Niranjan Prakashrao Patil1*, Avinash D. Bholay2,
Balu P. Kapadnis3 and Vishwas B. Gaikwad2
1Department of Microbiology, Abasaheb Garware College, Pune, Maharashtra, India.
2Department of Environmental Sciences, KTHM College, Nashik, Maharashtra, India.
3Department of Microbiology, Savitribai Phule Pune University, Pune, Maharashtra, India.
ABSTRACT
The synthetic dyes are complex and resist microbial degradation, posing serious threat to the environment. The present study investigate methyl red (MR) decolorization and degradation potential of Bacillus circulans NPP1 isolated and identified from textile effluent treatment plant. The Bacillus circulans NPP1 decolorized greater than 98% of 50 ppm MR under optimal conditions (static, 4.5 hr, pH 7.5, 35°C temperature and 1% inoculum size of OD600=0.1).The isolate exhibited efficient decolorization of broad spectrum of dyes (nine dyes other than MR). The combination of glucose and yeast extract (YE) cosubstrates were found to be best for decolorization. The comparative analysis of decolorized broth and control confirmed azoreduction of MR into component metabolites by employing UV/vis spectrometry, FTIR and GCMS techniques. The induction of various oxidative (lignin peroxidases, laccases and tyrosinases) and reductive enzyme (azoreductase) indicated their involvement in decolorization and biodegradation process. The results of phytotoxicity test on seeds of Sorghum bicolor and Pennisteum americanum, revealed less toxic nature of degraded product as compare to parent MR dye. This study emphasizes the potential of Bacillus circulans NPP1 for bioremediation of dye contaminated effluent.
Keywords:Methyl red; Bacillus circulans; Biodegradation; Textile effluent; Decolorization.
INTRODUCTION
The synthetic dyes are the aesthetic requirement of human civilization and it’s demand and usage is continually increasing. It was worldwide estimated that 280,000 tons of textile dyes find its way in to the effluent due to inefficiencies in dyeing process every year1. Color is the first contaminant observed in wastewater and has to be removed before discharging in to large water bodies. Many dyes are visible in water at concentrations as low as 1 ppm2.The azo dyes are most commonly used dyes and are resistant to chemical and biological degradation due to their complex chemical structure and synthetic origin. Many azo dyes and their breakdown products are toxic, carcinogenic, and mutagenic in nature. The mono azo dye MR used in this study show mutagenicity. Several physicochemical methods have been applied for the removal of dyes from wastewater effluent. However, physical/chemical methods have inherent drawbacks over eco-friendly microbial or enzymatic decolorization3,4. Recent research endeavors had reported wide variety of microorganisms capable of decolorizing various dyes3,5. The approaches to address dye decolorization problem by isolating microbes in pure culture4,6,7, developing consortium8,9,10 degrading dyes efficiently, broad dye decolorization profile,11,12, aerobically13,14,15, anaerobically4,16 are evident in scientific literature. The most frequent biological degradation of azo dyes was performed in static anoxic condition and few studies reported aerobic degradation15. MR is not practiced as a textile dye in industry but it is used as model azo dye to know underline decolorization and degradation attributes. Hsueh and Chen (2007) reported MR as the most difficult dye to degrade due to charged carboxyl group on Methyl red at ortho position to azo bond, inhibiting decolorization activity. Bacillus circulans was reported earlier as a member in an active microbial consortium (acclimatized activated sludge) with Pseudomonas aeruginosa and two unidentified bacteria8, but its standalone dye bioremediation potential was not investigated. The objective of this work was, to isolate and identify bacterial species from textile effluent and evaluate its performance in vitro with a view to find solution to textile dye effluent disposal problems. This work investigates possibility of isolate Bacillus circulans NPP1 for removal of textile dyes used in the present study.
MATERIALS AND METHODS
Dyes, Chemicals and Media
Acid Red 2 (MR) Acid orange 52, Acid red 27 and Acid blue 74 were purchased from Qualigen (India) and other textile dyes i.e. Acid orange 7, Direct yellow 12, Acid black 210, Reactive black 5, Reactive red 195, Reactive yellow 145 from M/s. Oswal dyechem (Mumbai, India). Microbiological media and components were purchased from HiMedia (India). The dyes were used without purification. The respective stock solutions of dyes were prepared by dissolving 0.5g of powder in 100ml of distilled water autoclaved after mixing. The lmax of each dye was determined in nutrient broth using spectrophotometer. All the other chemicals were reagent grade.
