pythium by the Cetyl Trimethyl Ammonium Bromide (CTAB) method as described by Lee and Taylor.18 The isolate of Pythium was grown at room temperature (28 ± 2°C), and transferred into 250 ml conical flasks containing 150 ml potato dextrose broth (PDB). It was incubated at 28 ± 2°C for 5 days. After complete colonization of the medium, the mycelium was harvested by filtration through sterile filter paper and stored at -80°C until used for DNA extraction. DNA was extracted from the harvested mycelia according to the procedure described by Mahuku20 . Mycelia were ground to a fine powder in liquid nitrogen and suspended in CTAB buffer.
The mixture was incubated at 65°C for 30 min. DNA was precipitated using ice-cold isopropanol and the pellet was washed with 70% ethanol, dried and dissolved in TE buffer.
Identification of Pythium sp.
To identify the species of Pythium isolates of 16S rDNA intervening sequence specific Pa1-(5’TCCACGTGAACCGTTGAAATC3’);ITS2-(5’GCTGCGTTCTTCATCGATGC-3’) primers were used to get an amplicon of 210 bp size.13 PCR amplification reactions were performed in a 50 ¼l mixture containing 50 mM KCl, 20 mM Tris HCl (pH 8.4), 2.0 mM MgCl2, 200 ¼M of each of the four deoxynucleotide triphosphates (dNTPs), 0.2 ¼M of each primer, 40 ·g/¼l of template and 2.5 U of Taq polymerase. Amplification was conducted with a total reaction volume of 50µl in Eppendorf Master Cycler, German. The PCR settings used were as follows: a hold of 2 min at 95°C, 30 cycles of 1min at 94°C, 30 sec at 54°C and 1min at 72°C and a final extension of 10min at 72°C. The PCR products were resolved on 1% agarose gel at 50 V, stained with ethidium bromide (0.5µg/ml) and analyzed using gel documentation system.
Screening of Trichoderma spp against P. aphanidermatum
The antifungal activity of Trichoderma spp. was tested by dual culture technique.7 The pathogen and Trichoderma were grown on PDA for a week at room temperature (28 ± 2°C), about nine mm diameter mycelial disc of the pathogen (Pythium aphanidermatum) was cut from the periphery and transferred to the Petri plate with PDA and nine mm diameter mycelial disc of Trichoderma was placed simultaneously at opposite sides of same Petriplate aseptically and incubated at room temperature 28 ± 2°C with alternate light and darkness for 7 days and observed periodically. The experiment was replicated thrice and per cent growth inhibition was calculated by the formula of I = (C-T)/C x 100, where C is mycelial growth in control plate, T is mycelial growth of test organisms in inoculated plate and I is inhibition of mycelial growth. Hyperparasitism was calculated by measuring the overgrowth of Trichoderma isolates on the pathogen from the zone of interaction of Trichoderma with pathogen in centimeter.
Testing the efficacy of Trichoderma spp. against P. aphanidermatum in green house
The efficacy of Trichoderma spp. against damping off pathogen was evaluated with the four effective Trichoderma isolates viz.,T. virens (TRI 37), T. harzianum isolates (TRI 36, TRI 35) and T. asperellum isolate (TRI 9) in pot culture. Treatment details include T1- Biopriming (BP) with TRI 37 @ 10g/kg of seeds, T2- BP with TRI 36 @ 10g/kg of seeds, T3- BP with TRI 35 @ 10g/kg of seeds,T4- BP with TRI 9 @ 10g/kg of seeds,T5- BP with (TRI 37+TRI 36+TRI 35+TRI 9) @ 10g/kg of seeds, T6- Soil application (SA) with TRI 37 @ 2.5kg/ha at 15 and 30th days after seeding, T7 – SA with TRI 36 @ 2.5kg/ha at 15 and 30th days after seeding,T8- SA with TRI 35 @ 2.5kg/ha at 15 and 30th days after seeding, T9- SA with TRI 9 @ 2.5kg/ha at 15 and 30th days after seeding, T10- SA with (TRI 37+TRI 36+TRI 35+TRI 9) @ 2.5kg/ha at 15 and 30th days after seeding, T11- BP+SA with TRI 37 @ 10g/kg of seeds+@ 2.5kg/ha at 15 and 30th days after seeding,T12- BP+SA with TRI 36 @ 10g/kg of seeds+@ 2.5kg/ha at 15 and 30th days after seeding,T13- BP+SA with TRI 35 @ 10g/kg of seeds+@ 2.5kg/ha at 15 and 30th days after seeding, T14- BP+SA with TRI 9 @ 10g/kg of seeds+@ 2.5kg/ha at 15 and 30th days after seeding, T15- BP+SA with (TRI 37+TRI 36+TRI 35+TRI 9) @ 10g/kg of seeds+@ 2.5kg/ha at 15 and 30th days after seeding ,T16- BP+SA with Metalaxyl 2g/kg of seeds + 0.1% @ 15 and 30th days after seeding,T17- Un treated control. The treatments were replicated thrice and pathogen inoculated control was maintained. Five cucumber seeds were planted in each pot containing sterile potting medium (red soil: sand: FYM at 1:1:1 w /w/w). The pathogen was multiplied in sand maize medium and incorporated @ 10g per pot up to the depth of 10cm @105cfu/g. Trichoderma was delivered through bio priming of seed and soil application with different combinations. Seeds were bioprimed with talc based bio formulation @ 10 g/ kg followed by two soil applications on 30 and 45 days after sowing @ 2.5 kg/ha. Plants inoculated with the pathogen alone served as control. Healthy controls were also maintained. Disease incidence was recorded after 20 days of sowing and per cent disease incidence was calculated as follows.
