Industrialization and anthropogenic activity represent significant environmental hazards. Emerging pollutants in nature pose a major risk and are linked to some immediate and long-term negative effects on the ecosystem. Traditional methods of excluding pollution are futile and lead to the creation of secondary contaminants that cause diseases, cancer, mental and cardiovascular issues, allergies, and other conditions. Microbes and their enzymes are key players in reducing and removing hazardous contaminants through bioremediation by their catalytic action under ideal settings (temperature/pH/contact time/concentration). Laccases, dehalogenases, proteases, cytochrome P450s, dehydrogenases, and lipases are the primary enzymes used in bioremediation. These enzymes have demonstrated encouraging potential in the breakdown of dangerous pollutants. These enzymes use oxidation, elimination, reduction, and other numerous mechanisms to biodegrade various pollutants. Recombinant enzymes produced from genetically modified microorganisms also enhance the breakdown of pollutants. Recent developments and opportunities for microbial enzymes in the sustainable breakdown of hazardous pollutants such as dyes, polyaromatic hydrocarbons, plastics, heavy metals, pesticides, etc. in the environment due to industrial pollution are the major focus of this review.
Bioremediation, Industrial Pollutants, Microbial Enzymes, Sustainable Solution
Uncontrolled release of pollutants and untreated effluents into the environment, environmental pollution is one of the biggest issues faced globally. Environmental contamination is mostly caused by population increase, industrialization, exploration, urbanization, and mining.1 The survival of humanity is seriously threatened by the significant amount of toxins that have been dumped into the environment, ranging from untreated sewage to nuclear waste.2 Anthropogenic activities including industrialization, farming methods, population growth, and unhealthy competition for dominance are having severely negative effects on Earth. Pollutants, mostly phenols, polyaromatic hydrocarbons, pesticides, azo dyes, polychlorinated chemicals, heavy metals, and other hazardous substances, are produced as a result of these processes. These chemicals endanger the biotic components of ecosystems because they are resistant to biodegradation.3 These contaminants have a profound impact on every area of the planet, having carcinogenic, mutagenic, and toxic consequences on people and other living things.4
Clean water is essential for life, but it is often contaminated by industrial activities, leading to wastewater. This pollution, containing pesticides, pharmaceutical wastes, plastics, heavy metals, organic solvents, acids, alkali, biological wastes, and xenobiotic compounds, poses significant environmental and health risks. To address these issues, conventional methods like physical, chemical, and biological treatments are insufficient. Nanotechnology and nanofiltration are promising alternatives for industrial wastewater treatment due to their high surface area, eco-friendliness, and effectiveness in eliminating pollutants.5
Micropollutants (MPs) are a growing concern in wastewater due to human activities. Despite their low presence, MPs are highly toxic and non-degradable, causing global water quality and health hazards. Conventional wastewater treatment is inefficient and produces hazardous sludge. Alternative sustainable approaches are needed, and recent micro-biotechnological methods, such as bio-adsorption, membrane filtration, biocatalysts, bioreactors, and nanotechnology, have shown effectiveness in removing MPs. Bacteria, fungi, and microalgae can degrade MPs through biosorption, biotransformation, bioaccumulation, and bioconversion mechanisms. Microbial-based nanoparticles (MNPs) can be integrated into existing membrane or adsorption technologies, but systematic studies are still in its infancy.6 Micropollutants, including pesticides, pharmaceuticals, and personal care products, have improved human quality of life but are increasingly contaminating aqueous systems due to seepage, run-off, effluent discharge, and environmental uptake. These pollutants have significant ecotoxicities and adverse effects on human health, making them a critical environmental concern. Current remediation methods