Sampling, Isolation and Identification of dye decolorizing bacteria
The sludge samples were collected from the different stages of the Naroda common effluent treatment plant (Ahmadabad, India). In order to obtain potential bacterial azo dye degrader, enrichment and acclimatization was performed in mixture of dyes (100ppm concentration) in Nutrient Broth maintained under stationary conditions. Aliquots were withdrawn and microbiological culture plating (Streak plate and Spread plate method) were performed on 100ppm dye mixture containing Nutrient agar. The colorless colonies surrounded by a decolorized zone were selected further. The isolates were assessed for their decolorization ability in Nutrient broth containing dye. The promising bacterial isolate having greatest decolorization potential was characterized using routine microbial identification test (morphology, cultural & biochemical) in our laboratory. The selected bacterial isolates were further characterized by automated Vitek® 2 System version 03.01 (BioMerieux) and partial sequencing of 16S rRNA gene at Molecular Diagnostic Center ( Pune, India). The gene sequence of the isolate was submitted to the Genebank.
MR and other dyes decolorization
Unless otherwise stated, the decolorization system consists of 1% inoculum size of 0.1OD at 600nm, 50ppm MR in nutrient medium ( 0.5% glucose + 0.5%YE+ 0.5% NaCl), pH-7.5, 35oC temperature and static anoxic condition. The decolorization of other dyes was done by replacing MR in previously mentioned set of conditions. The % decolorization was calculated using formula given by Zhao et al., (2014).
Optimization of physicochemical parameters
For optimization of inoculum, 18 hrs old culture having 0.1 OD at 600nm inoculated with varying size from 0.01- 10 %(v/v) in Liquid culture medium containing 50 mg/L concentration of MR. The pH was varied from 4 -11 and temperature was varied in the range 20 to 50oC. The effect of oxygen on the decolorization was studied at static and shaking/agitated conditions (150 rpm) on orbital shaking incubator. The effect of MR concentration was studied in the range 50-600 mg/L. The effect of salt i.e. NaCl was studied from 0-10 g % .Decolorization was studied using various co-substrates like glucose, YE, peptone, urea, casein, sucrose, lactose, whey and combination of two co-substrate in Bushnell Hass medium5(BHM). The whey was filtered to remove particulate matter, neutralized using 0.1N NaOH solution and used 4% in BHM. % decolorization was calculated at a time when any tube in experimental set showed maximum visible decolorization. The MR decolorization rate was calculated by measuring the amount of decolorization or removal of MR(mg)/L/hr.
MR degradation analysis
100 ppm MR decolorized broth was centrifuged at 4000g for 20mins and supernatant was filtered through 0.02 micron filter (Pall Make). The culture filtrate was treated with equal volume of ethyl acetate in separating funnel and organic phase was collected. The organic phase was dried over anhydrous Na2S04. The un-inoculated tube served as a control and given similar treatment. The extract was concentrated in rotary evaporator and used as sample for UV/vis, FTIR and GCMS analysis. The instrumentations methods reported earlier19 were followed.
Enzymes involved in degradation of MR
The cell free extract of induced (MR) and un-induced cells were prepared according to method give by Gomare et al., (2009). The protein content of cell extract was determined by the method of Lowry20 using bovine serum albumin as a standard Laccase was assayed using Guaicol21, Lignin peroxidases22 using n-propanol, azoreductase23 using MR and tyrosinase24 using catechol as substrate.
Toxicity assay
The ethyl acetate extracted products of MR decolorization were dried and dissolve in 10ml sterile distilled water to make final concentration of 300ppm to be used in phytotoxicity assay11 on seeds of Sorghum bicolor and Pennisteum americanum.
Statistical Analysis
The data was analyzed by one-way analysis of variance ( ANOVA) using a Tukey-Kramer multiple comparison test. The observations were considered significant when P was £ 0.05.