The experimental design was completely randomized with three replicates (pots) for each treatment and repeated twice.
Isolation of Trichoderma spp.
A total of 34 isolates of Trichoderma were isolated from different rhizosphere soil samples of different crop plants. Isolate code, species identification, location, NCBI accession numbers, TrichOKEY identification and isolation details of Trichoderma strains are furnished in Table 1.
Table (1):
Identification, NCBI Genebank accession number and isolation details of different isolates of Trichoderma.
| S. No. | Isolate code | Area | GPS Location Longitude | Latitude | Source of culture | Genebank accession number | Identity of Species | |
|---|---|---|---|---|---|---|---|---|
| NCBI | TricHOKEY | |||||||
| 1 | TRI 1 | Coonoor | 11.3530 f N | 76.7959 f E | Lillium | KX533988 | T. asperellum | T. koningiopsis |
| 2 | TRI 2 | Coonoor | 11.3530 f N | 76.7959 f E | Lillium | KX533979 | T. asperellum | T. koningiopsis |
| 3 | TRI 3 | Gudalur | 11.5029 f N | 76.4917 f E | Tea | KX533983 | T. asperellum | T. koningiopsis |
| 4 | TRI 4 | Gudalur | 11.5029 f N | 76.4917 f E | Tea | KX533985 | T. asperellum | T. koningiopsis |
| 5 | TRI 5 | Baraliayur | 11.3429 f N | 76.8500 f E | Tea | KX533991 | T. asperellum | T. koningiopsis |
| 6 | TRI 6 | Chinnaka- rumpalam | 11.3347 f N | 76.7705 f E | Silver oak | KT462693 | T. asperellum | T. koningiopsis |
| 7 | TRI 7 | Chinnaka- rumpalam | 11.3347 f N | 76.7705 f E | Tea | KX533980 | T. asperellum | T. koningiopsis |
| 8 | TRI 8 | Baraliayur | 11.3429 f N | 76.8500 f E | Chrysanthemum | KX533992 | T. asperellum | T. koningiopsis |
| 9 | TRI 9 | Kaikatti | 11.3128 f N | 76.6909 f E | Tea | KX533993 | T. asperellum | T. koningiopsis |
| 10 | TRI 10 | Edavanalli | 12.5207 f N | 77.9855 f E | Lillium | KX533994 | T. asperellum | T. koningiopsis |
| 11 | TRI 11 | Edavanalli | 12.5207 f N | 77.9855 f E | Lillium | KX533986 | T. asperellum | T. koningiopsis |
| 12 | TRI 12 | Kallar | 11.3390 f N | 76.8652 f E | Tea | KU361372 | T. asperellum | T. koningiopsis |
| 13 | TRI 13 | Kallar | 11.3390 f N | 76.8652 f E | carnation | KX533978 | T. asperellum | T. koningiopsis |
| 14 | TRI 14 | Kasolai | 11.3151 f N | 76.6992 f E | Carnation | KX533987 | T. asperellum | Unidentified |
| 15 | TRI 15 | Kasolai | 11.3151 f N | 76.6992 f E | Gerbera | KX533984 | T. asperellum | T. asperellum |
| 16 | TRI 16 | Kasolai | 11.3151 f N | 76.6992 f E | Tea | KX533995 | T. asperellum | T. koningiopsis |
| 17 | TRI 19 | Katteri | 11.3341 f N | 76.7879 f E | Chrysanthemum | KX533996 | T. asperellum | T. koningiopsis |
| 18 | TRI 20 | Kunnakombai | 11.3149 f N | 76.7111 f E | Tea | KX523262 | T.asperellum | T.koningiopsis |
| 19 | TRI 21 | Kunnakombai | 11.3149 f N | 76.7111 f E | Silver oak | KX533997 | T. asperellum | Unidentified |
| 20 | TRI 22 | Kothagiri | 11.4086 f N | 76.8720 f E | Rose | KX523263 | T.asperellum | T.koningiopsis |
| 21 | TRI 23 | Kothagiri | 11.4086 f N | 76.8720 f E | Lillium | KX147092 | T. asperellum | T. koningiopsis |
| 22 | TRI 24 | Kodanad | 11.5015 f N | 76.9063 f E | Lillium | KX084067 | T. asperellum | T. koningiopsis |
| 23 | TRI 25 | Kodanad | 11.5015 f N | 76.9063 f E | Chrysanthemum | KX533998 | T. asperellum | Unidentified |
| 24 | TRI 26 | Kodanad | 11.5015 f N | 76.9063 f E | Carnation | KX533999 | T. asperellum | T. koningiopsis |
| 25 | TRI 27 | Denad | 11.44906916 | 76.9420 f E | Gerbera | KX533981 | T. asperellum | T. koningiopsis |
| 26 | TRI 28 | Masiangudi | 11.5634 f N | 76.6338 f E | Tea | KX533982 | T. asperellum | T. koningiopsis |
| 27 | TRI 29 | Masiangudi | 11.5634 f N | 76.6338 f E | Chrysanthemum | KX5334000 | T. asperellum | Unidentified |
| 28 | TRI 35 | Devasola | 11.3204 f N | 76.6868 f E | Lillium | KX533989 | T. harzianum | T. harzianum |
| 29 | TRI 36 | Devasola | 11.3204 f N | 76.6868 f E | Silver oak | KX533990 | T. harzianum | T. harzianum |
| 30 | TRI 37 | Devasola | 11.3204 f N | 76.6868 f E | Tea | KU666466 | T. virens | T. virens |
| 31 | TRI 38 | Chinnaka- rumpalayam | 11.3347 f N | 76.7705 f E | Tea | KX523264 | T. asperellum | T. koningiopsis |
| 32 | TRI 50 | Kallar | 11.3390 f N | 76.8652 f E | Chrysanthemum | KX555650 | T. asperellum | T. koningiopsis |
| 33 | TRI 60 | Maniyapuram | 11.3157 f N | 76.7080 f E | Carnation | KX147094 | T. asperellum | T. koningiopsis |
| 34 | TRI 70 | Maniyapuram | 11.3157 f N | 76.7080 f E | Gerbera | KX147093 | T. asperellum | T. koningiopsis |
Molecular characterization of Trichoderma spp.