are costly, consume excess chemicals, require high energy, and create harmful by-products. However, oxidative laccases and peroxidases, microalgae, and microalgae-bacteria consortia are emerging as green alternatives for remediating micropollutants in contaminated systems. These methods can be used as secondary treatment techniques for wastewater plant effluents or as tertiary treatments in conjunction with other chemical and biological technologies.7 Pollutants have been cleaned up using a variety of physical and chemical techniques, including oxidising agents, electrochemical processes, pollutant adsorption, ion exchange, and membrane filtering. Traditional procedures were adequate for the high concentration of contaminants, but they were insufficient to reduce the pollution to allowable levels.8 Traditional methods for cleaning up pollutants have several drawbacks, including their high cost, difficult procedures, stringent international regulations imposed on decontamination, general public rejection, non-specificity, space limitations, and potential for secondary pollution creation.9 As a result, interest has grown in bioremediation, eco-friendly, and biological procedures. Plastic, introduced in the mid-20th century, has been crucial for modern civilization due to its convenience and cost-effectiveness. However, its extensive use has led to significant environmental issues, including waste management issues in landfills and natural ecosystems.9 Microplastics, particularly those from polyethylene (PE), polyethylene terephthalate (PET), polyurethane (PU), polystyrene (PS), polypropylene (PP), and polyvinyl chloride (PVC), have a profound negative impact on ecosystems and species health.10 Enzyme technology is crucial in promoting environmental conservation by converting plastic waste into less harmful compounds. Research on Thermobifida fusca hydrolase has led to the discovery of various enzymes that can break down PET, with PETase enzyme from Ideonella sakaiensis being particularly noteworthy for its ability to effectively degrade PET into intermediates like BHET, MHET, and TPA. MHETase further processes MHET to produce terephthalic acid and ethylene glycol.11 Significant progress has been achieved in the domain of enzymatic microplastic degradation, specifically regarding polystyrene (PS). Advancements in this domain are apparent, as numerous microbes, including bacteria and fungi, possess the capability to decompose PS. Nonetheless, the specific enzymes that initiate this breakdown process are not yet well comprehended.12
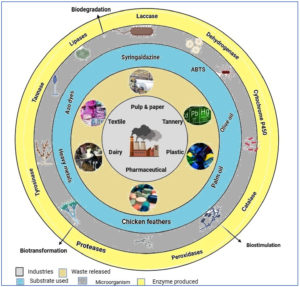
Microorganisms that break down many contaminants enzymatically and convert them into less harmful compounds or metabolites that could be beneficial products are the mainstay of the bioremediation process.13 Reducing environmental pollution through the use of biological agents is an affordable process and reduces the risks they impose on people’s health and threats to the environment. To break down and detoxify pollutants, it mostly employs intracellular accumulation or enzymatic transformation.9 Microbial enzymes are regarded as innovative, economical, and promising when used in the bioremediation of persistent pollutants (Figure 1). Microbes are an abundant source of biocatalytic enzymes, essential in industries like textiles, agriculture, and pharmaceuticals. They reduce environmental pollution through biodegradation and bioremediation. The global enzyme market is growing due to their eco-friendliness, stability, and easy modification. Alternatives like recombinant DNA technology and protein engineering are used to create novel products.14 Because of their metabolic activity and their ability to thrive in various environmental conditions, bacteria, fungi, etc. are present throughout the biosphere and produce enzymes. Among the various microbial enzymes, such as those from Mycobacterium, Pseudomonas, Sphingomonas, Rhodococcus, Mycobacterium has already been shown to break down pesticides and hydrocarbons. Cytochrome P450s, dehydrogenases, laccases (Lac), proteases, hydrolases, lipases, and dehalogenases are the most prevalent microbial enzymes involved in bioremediation.15 The bioremediation technique has some constraints. Only a few biological agents have the aptness to produce specialized enzymes with sufficient power to break