RESULTS AND DISCUSSION
Isolation and Identification of Bacillus circulans NPP1
A potential bacterial isolate having rapid dye decolorization activity and broad decolorization profile was chosen as screening and selection criteria. The special attention was given to the color of the cell pellet after centrifugation as dyes decolorization proceed through two steps i.e. adsorption of dyes on cell(biosorption) and their degradation further. The colored pellet indicated first reaction, while colorless cell pellet pointed towards biosorption followed by degradation of dyes. The isolate NPP1 showed promising decolorization capability during screening and isolation program as per the above mentioned selection criteria. The complete (visible) decolorization of 50 ppm MR was achieved by isolate NPP1 under static anoxic condition. The decolorization of MR in tube was initiated from the bottom of tube and after centrifugation pellet was colorless. The concerned bacterium was Gram positive, rod shaped, motile, catalase positive, endospore bearing and typical colony morphology of Bacillus sp. on nutrient agar. The commercial biochemical identification tests (bioMérieux’s API®)) for Gram positive (GP2 MicroPlateTM) bacteria revealed 93% probability for Bacillus circulans. The 16S rRNA gene sequence BLASTN search matched 100% identity with Bacillus circulans strain D1(accession number EU116046) and Bacillus circulans strain I1(accession number FJ009417).The sequence was deposited to GenBank sequence database under accession number GQ478244.In our opinion the combined results of phenetic characters, biochemical characters and 16S rRNA gene sequence similarity sufficient for assuming isolate NPP1 taxonomic identity as Bacillus circulans. The isolate was evaluated for decolorization of dyes other than MR as shown in Table 1.The decolorization of various groups of dyes by bacterium offer flexibility in decolorization of mixture of dyes encountered in real time scenario. The Table 1 confirmed broad decolorization capacity and rapid decolorization of dyes.
Table 1. Details of dyes used in study and their % decolorization by Bacillus circulans NPP1.
Sr. |
Dye |
Class |
λmax( nm) |
% Decolorization* |
1 |
Acid red 27 |
Monoazo sulphone |
520 |
87±1.39 |
2 |
Acid blue 74 |
Indigoid |
586 |
85±1.68 |
3 |
Acid red 2(MR)** |
Acid monoazo |
430 |
98±0.53 |
4 |
Acid red 52 |
Acid sulphone monoazo |
465 |
95±0.87 |
5 |
Acid orange 7 |
Vinylsulphone monoazo |
483 |
84±1.61 |
6 |
Acid black 210 |
Vinyl sulphone triazo |
604 |
89±1.28 |
7 |
Direct yellow 12 |
Diazo sulphone |
403 |
96±1.13 |
8 |
Reactive red 195 |
Diazo sulphone |
540 |
93±1.88 |
9 |
Reactive black 5 |
Vinyl sulphone diazo |
598 |
92±2.32 |
10 |
Reactive yellow 145 |
Monoazo sulphone |
420 |
97±0.6 |
*values are represented as arithmetic mean ±standard deviation, * MR decolorization was measured at 4.5 hrs and rest of dyes at 12 hrs.
Optimization of Physicochemical factors
The optimization experiments were followed to know a culture condition that enhances decolorization of MR by employing one factor at a time approach. The various optimized factors are shown in Table 2 in relation to broad range in which decolorization achieved (greater than 75%). Bacillus circulans NPP1 offered advantages over other reported strains11,19,,26 on small inoculum size required for decolorization. The decolorization time did not reduced despite increase in inoculum size. This may be due to initial acclimatization time and building up of substantial NAD(P)H pool required for decolorization. Under oxygen limited condition (anoxic) azo dyes act as terminal electron acceptors25 that leads to the azo reduction of dyes. In oxygenic/shaking condition turbidity was developed in the MR containing medium but upon broth centrifugation red color pellet was observed (biosorption). This was due to competition between MR and oxygen for acting as terminal electron acceptor. Sulfonic azo dyes are impermeable to the cell membrane and its permeation into cell is the rate limiting step in the dedgradation28 this may be reason of rapid decolorization of MR over sulfonated dyes used in this work.