PCR amplification with the conserved primer (ITS 1 -5’TCTGTAGGTGAACCTGCG 3’) and ITS 4-5’TCCTCCGCTTATTGATATGC 3’) of ITS region yielded the genomic product of 600 bp (Fig 1) in the reactions performed with 34 isolates of Trichoderma species. Absence of size variation among the isolates collected suggest that, majority of the isolates belong to Trichoderma.
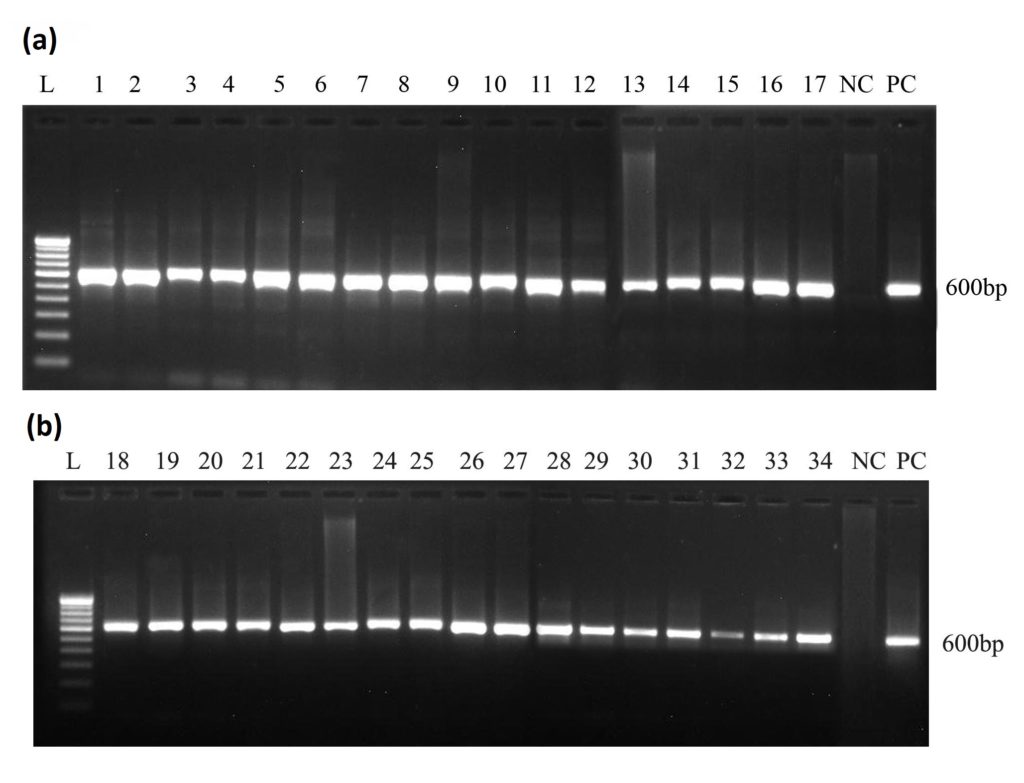 Fig. 1. Molecular confirmation of Trichodema isolates with ITS primers
Fig. 1. Molecular confirmation of Trichodema isolates with ITS primersDNA sequencing
The size of the amplicon containing the ITS1, ITS2 and 5.8S r RNA was around 600 bp. In order to ascertain the Trichoderma orgin of sequence, the sequences were initially analyzed in BLAST. In the BLAST analysis isolates TRI 1 to TRI 16, TRI 19 to TRI 21, TRI 23- TRI 29, 38, 50, 60 and 70 had highest identity with T. asperellum. The query coverage was between 93-100% and identity was between 96-100%. In the case of other next three isolates TRI 35, 36 and TRI 37, isolate TRI 35 and 36 exhibited maximum identities with T. harzianum isolate TRI 37 with T. virens. On the basis of identity search in BLAST the isolates collected in the present study could be clearly categorized into three groups.1. Major group comprising isolates were identified as T. asperellum and another small group of two isolates belonging T. harzianum and one belonging to T. virens(table) To overcome this problem the International Commission of Taxonomy of Fungi has recommended the use of DNA barcode tools for correct identification of Hypocrea and Trichoderma species. Therefore the ITS neucleotide sequence of the 34 isolates of the present study were analysed in TrichOKEY programme (www.isth.info) contrasting to results obtained in BLAST search in TrichOKEY analysis. The 34 isolates could be differentiated into 4 groups, one major group comprise isolate TRI 2-13, 16, 19, 20, 24, 26, 27, 28, 38, 50, 60 and 70 belonging to T. koningiopsis pertaining to the Rufa clade. The second group consists of isolates TRI 35 and TRI 36 which were confirmed as T. harzianum under catopteron clade, third group comprised isolate TRI 37 which was identified as T. virens belonging to virens clade, fourth group comprised of isolate TRI 15 which was identified as T. asperellum belonging to pachybasium A clade. Comparison of the results of TrichOKEY with BLAST indicate that the isolates TRI 2-13, 16, 19, 20, 24, 26, 27, 28, 38, 50, 60 and 70 under BLAST search were identified as T. asperellum. The other two group of isolates were identified as T. harzianum and T. virens by BLAST and appeared to be similar to TrichOKEY analysis. The six isolates (TRI 1, 14, 21, 23, 25 and 29) identified as T. asperellum in BLAST were found to have only genus specific hall mark sequences in TrichOKEY. However further species identification was not possible in TrichOKEY since species specific hall mark were not detectable, the isolates are therefore considered as unidentified species under the genus Trichoderma.