down contaminants, and the process is exceedingly slow and constant. Genetically engineered microbes produce required enzymes in huge quantities under ideal conditions; we choose them for bioremediation. Industrial microbial enzymes currently have a very competitive global market. Worldwide recognized companies in the enzyme production sector are DSM, Novozymes, Amano Enzymes Inc., and DuPont. By employing enzymes, we can safeguard our planet against the accumulation of hazardous and poisonous waste in the environment.16 Numerous novel microbial enzymes are been developed for various bioprocesses using rDNA technology, meta-genomics, and protein engineering. To increase the production and efficacy of various microbial enzymes, a range of molecular approaches are used in the food, paper, pharmaceutical, biotechnological, textile, leather, and other industries.17,18 The major focus of this review are the function and mechanism of microbial enzymes in the sustainable bioremediation of industrial pollutants. Additionally, there is a gap in suitable approaches considering the bioremediation of toxic multi-pollutants using enzymatic applications. Most of the research remains lab-scale, with few studies on pilot or field applications. Enzyme stability and activity under real environmental conditions (e.g., temperature fluctuations, mixed contaminants, pH changes) are underexplored. The mechanisms of enzymatic degradation for complex pollutants (e.g., microplastics, PFAS, pharmaceutical residues) are not fully understood. The mechanisms of enzymatic degradation for complex pollutants (e.g., microplastics, PFAS, pharmaceutical residues) are not fully understood. Few sustainable, low-cost carriers (e.g., biochar, waste-derived supports) should have been optimized for enzyme immobilization. Focus is often on well-known microbes; the microbiomes of extreme environments (e.g., deep-sea vents, deserts, Antarctic soils) remain untapped for novel enzymes.
Figure 1. Microbial enzymes’ use in the bioremediation of hazardous industrial wastes into non-toxic substances
Production, optimization, and application of microbial enzymes for industrial pollution
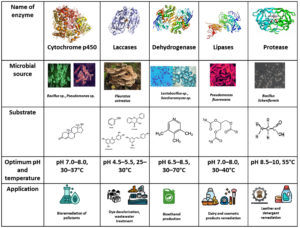
Enzymes are efficient biocatalysts for various biochemical reactions, producing few by-products compared to chemical catalysts. Industrial enzymes, such as proteases, amylases, cellulases, and lipases, can be obtained from microorganisms through optimization and enzyme engineering. These enzymes are essential for industrial conditions, as wild microbial strains produce less enzyme than engineered microorganisms. Most industrial enzymes are derived from plants, animals, and microorganisms due to better yields, cost reduction, and reduced labor. Industrial enzymes are essential for various industries, used for commercial purposes (Figure 2). They are produced by microorganisms, which involve various steps such as isolation, screening, optimization of process parameters, fermentation, purification, characterization, formulation for sale, customer liaison, and working with regulatory authorities. Most bacteria and fungi used to produce industrial enzymes are genetically modified to overproduce them. Solid state and submerged fermentation are commonly used, but submerged fermentation is preferred due to the extracellular nature of the enzyme. Criteria such as pH and temperature stability, specificity, influence of activators and inhibitors, and reaction velocity are used in selecting industrial enzymes. Industrial enzymes are typically produced under controlled conditions using microorganisms, mainly bacteria and fungi being the main producers (Table 1 and 2). Industrial enzymes are frequently produced by submerged and solid-state fermentation. However, due to the extracellular character of the industrial enzyme that is released into the production medium, submerged fermentation has been consistently described as the preferred technique for industrial enzyme secretion from microorganisms. According to Sarrouh et al.,19 some of the factors taken into consideration when choosing which industrial enzymes to manufacture from microbes include pH and temperature stability, specificity, the impact of activators and inhibitors, and reaction velocity. Figure 2 demonstrates the comparative study between various microbial enzymes.