Table 2. The investigated range of the decolorization factors selected for the optimization using Bacillus circulans NPP1.
Factors |
Range investigated |
Operational range* |
Optimized factor |
pH |
3-10 |
6-8.5 |
7.5 |
Temperature(oC) |
20-50 |
25-45 |
35 |
MR concentration(mg/L) |
50-600 |
50-250 |
150** |
NaCl(g %) |
0-10 |
0.5-4% |
0.5 |
Inoculum size(%) |
0.1-10% |
0.5-1 |
1 |
Oxygen relationship |
Static and agitated |
– |
Static |
Carbon sources(1g %) |
Starch, glucose, Lactose, sucrose & whey(4%) |
All |
Starch & glucose |
Nitrogen sources (0.5g %) |
Peptone, urea, casein, YE |
Peptone & YE |
YE |
Combination of N and C source |
Glucose+YE
Starch+YE Peptone+YE |
All |
Glucose+YE
|
* > 75% decolorization of 50 ppm MR, **highest decolorization rate(13.66 mg/L/h)obtained.
Analysis of MR degradation
The dissimilarity in UV/VIS spectral pattern of treated and untreated MR as shown in Figure 1 suggested azo reduction. The hypochromic shift (lower absorbance) at 430nm and appearance of new peak at 308nm (hypsochromic shift) indicated reduction. The peak at 308nm may be a combined peak of anthranilic acid and N,N-dimethyl-p-phenylenediamine intermediate and further incubation leads to the hypochromic shift indicating its probable mineralization. In treated MR absorbance was not reduced zero because, the azo reduction products of MR contributed absorbance at wavelength 430 nm18. The GCMS spectra revealed two major intermediates of MR degradation at retention time 7.754(anthranilic acid) and 11.997 (N,N’dimethyl aniline) as shown in Tables 3. The immediate intermediate of azo reduction of MR i.e. N,N’dimethyl-p- phenylenediamine was not detected in degraded extract, instead closer chemical species N,N-dimethyl aniline was detected. This may have been formed by deamination of N,N-dimethyl-p-phenylenediamine. The FTIR spectra (Figure 2) obtained after 24 hrs of treated MR exhibited significant changes in position of peak when compared to untreated MR confirming its degradation. The control MR spectra displayed peak at 1600.97 cm-1 for azo bond, a peak at 1725.38 cm-1 for C=O stretch and peak at 2923.22 cm-1 for asymmetric CH3 stretch, a peak at 1273.06 cm-1 for C-N stretch of aromatic amines, a peak at 1487.17 cm-1 designate C-C stretch in aromatics, a peak at 940.33 cm-1 for O-H bend of carboxylic acid that confirmed the structure of MR as previously reported19. The FTIR spectra of treated MR showed absence of peak at 1600.97 cm-1 indicates breakdown of azo bond, due to action of azoreductase. The absence of peak at 686.68 cm-1, 727.19 cm-1,762.87 cm-1 indicates loss of aromaticity, lack of peak at 1273.06 cm-1 represent absence of C-N stretch of aromatic amines. The appearance of strong peak at 1374.33 cm-1 indicated CH2, CH3 bending. The FTIR spectra of treated MR indicate biodegradation. The azo reduction products were further degraded into aliphatic amines possibly through the mediation of oxidative enzymes such as lignin peroxidase and laccase29.
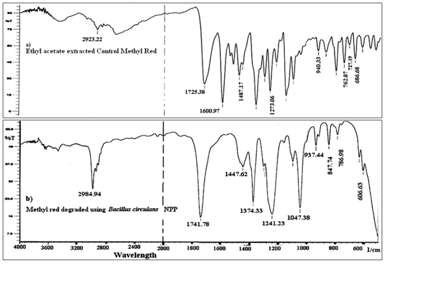
Fig. 1. Uv/vis.spectral changed during decolorization of MR by Bacillus circulans NPP1. Observations were recorded after 0 hr(MR control), 4.5 hrs(I) and 8 hrs(II).