Diversity study of Trichoderma spp.
The result of the phylogenetic analysis based on the 18S-28S-rRNA gene sequences of different species of Trichoderma isolates were analyzed and results revealed that three different clusters were formed in phylogenetic tree (Fig 2). The evolutionary history was inferred using the Neighbor-Joining method Saitou and Nei.25 The optimal tree with the sum of branch length = 1.69400390. The difficulty in identification of species using NCBI similarity search tool, BLAST (http://blast.ncbi.nlm.nih.gov). has been expressed by several workers.8 The lacunae in identifying species on the basis of similarity search in BLAST are absence of quality control of species authentification, sequences deposited under the original names and not under the names after verification. Kredics et al16 suggested that more than 40% of Hypocreae and Trichoderma sequences available in Genbank database are unidentified or misidentified at the species level. In the present study the isolate which had maximum hit with T. asperellum were identified as T. koningiopsis in TrichOKEY. However identification of the other isolates as T. virens, T. harzianum tally both in BLAST and TrichOKEY search.
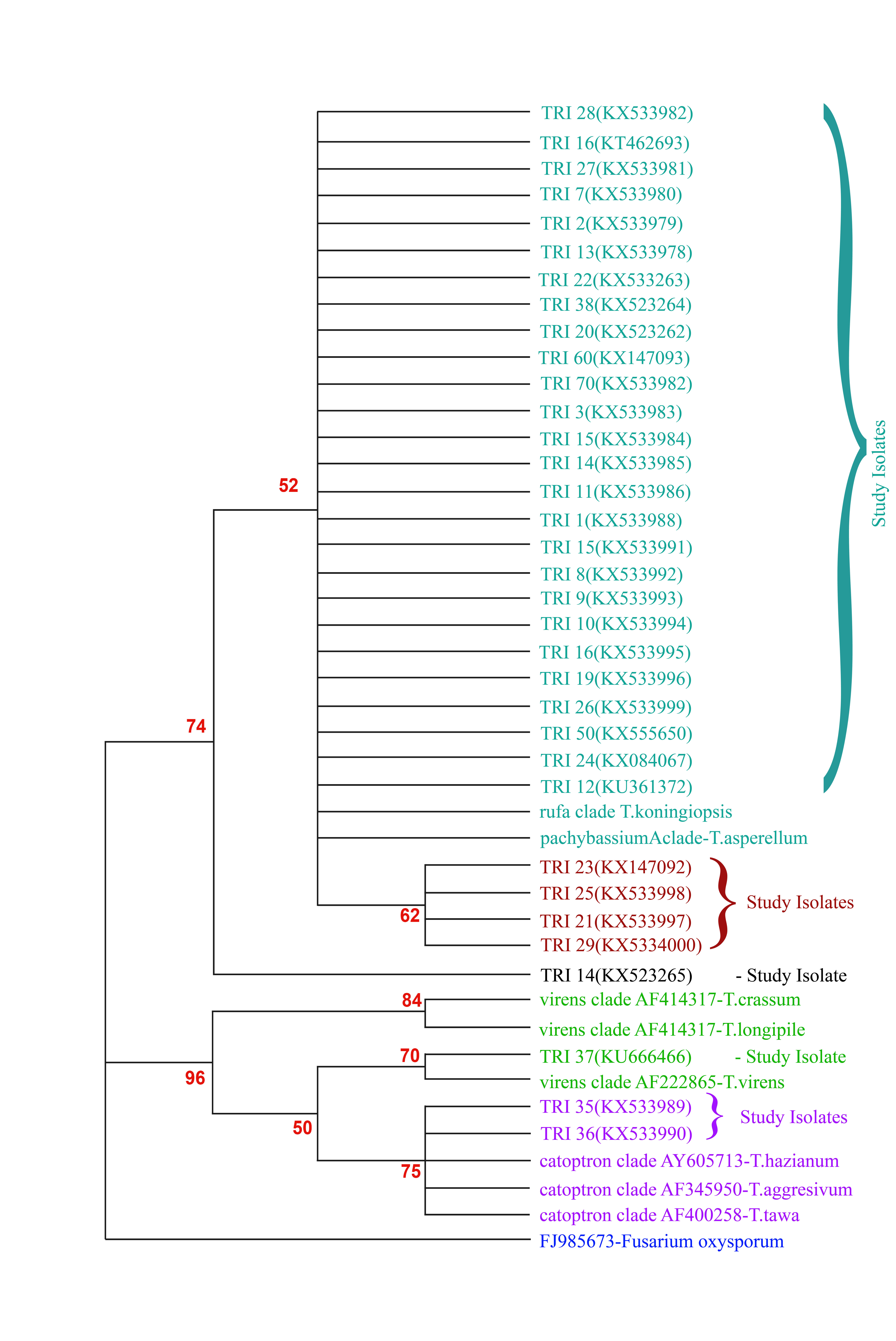 Fig. 2. Phylogenetic tree of the Trichoderma isolates. The numbers given over branches indicate bootstrap co-efficient
Fig. 2. Phylogenetic tree of the Trichoderma isolates. The numbers given over branches indicate bootstrap co-efficientThe relationship between the Trichoderma isolates of the present study and the established species in the TrichOKEY programme were assessed further. The type sequences of the identified species, T. koningiopsis, T. asperellum, T. virens and T. harzianum belonging to clade Rufa, Pachybasium A,Virens and Catopteron (harzianum clade) were retrived from database and analyzed in multiple alignment in CLUSTAL-W programme. A phylogenetic tree was constructed based on alignment clearly revealed the species identity of the isolates under study. The TRI isolates 2-13, 15, 16, 19, 20, 22, 24, 26-28, 38, 50, 60 and 70 occupied the same position as the T. koningiopsis the member of rufa clade, it is intresting to note here that T. asperellum which belong to pachybasium clade also occupy the same branch as that of rufa clade which suggest that on the basis of nucleotide identity these two species are not easily distinguishable. The isolate TRI 37 was grouped with virens clade, TRI 35 and TRI 36 was aligned with catopteron clade. Very clearly these three rufa,virens and catopteron clade branch off distinctly. Interesting results are observed with TRI isolates TRI 23, 25, 21, 29 and 14 which could not be identified at species level in TrichOKEY. They have origin along with rufa and pacybacium clade but branch off distinctly. They may represent a new species group which need to be validated by taking more phylogenetic marker genes.