Table (1):
Bioremediation of industrial pollutants through various bacterial enzymes
| No. | Name of enzyme | Name of bacteria | Substrate used | Mechanism Involved | Industry | Application | Ref. |
|---|---|---|---|---|---|---|---|
| 1 | Laccase (EC 1.10.3.2.) | Streptomyces maltophilia | Mainly Synthetic dyes e.g. Congo red, Methylene blue, Toluidine blue, Methyl pink, green, and methyl orange | Ring cleavage in aromatic compounds and reduce one molecule of oxygen in the water and produce free radicals | Textile | Degradation as well as decolorization of synthetic dyes in Textile effluents | 78 |
| 2 | Laccase (EC 1.10.3.2.) | Streptomyces cyaneus | 2,2′-Azino-bis-(3-ethylbenzothiazoline -6-sulfonic acid (ABTS) | Plastic | Oxidation of Micro-pollutants like BPA (Bisphenol A), DFC (Diclofenac), and MFA (Mefenamic acid) | 79 | |
| 3 | Laccase (EC 1.10.3.2.) | Geobacillus thermocatenulatus | ABTS | Textile | Degradation and Decolorization of Textile dyes, especially Congo red and bromophenol blue | 80 | |
| 4 | Laccase (EC 1.10.3.2.) | Anoxybacillus gonensis | ABTS | Tannery | Bioremediation of tannery effluents | 15,80 | |
| 5 | Cytochrome P450 (EC 1.14.14.1) | Rhodococcus rhodochrous | Hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX) | Performs electron transfer reactions and catalysis by reduction or oxidation of heme iron | Pharmaceutical | Degradation of RDX | 24 |
| 6 | Lipase (EC 3.1.1.3) | Bacillus subtilis | Olive oil | Catalyzes the hydrolysis of mono-, di-, and triglycerides into fatty acids and glycerol as well as catalyse the esterification reactions | Food | Bioremediation of Wastewater, Cleaning detergent of tough oil or grease stains | 15,81,52 |
| 7 | Lipase (EC 3.1.1.3) | Bacillus pumilus | Palm oil | Detergent, food, cosmetic | Degradation of palm oil containing Industrial wastewater | 82 | |
| 8 | Dehydrogenase (EC 1.1.1.1) | Pseudomonas putida | 4-Hydroxybenzaldehyde and 4-hydroxy-3-methylbenzaldehyde | Oxidizing organic compounds and generating energy | Tannery | Breakdown of 2,4- xylenol | 83,84 |
| 9 | Dehydrogenase (EC 1.1.1.1) | Stenotrophomonas rhizophila | Vinyl alcohol oligomer and polyvinyl alcohol | Textile, paper, food | Polyvinyl alcohol degradation | 83,84 | |
| 10 | Protease (EC 3.4.21.12) | Bacillus subtilis | Feather culture medium | Assist in the breaking of protein peptide bonds | Poultry, tannery | Deterioration of casein as well as feathers and degradation of proteins like keratin, casein, etc., leather dehairing, and wastewater treatment | 85 |
| 11 | Protease (EC 3.4.21.12) | Chryseobacterium sp. strain kr6, Bacillus pumilus | Chicken feathers | Poultry | Deterioration of feathers | 85 | |
| 12 | Protease (EC 3.4.21.12) | Streptomyces thermoviolaceus | Hair, collagen, Muscle, nail, feathers | Poultry | Hydrolyze the fibrin, collagen, muscle, nail, and hair | 85 | |
| 13 | Protease (EC 3.4.21.12) | Thermoanaerobacter keratinophilus | Complex medium without oxygen having merino wool, human hairs, chicken feathers | Poultry | Breakdown of keratin fibers | 85 | |
| 14 | Dehalogenase (EC 3.8.1.5) | Bacillus sp. | 2,4,6- Trinitrobromophenol (TBP) | Cleaves the carbon-halogen bond and eliminates the halogens | Pesticides | Degradation of TBP | 21 |
Table (2):
Bioremediation of industrial pollutants through various fungal enzymes
| No. | Name of enzyme | Name of fungus | Substrate used | Mechanism involved | Industry | Application | Ref. |
|---|---|---|---|---|---|---|---|
| 1 | Laccase (EC 1.10.3.2) | Coniophora puteana | Syringaldazine (SGZ) | Cleavage of ring in aromatic compounds as well as reduction of one molecule of oxygen in the water molecule and production of free radicals | Textile | Deterioration of artificial dye | 80 |