Table 3.GCMS spectra details of important peaks of MR degraded byBacillus circulans NPP1 after 12 hrs
Major peak number |
Retention
Time |
m/z (Predominant ion Fragment) |
Base
Peak |
Mass
peak |
Identified intermediate |
1 |
7.754 |
50,65,73,92,105,119,137 |
119 |
137 |
Anthranilic acid |
2 |
11.997 |
39,42,51,77,105,120,121 |
120 |
121 |
N,N-dimethyl aniline |
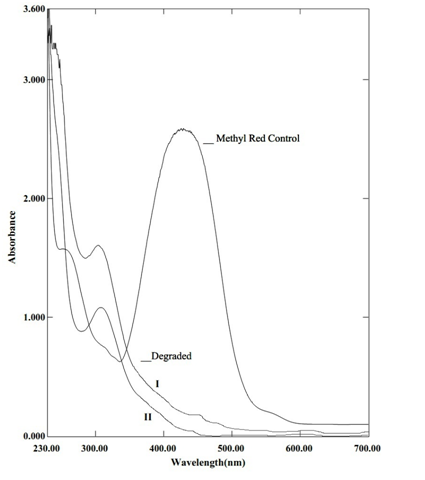
Fig. 2. The comparative FTIR spectra of treated and untreated MR usingBacillus circulans NPP1 after 24 hrs of incubation
Enzymes involved in degradation
Cell free intracellular content of Bacillus circulans NPP1 had shown the presence of significant activities of lignin peroxidase, azoreductase, laccase and tyrosinase. The results of the specific activity of different enzyme assessed are presented in Table 4. The azoreductase activity induced 230%, Lignin peroxidase activity induced 227%. Laccase and Tyrosinase activity was induced 142% and 166 % respectively. All four enzymes are important candidates for degradation of various synthetic chemical compounds. The presence and induction of enzymes activities in the intracellular content of cell indicate harmless fate of primary aromatic amines produced after reductive cleavage of azo bond (-N=N-) in the dyes.
Table 4. Effect of MR on enzyme activities in Bacillus circulans NPP1
Enzymes |
Enzyme activity# Without MR |
Enzyme activity# With MR |
Azoreductasea |
1.140**±0.035 |
2.626** ± 0.06 |
Lignin peroxidaseb |
0.011*±0.002 |
0.025* ± 0.003 |
Laccaseb |
0.018**± 0.001 |
0.030**± 0.002 |
Tyrosinaseb |
0.050*±0.019 |
0.071*±0.022 |
# values are mean of three experiments(±) SEM, Significantly different from the without MR(Un-induced) at *P<0.05, **P<0.01 by paired t-test. a μg MR reduced min-1mg protein-1
b Umin-1mg protein-1
Table 5. Toxicity of MR and its degradation product on seeds of Sorghum bicolor and Pennisteum americanum
| Parameters studied | Sorghum bicolor | Pennisteum americanum | ||||
| Water | MR# | Degraded MR | Water | MR | Degraded MR | |
| Germination rate(%) | 100 | 83 | 100 | 100 | 65 | 100 |
| Root length(cm) | 6.50±0.07 | 3.09±0.12* | 6.06±0.12$ | 5.87±0.06 | 2.84±0.12* | 5.48±0.13$ |
| Shoot length(cm) | 6.26±0.05 | 1.71±0.11* | 5.90±0.06$ | 5.36±0.05 | 1.39±0.09* | 4.84±0.08$ |
# 300 ppm concentration, Values are mean of three experiments ±SEM(Standard Error of Mean), significantly different from the control(seeds germinated in water) at *P < 0.001, $P<0.05 and non significant at ns P>0.05 by one way analysis of variance(ANOVA) with Tukey Kramer multiple comparison test.
Toxicity studies
The impact of treated MR wastewater on economically important vegetation was evaluated to explore possible reuse in irrigation. The relative sensitivity of two plant seeds (Sorghum bicolor and Pennisteum americanum) against the untreated dye presented in Table 5. There was no germination inhibition of seeds of both the plant seeds by MR metabolites at 300 ppm concentration while untreated MR showed 83% germination of Sorghum bicolor and 65% germination of Pennisteum americanum. Overall, the extracted metabolites of MR showed insignificant toxic effect on roots and shoots length in seed assay compare to seeds grown in presence of MR (significant length reduction).