The shortcoming in identification of species by using only ITS marker has been reported by several workers.30,17 Druchian and kubich9 evaluated along 11 gene loci and formed that the 4th and 5th introns of translation elongation factor 1 alpha (tef1-EF-1±) and the coding region of endochitinase -42(ech 42) aid in resolving the species
Antagonistic activity
Dual culture assay revealed that all the isolates of Trichoderma spp inhibited the mycelial growth of P. aphanidermatum more than 50% over control (Table 2, Fig 3). However, the maximum inhibition of 87.78% of the mycelia growth of P. aphanidermatum was observed with the T. virens isolate TRI 37. It was followed by the T. harzianum isolates TRI 35 and TRI 36, which inhibited the mycelial growth to an extent of 85.5% over control. The next best isolates TRI 7, TRI 9, TRI 26 and 38 which inhibited the growth of pathogen to an extent of 81.5, 81.3, 80.0 and 80.0 per cent over the control were T. asperellum respectively. Similarly Anita et al2 reported that, interaction between Trichoderma and isolated Pythium species in dual culture technique, range of inhibition was observed ranging from 56.92-86.67%. The significant inhibition was observed in case of T. viride against P. viniferum 86.67% . Studies on hyperparasitism indicated that the T.virens isolate (TRI 37) overgrew on P. aphanidermatum up to 2.16 cm from the zone of interaction, indicating the hyperparasite nature of the T. virens isolate TRI 37. Hyperparasitism by the T. virens indicate the capability of these isolate produce hydrolytic enzyme followed by lysis of pathogen. Similarly hyperparasitism nature reported by Yang et al,36 In co-culture in vitro, isolates of Trichoderma spp., including Tri01003, Tri01090 and Tri01091, displayed the ability to steadily colonize and aggressively attack the mycelia of P. ultimum, and finally produce conidia on the Pythium colony.
 Fig. 3. Antagonistic efficacy of Trichoderma spp. against cucumber damping off pathogen under in vitro condition
Fig. 3. Antagonistic efficacy of Trichoderma spp. against cucumber damping off pathogen under in vitro conditionTable (2):
In vitro efficacy of Trichoderma spp. against Pythium aphanidermatum by dual culture method.