| 2 | Laccase (EC 1.10.3.2) | T. versicolour | Pellets of T. versicolour | Textile | Detoxifying and reducing the colour, aromatic compounds, and chemical oxygen demand (COD) were reduced up to 70–80% and COD was reduced up to 60%. | 42,80 | |
| 3 | Laccase (EC 1.10.3.2) | Aspergillus flavus | Dyes | Textile | Removing surfactants and dyes | 86 | |
| 4 | Laccase (EC 1.10.3.2) | Cerrena unicolor | Sugarcane bagasse | Paper & pulp | Reduce lignin | 87 | |
| 5 | Laccase (EC 1.10.3.2) | Flavodon flavus, Pycnoporus sanguineus, Trichosporon beigelii NCIM-3326 | Bromophenol blue, malachite green | Textile | Dye Decolorization (Azure B, Brilliant Blue R, bromophenol blue and malachite green) | 88,89 | |
| 6 | Laccase (EC 1.10.3.2) | Phanerochaete chrysosporium | Azo black reactive dye | Tannery | Degradation of Azo Black Reactive 5 Dye | 24 | |
| 7 | Laccase (EC 1.10.3.2) | Phanerochaete chrysosporium URM 6181 and Curvularia lunata URM 6179 | Indigo dye | Textile | Decolourize effluent containing textile indigo dye by approximately 95% | 26,27 | |
| 8 | Laccase (EC 1.10.3.2) | Flavodon flavus, Coriolopsis gallica | Sugar cane bagasse | Pulp and paper | Decolourize the effluent from a Kraft paper mill bleach plant | 25 | |
| 9 | Laccase (EC 1.10.3.2) | Phanerochaete chrysosporium | Polyaromatic hydrocarbon (PAHs) | Pharmaceutical | Catabolize PAHs including anthracene and the endocrine disrupting alkylphenols, a variety of pharmaceutical compounds, antibiotics, anti-inflammatories, and β-blockers are detoxified | 90 | |
| 10 | Lipases (EC 3.1.1.3) | Aspergillus terreus | Disposed engine oil | Catalyses the hydrolysis of mono-, di-, and triglycerides into fatty acids and glycerol as well as catalyse the esterification reactions | Petroleum | Remediate oily polluted soils | 51 |
| 11 | Lipases (EC 3.1.1.3) | Aspergillus terreus | Dairy effluent | Oxidizing organic compounds and generating energy | Dairy | Bioremediation of dairy effluent to reduce over 90% oil and grease | 45 |
| 12 | Dehydrogenase (EC 1.1.1.1) | Phanerochaete chrysosporium (acidic conditions) and Humicola insolens (alkaline conditions) | Pulp mill effluent | Catalyses the hydrolysis of mono-, di-, and triglycerides into fatty acids and glycerol as well as catalyse the esterification reactions | Pulp and paper | Used to treat the acid effluent stream discharged from a pulp plant and remove colour | 91 |
| 13 | Dehydrogenase (EC 1.1.1.1). Laccase (EC 1.10.3.2) | Funalia trogii | Textile Dyes | Oxidizing organic compounds and generating energy | Textile | Applicable to the effluent released from the caustic sewer of bleach plant and for the decolorization of textile dyes | 92 |
| 14 | Catalase (EC 1.11.1.6) | Neurospora crassa | Hydrogen Peroxide | Convert hydrogen peroxide to water and oxygen | Tannery | Bioremediation of heavy metals from tannery effluent | 93 |
| 15 | Peroxidases (EC 1.11.1.7) | Thanatephorus sp., Auricularia sp., Penicillium geastrovirus, Candida tropicalis | Phenols, hydroquinone, dyes, amines, aromatic alcohols and xenobiotic | Catalyze the oxidation of various organic and inorganic substrates by reacting with hydrogen peroxide and similar molecules | Textile | Degradation of synthetic dyes such as azo, remazol blue, Cibacron red, remazol brilliant blue, anthraquinone | 41 |
| 16 | Proteases (EC 3.4.21.12) Keratinase (EC 3.4.99.11) |
Aspergillus flavus, A. oryzae, A. niger, Cladosporium herbarum, Fusarium solani, Penicillium chrysogenum | Hides and skins | Catalyze hydrolytic reactions that help in the degradation of protein molecules firstly into peptides and finally to free amino acids | Tannery | Widely used in the leather processing industries, waste management and wastewater treatment | 94,95 |