CONCLUSION
The bacterium Bacillus circulans NPP1 isolated from dye polluted industrial effluent has ability to degrade MR, decolorize various textile dyes. The broad dye decolorization activity of isolate is beneficial to claim commercial application. The physiological requirements of isolate to achieve dye decolorization were moderate. The isolate demonstrated advantage over other reported strain on criteria of fast decolorization achieved by inoculating minuscule biomass. The induction in the activities of various dye degradation and mineralization enzymes indicates significance of isolate. The degradation of MR into less toxic metabolite ascertains its use in irrigation. In our opinion, based on the result of present study, further it is remarkable to evaluate isolates applicability in suitable decolorization process.
ACKNOWLEDGEMENTS
The first author gratefully acknowledges financial support provided by Board of Studies for Colleges and University Departments of University of Pune via grant number BCUD/578.
REFERENCES
- Maas, R., Chaudhari, S. Adsorption and biological decolorization of azo dye reactive red 2 in semicontinuous anaerobic reactors. Process Biochem., 2005;40(2): pp 699-705.
- Banat, I.M., Nigam, P., Singh, D., Marchant, R. Microbial decolorization of textile dye containing effluents: A review. Bioresour., Technol., 1996;58(3): pp 217-227.
- Saratale, R.G., Saratale, G.D., Parshetti, G.K., Chang, J.S., Govindwar, S.P. Bacterial Decolorization and Degradation of Azo Dyes: A Review. J. Taiwan Inst. Chem. Eng., 2011;42(1): pp 138-157.
- dos Santos, A.B., Cervantes, F.J., van Lier, J.B. Review Paper on Current Technologies for Decolorization of Textile Wastewaters: Perspectives for Anaerobic Biotechnology. Bioresour. Technol., 2007;98(12): pp 2369-85.
- Bhatt, N., Patel, K.C., Keharia H., Madamwar, D. Decolourisation of Diazo-Dye Reactive Blue 172 by Pseudomonas aeruginosa NBAR12. J. Basic Microbiol., 2005:45(6); pp 407-418.
- Dave, S.R., Patel, T.L., Tipre, D.R. Bacterial Degradation of Azo Dye Containing Wastes. In: Microbial Degradation of Synthetic Dyes in Wastewaters (Singh SN, ed). Springer International Publishing, 2015; pp 57-83.
- Murugesh, S., Ponmurugan, P., Murugesh, S., Priyadarsini, I. Molecular Characterization of Isolated Bacteria and Application of RSM for Decolorization of Acid Black 1. J. Pure Appl. Microbio., 2011;5(1): pp 77-85.
- Dafale, N., Rao, N.N., Meshram, S.U., Wate, S.R. Decolorization of Azo Dyes and Simulated Dye Bath Wastewater Using Acclimatized Microbial Consortium biostimulation and halo tolerance. Bioresour. Technol., 2008;99(7): pp 2552-8.
- Jadhav, S.U., Jadhav, M.U., Kagalkar, A.N., Govindwar, S.P. Decolorization of Brilliant Blue G Dye Mediated by Degradation of the Microbial Consortium of Galactomyces geotrichum and Bacillus sp. J. Chin. Inst. Chem. Engrs., 2008;39(6): pp 563-70.
- Patel, D.K., Tipre, D.R., Dave, S.R. Selection and Development of Efficient Consortia for Decolorization of Metal Complex Dyes. Toxicol. and Environ.Chem., 2016; pp 1-13.
- Jadhav, U.U., Dawkar, V.V., Ghodake, G.S., Govindwar, S.P. Biodegradation of Direct Red 5B, a Textile Dye by Newly Isolated Comamonas sp. UVS. J. Hazard. Mater., 2008;158(2-3): pp 507-516.
- Deng, D., Guo, J., Zeng, G, Sun, G. Decolorization of Anthraquinone, Triphenylmethane and Azo Dyes by a New Isolated Bacillus cereus Strain DC11. Int.Biodeter.Biodegr., 2008;62(3): pp 263–269
- Kodam, K.M., Soojhawon, I., Lokhande, P.D., Gawai, K.R. Microbial Decolorization of Reactive Azo Dyes under Aerobic Conditions. World J. Microbiol. Biotechnol., 2005;21(3): pp 367-370 .