S. No. |
Isolates No. |
Mycelia growth (cm) Pythium |
Mycelia growth (cm) Trichoderma |
Hyperpar- asitism (cm) |
Inhibition over control (%) |
|---|---|---|---|---|---|
1 |
TRI 1 (T. asperellum-KX533988) |
2.67 |
6.3 |
1.8 |
70.33n (56.99) |
2 |
TRI 2 (T. asperellum -KX533979) |
2.20 |
6.8 |
1.7 |
75.56h (60.37) |
3 |
TRI 3 (T. asperellum -KX533983) |
2.37 |
6.6 |
1.8 |
73.67i(59.78) |
4 |
TRI 4 (T. asperellum -KX533985) |
2.70 |
6.3 |
1.9 |
70.00o (56.78) |
5 |
TRI 5 (T. asperellum -KX533991) |
2.63 |
6.4 |
1.62 |
70.78m (57.27) |
6 |
TRI 6 (T. asperellum-KT462693) |
3.60 |
5.4 |
1.60 |
60.00s (50.76) |
7 |
TRI 7 (T. asperellum-KX533980) |
1.40 |
7.6 |
1.98 |
84.44e (66.76) |
8 |
TRI 8 (T. asperellum-KX533992) |
2.10 |
6.9 |
1.8 |
76.67f (61.12) |
9 |
TRI 9 (T. asperellum-KX533993) |
1.37 |
7.6 |
1.08 |
84.81d (67.06) |
10 |
TRI 10 (T. asperellum-KX533994) |
2.70 |
6.3 |
1.08 |
70.00o (56.78) |
11 |
TRI 11 (T. asperellum-KX533986) |
2.50 |
6.5 |
1.98 |
72.22l (58.19) |
12 |
TRI 12 (T. asperellum- KU361372) |
2.10 |
6.9 |
1.44 |
76.67f (61.11) |
13 |
TRI 13 (T. asperellum -KX533978) |
3.57 |
5.4 |
0.72 |
60.37r (50.98) |
14 |
TRI 14 (T. asperellum -KX533987) |
3.63 |
5.4 |
1.8 |
59.63t (50.55) |
15 |
TRI 15 (T. asperellum -KX533984) |
2.37 |
6.6 |
1.44 |
73.70i (59.14) |
16 |
TRI 16 (T. asperellum -KX533995) |
3.07 |
5.9 |
1.98 |
65.93p (54.28) |
17 |
TRI 19 (T. asperellum -KX533996) |
2.37 |
6.6 |
1.44 |
73.70i (59.14) |
18 |
TRI 20 (T. asperellum -KX523262) |
4.30 |
4.7 |
0.36 |
52.22w (46.27) |
19 |
TRI 21 (T. asperellum -KX533997) |
2.13 |
6.9 |
1.8 |
76.30g (60.86) |
20 |
TRI 22 (T. asperellum- KX523263) |
3.63 |
5.4 |
1.08 |
59.63t (50.55) |
21 |
TRI 23 (T. asperellum- KX147092) |
2.40 |
6.6 |
1.98 |
73.33j (58.90) |
22 |
TRI 24 (T. asperellum- KX084067) |
4.30 |
4.7 |
0.36 |
52.22w (46.27) |
23 |
TRI 25 (T. asperellum- KX533998) |
2.67 |
6.3 |
1.98 |
70.37n (57.02) |
24 |
TRI 26 (T. asperellum- KX533999) |
2.43 |
6.6 |
1.98 |
72.96k (58.66) |
25 |
TRI 27 (T. asperellum- KX533981) |
4.20 |
4.8 |
1.08 |
53.33v (49.90) |
26 |
TRI 28 (T. asperellum- KX533982) |
3.70 |
5.3 |
1.98 |
58.89u (50.11) |
27 |
TRI 29 (T. asperellum- KX5334000) |
2.63 |
6.4 |
1.8 |
70.74m (57.25) |
28 |
TRI 35 (T. harzianum- KX533989) |
1.33 |
7.7 |
1.98 |
85.19c (67.36) |
29 |
TRI 36 (T. harzianum- KX533990) |
1.30 |
7.7 |
1.98 |
85.56b (67.66) |
30 |
TRI37 (T. virens -KU666466) |
1.10 |
7.8 |
2.16 |
87.78a (69.53) |
31 |
TRI 38 (T. asperellum- KX523264) |
2.20 |
6.8 |
1.98 |
75.56h (60.37) |
32 |
TRI 50 (T. asperellum- KX555650) |
2.13 |
6.9 |
1.8 |
76.30g (60.86) |
33 |
TRI 60 (T. asperellum- KX147094) |
3.40 |
5.6 |
1.97 |
62.22q (52.07) |
34 |
TRI 70 (T. asperellum -KX147093) |
3.57 |
5.4 |
1.8 |
60.37r (50.98) |
35 |
Control |
9.0 |
– |
– |
– |
Means followed by a common letter are not significantly different at the 5% level by DMRT; Figures in parentheses are square root transformed values
Bioefficacy of Trichoderma formulation on the management of cucumber damping- off
The effective isolates of T. virens, T. harzianum isolates (TRI 35 and 36) and T. asperellum isolate (TRI9) were evaluated for the management of cucumber damping off under pot culture in green house through biopriming of seeds and soil application either as individual isolate or as consortia. Results of the investigation emphasized ingeneral that bio-priming, soil application and bio priming coupled with soil application with consortia of Trichoderma isolates comprising of T. virens (TRI 37), T. harzianum isolates (TRI 35 and 36) and T. asperellum (TRI 9) were effective in the suppression of damping off rather than the application of individual isolates of Trichoderma compared to untreated control. However, bio priming and soil application with the consortia comprising of T. virens (TRI37), T. harzianum isolates (TRI 35 and 36) and T. asperellum (TRI 9) suppressed damping off to an extent of 76.82% over untrated control and was followed by the soil application of consortia comprising of (TRI 37+TRI 36+TRI 35+TRI 9), which was applied on 15 and 30th days after seeding (74.08% reduction over control).