- Adedayo, O., Javadpour, S., Taylor, C., Anderson, W.A., Moo-Young, M. Decolorization and Detoxication of Methyl Red by Aerobic Bacteria from a Wastewater Treatment Plant. World J. Microbiol. Biotechnol., 2004;20(6): pp 545-550 .
- Wong, P.K., Yuen, P.Y. Decolorization and Biodegradation of Methyl Red by Klebsiella pneumoniae RS-13. Water Res., 1996;30(7): pp 1736-1744.
- Dawkar, V.V., Jadhav, U.U., Jadhav, S.U., Govindwar, S.P. Biodegradation of disperse textile dye Brown 3 REL by newly isolated Bacillus sp. VUS. J. Appl. Microbiol., 2008;105(1): pp 14-24.
- Hsueh, C.C., Chen, B.Y. Comparative study on reaction selectivity of azo dye decolorization by Pseudomonas luteola. J. Hazard. Mater., 2007;141(3): pp 842-849
- Zhao, M., Sun, P.F., Du, L.N., Wang, G., Jia, X.M., Zhao, Y.H. Biodegradation of Methyl Red by Bacillus sp. strain UN2: decolorization capacity, metabolites characterization, and enzyme analysis. Environ. Sci. Pollut. Res., 2014;21(9): pp 6136-6145.
- Gomare, S.S., Govindwar, S.P. Brevibacillus laterosporus MTCC 2298: A potential azo dye degrader. J. Appl. Microbiol., 2009;106(3): pp 993-1004.
- Lowry, O.H., Rosebrough, N.J., Farr, A.L., Randall, R.J. Protein measurement with the Folin phenol reagent. J. Biol. Chem., 1951;193(1): pp 265–275.
- Bains, J., Capalash, N., Sharma, P. Laccase from a nonmelanogenic, alkalotolerant ã-proteobacterium JB isolated from industrial waste water drained soil. Biotechnol. Lett., 2003; 25(14): pp 1155-1159.
- Shanmugam, V., Kumari, M., Yadav, K.D. n-Propanol as a substrate for assaying the lignin peroxidase activity of Phanerochaete chrysoporium. Indian J. Biochem. Biophys., 1999;36(1): pp 39-43.
- Chen, H., Hopper, S.L., Cerniglia, C.E. Biochemical and molecular characterization of an azoreductase from Staphylococcus aureus, a tetrameric NADPH dependent flavoprotein. Microbiology, 2005;15(Pt 5): pp 1433-1441.
- Zhang, X., Flurkey, W. Phenol oxidase in Portabella mushrooms. J. Food Sci., 1997;62(1): pp 97-100.
- Jadhav, S.U, Kalme, S.D., Govindwar, S.P., Biodegradation of Methyl red by Galactomyces geotrichum MTCC 1360. Int. Biodeter. Biodegr., 2008; 62(2):135-142.
- Kalyani, D.C., Patil, P.S., Jadhav, J.P., Govindwar, S.P. Biodegradation of reactive textile dye Red BLI by an isolated bacterium Pseudomonas sp.SUK1. Bioresour. Technol., 2008;99(11): pp 4635–4641.
- Ramalho, P.A., Cardoso, M.H., Cavaco-Paulo, A., Ramalho M.T. Characterization of azo reduction activity in a novel ascomycete yeast strain. Appl. Environ. Microbiol., 2004;70(4): pp 2279–2288.
- Wuhrmann, K., Mechsner, K., Kappeler, T. Investigation on rate- Determining factors in the microbial reduction of azo dyes. European J. Appl. Microbiol. Biotechnol., 1980;9(4): pp 325-338.
- Jadhav, J.P., Parshetti, G.K., Kalme, S.D., Govindwar, S.P. Decolorization of azo dye, methyl red by Saccharomyces cerevisiae MTCC 463. Chemosphere, 2007;68(2): pp 394-400.