Comparison of Trichoderma consortia, delivered through biopriming and soil application with T. virens isolate (TRI 37) and T. harzianum isolates (TRI 35 and 36) was only next to seed treatment with metalaxyl coupled with soil application of metalaxyl 0.1% on 15 and 30th days after seeding, which reduced damping off upto 85.02% over control (Table 3). Similar results were also reported by Abd-El-Khair et al1 and Singh et al28 that confirms our findings. They reported that the incidence of damping-off was found maximum in the pathogen inoculated control (54.67%) and lowest in the plants treated with the consortium of Trichoderma isolates BHU51+BHU105 (22.00%) rather than the individual application of Trichoderma isolate BHU51 and BHU105 on to seeds. Singh and Singh27 also reported that the use of mixture of Trichoderma, increase the level of defense related enzymes in the plant that protect the plant from the infection caused by Macrophomina.
Table (3):
Effect of bioformulations of Trichoderma spp. on the incidence of cucumber damping off under glasshouse conditions.
S. No |
Treatments |
Damping off incidence (%) |
Per cent reduction over control |
|---|---|---|---|
T1 |
BP with TRI 37 @10g/kg of seed |
38.00k (37.16) |
53.44 |
T2 |
BP with TRI 36 @10g/kg of seed |
39.10l (38.70) |
53.15 |
T3 |
BP with TRI 35 @10g/kg of seed |
39.07l (38.68) |
53.19 |
T4 |
BP with TRI 9 @10g/kg of seed |
39.20l (38.76) |
53.03 |
T5 |
BP with TRI37+TRI 36+TRI 35+TRI9 10g/kg of seed |
37.17j (37.56) |
55.46 |
T6 |
SA with TRI 37 @ 2.5 kg/ha at 15 and 30th days after seeding |
29.66g (32.99) |
64.46 |
T7 |
SA with TRI 36 @ 2.5 kg/ha at 15 and 30th days after seeding |
32.71h (34.88) |
60.81 |
T8 |
SA with TRI 35 @ 2.5 kg/ha at 15 and 30th days after seeding |
32.69h (34.87) |
60.83 |
T9 |
SA with TRI 9 @ 2.5 kg/ha at 15 and 30th days after seeding |
35.27i (36.43) |
57.74 |
T10 |
SA with TRI37+TRI 36+TRI 35+TRI 9 @ 2.5 kg/ha at 15 and 30th days after seeding |
21.13c (27.36) |
74.68 |
T11 |
BP +SA with TRI 37 @10g/kg of seed+2.5 kg/ha at 15 and 30th days after seeding |
23.04d (28.68) |
72.39 |
T12 |
BP +SA with TRI 36 @10g/kg of seed+2.5 kg/ha at 15 and 30th days after seeding |
23.00d (28.65) |
72.44 |
T13 |
BP +SA with TRI 35 @10g/kg of seed+ 2.5 kg/ha at 15 and 30th days after seeding |
26.45e (30.95) |
68.31 |
T14 |
BP +SA with TRI 9 @10g/kg of seed+ 2.5 kg/ha at 15 and 30th days after seeding |
27.24f (31.46) |
67.36 |
T15 |
BP +SA with TRI37+TRI 36+TRI 35+TRI9 @10g/kg of seed+ 2.5 kg/ha at 15 and 30th days after seeding |
19.34b (26.09) |
76.82 |
T16 |
BP +SA with Metalaxyl 2g/kg of seed+ 0.1% at 15 and 30th days after seeding |
12.50a (20.70) |
85.02 |
T17 |
Control |
83.47m (66.01) |
– |
ACKNOWLEDGMENTS
I would like to duly acknowledge UGC grant and DST-FIST for providing the facilities of instrumentation in Department of Plant Pathology, Tamil Nadu Agricultural University.
- Abd-El-Khair, H., Khalifa, R. Kh. M., Haggag, K. H. E. Effect of Trichoderma species on damping off diseases incidence, some plant enzymes activity and nutritional status of bean plants. J. Amer. Sci. 2010; 6(9): 486-497.
- Anita, P., Aarti, L., Ashwin, L., Hariprasad, P., Shubhada, M. In vitro antagonistic properties of selected Trichoderma spp. against tomato root rot causing Pythium species. Int. J. Sci. Environ. Technol., 2012; 1(4): 302-315.
- Anke, H., Kinn, J., Bergquist, K.E., Sterner, O. Production of siderophores by strains of the genus Trichoderma: Isolation and characterization of the new lipophilic coprogen derivative, palmitoylcoprogen. Biol.Met., 1991; 4(3): 176 180.
- Bailey, B.A., Bae, H., Strem, M.D., Roberts, D.P., Thomas, S.E., Crozier, J., Samuels, G.J., Choi, I.Y., Holmes, K.A. Fungal and plant gene expression during the colonization of cacao seedlings by endophytic isolates of four Trichoderma species. Planta., 2006; 224: 1449–1464.
- Benítez, T., Rincón, A. M., Limón, M. C., Codón, A. C. Mecanismos de biocontrol de cepas de Trichoderma. Int. Microbiol., 2004; 7(4): 249-260.
- Bisset, J. A. revision of the genus Trichoderma. II. Infrageneric classification. Can. J. Bot., 1991; 69: 2357-2372.
- Chernin, L., Chet, I.: Microbial enzymes in the bio control of plant pathogens and pests. In: Enzyme in the environment (Dick, R.P. and Burns, R.G. eds). Marcel Dekker,New York, 2002; pp171-225
- Dennis, C. Webster, J. Antagonistic properties of species groups of Trichoderma III. Hyphal interaction. Transaction of British mycological Society.1971; 57: 363-369.
- Druzhinina, I., Koptchinski, A., Komon, M., Bissett, J., Szakacs, G., Kubicek, C.P. An oligonucleotide barcode for species identification in Trichoderma and Hypocrea. Fungal Genet. Biol., 2005; 42: 813–828.
- Druzhinina, I., Kubicek, C.P. Species concepts and biodiversity in Trichoderma and Hypocrea: from aggregate species to species clusters. J. Zhejiang Univ. Sci., 2005; 6: 100–112.
- Elad, Y., Chet, L., Henis, Y. A selective medium for improving quantitative isolation of Trichoderma spp. from soil. Phytoparasitica, 1981; 9 (1): 59-67.
- Hall, T.A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98NT. Nucleic Acids Symposium Series , 1999 ; 41: 95-98.
- Harman, G.E., Howell, C.R., Viterbo, A., Chet, I. Lorito, M. Trichoderma species opportunistic, avirulent plant symbionts. Nat. Rev. Microbiol., 2004; 2: 43–56.
- Jarvis, W. R.(ed): Managing diseases in green house crops. Saint Paul, Minnesota: APS Press. 1992; 122-7.
- Jash,S., Pan, S. Evaluation of mutant isolates of T.harzianum against R.solani causing seedling blight of green gram. Ind. J. Agric. Sci., 2004; 74: 190-193
- Kredics, L., Antal, Z., Doczi, I., Manczinger, L., Kevei, F., Nagy, E. Clinical importance of the genus Trichoderma. A review. Acta. Microbiol. Immunol. Hung., 2003; 50:105–117.
- Kullnig-Gradinger, C.M., Szakacs, G., Kubicek, C.P. Phylogeny and evolution of the fungal genus Trichoderma: a multigene approach. Mycol. Res., 2002; 106: 757–767.
- Lee, S.B., Taylor, J.W. Isolation of DNA from fungal mycelia and single spores. In: PCR protocols: A guide to method and applications (Innis, M.A., Gelfand, D. H., Sninsky, J.J., White, T.J. eds). New York, USA, Academic press,1990; pp 282-287.
- Lieckfeldt, E., Samuels, G.J., Helgard, H.I., Petrini, O. A morphological and molecular perspective of Trichoderma viride: is it one or two species. Appl. Environ. Microbiol., 2002; 65: 2418-2428.
- Mahuku, G. A Simple Extraction Method Suitable for PCR-Based Analysis of Plant, Fungal and Bacterial DNA. Plant Mol. Biol. Rep., 2004; 22: 71-81.
- Meyer, R. J. Mitochondrial DNAs and plasmids as taxonomic characters in Trichoderma viride. Appl. Environ. Microbiol., 1991; 57: 2269-2276.
- Osburn, R.M., Schroth, M.N., Hancock, J.G., Hendson, M. Dynamics of sugarbeet colonization by Pythium ultimum and Pseudomonas species: Effects on seed rot and damping-off. Phytopathol.,1989 ; 79: 709–716.
- OzbayNusret, S. E. N. Fusarium crown and root rot of tomato and control methods. J Plant Pathol., 2004; 2: 1-4.
- Raeder, U., Broda, P. Rapid preparation of DNA from filamentous fungi. Lett. Appl. Microbiol .,1985; 1: 17–20
- Saitou, N., Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. and Evol., 1987; 4:406-425.
- Sinclair, J.B., Dhingra, O.D. (ed): Basic plant pathology methods, 2nd edn. CRC press, 1995; pp 448.
- Singh, S. P. and Singh, H. B. 2014. Effect of mixture of Trichoderma isolates on biochemical parameters in leaf of Macrophomina phaseolina infeceted brinjal. J. Environ. Biol. 35: 871-876.
- Singh, S.P., Singh, H. B., Singh, D. K. Biocontrol potential mixture of Trichoderma isolates on Damping off and collar rot of tomato. The Bioscan, 2014; 9(3): 1301-1304.
- Srivastava, M., Tiwari, R., Sharma, N. Effect of different cultural variables on siderophores produced by Trichoderma spp., 2013; 1(7): 1- 6.
- Taylor, J.W., Jacobson, D.J., Kroken, S., Kasuga, T., Geiser, D.M., Hibbett, D.S., Fisher, M.C. Phylogenetic species recognition and species concepts in fungi. Fungal Genet. Biol., 2000; 31: 21–32.
- Tamura, K., Stecher, G., Peterson, D., Filipski, A., Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. and Evol., 2013; 30: 2725-2729.
- Vinale, F., Sivasithamparam, K., Ghisalberti, E.L., Marra, R., Woo, S.L., Lorito, M. Trichoderma–plant–pathogen interactions. Soil Biol. Biochem., 2008; 40: 1–10.
- Whipps, J.M., Lumsden, D.R. Biological control of Pythium species. Biocontrol Sci. and Techn., 1991; 1: 75–90.
- White, T.J., Bruns, T., Lee, S.,Taylor, J.W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: PCR protocols: a guide to methods and applications. (Innis, M.A., Gelfand, D.H., Shinsky, J.J., White, T.J., eds). Academic Press, San Diego, 1990; pp 315–322.
- Woese, C.R., Fox, G.E. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc. Natl. Acad. Sci., USA. 1977; 74: 5088-5090.
- Yang, Y., Chang, K. F., Hwang, S.F., Callan, N. W., Howard, R.J., Blade, S.F. Biological control of Pythium damping off in Echinacea angustifolia with Trichoderma species. J.Plnt.Dis.Protn.,2004; 111(2): 126-136.
© The Author(s) 2017. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.