| 17 | Proteases (EC 3.4.21.12) | Aspergillus niger, A. oryzae, A. japonicus, A. gallonyces, A. awamori | Tannin | Catalyzes the hydrolysis of ester bonds | Tannery | Used in the degradation of tanneries effluents containing tannins | 55 |
Optimization of parameters for enzyme production by microorganisms
A number of factors, including incubation duration, agitation/shaking, pH, inoculum concentration, incubation temperature, carbon supply, metal ions, and nitrogen source, primarily affect the industrial enzymes produced by microorganisms. Improving the industrial enzyme yield is largely dependent on optimizing these parameters.20
Application of microbial enzymes for the bioremediation of industrial pollutants
In the present scenario, microbial enzyme-mediated bioremediation is a viable approach for reduction and removal of hazardous industrial pollutants. Enzymes are present in almost every naturally existing organism i.e., from prokaryotes to eukaryotes.15 Many microbial enzymes act as catalysts for producing a variety of products from a variety of substrates under controlled circumstances. Additionally, through bioconversion or biodegradation processes, several microbial enzymes can quickly convert hazardous substances into valuable by-products (Figure 1).
Because they lower the reaction activation energy without having any long-term effects, enzymes speed up a variety of biological processes that are essential to maintaining human life. The production of enzymes from microorganisms offers a number of benefits, such as simple gene manipulation, quick growth under regulated conditions, ease of handling, increased manufacturing yield, etc.16
Microbial enzymes are also being employed more frequently in industrial settings as a result of their catalytic activity, non-toxicity, specificity, eco-friendliness, stability, cost-effectiveness, and ease of manufacturing.9 The sources, mechanisms involved, and uses of microbial enzymes isolated from different bacterial and fungal species for bioremediation are described in Table 1 and 2, respectively.
Cytochrome P450 (EC 1.14.14.1)
A member of the heme enzyme superfamily, cytochrome P450 is found extensively throughout the three domains of life-bacteria, archaea, and eukaryote.21 It carries out a variety of tasks, such as the biotransformation of harmful compounds in our ecosystem and synthesizing intricate natural products in living systems. They combine molecular oxygen with NADH or NADPH to produce oxidized products and a carbon substrate. They function as a cofactor.22 P450s can degrade xenobiotics naturally.23 For catalytic activity, they also require ferredoxin and ferredoxin reductase as an electron source. Research on microbial P450s for the bioremediation of hydrocarbons and organic pollutants has been done utilizing both non-engineering and protein engineering approaches.24
One of the recognized microbial P450s is the Bacillus megaterium CYP102A1 (P450BM3) model, which has been proven through protein engineering research to have the ability to oxidize PAHs.22 Many microorganisms, mainly bacteria such as Rhodococcus, Gordonia, Mycobacterium, Pseudomonas, etc., have been found to include some catabolic genes and plasmids that express P450s for the reduction of POPs (persistent organic pollutants) and their elimination from our environment.25
Laccases (EC 1.10.3.2)
Laccases are extracellular enzymes composed of several glycoprotein subunits and contain multiple copper ions. Laccases oxidize certain phenolic and aromatic chemicals in bacteria, fungi, and plants, as well as certain amines, ethers, and esters, via a one-electron process.26 The possibility of laccases being used in many biotechnological processes has been explored because oxygen is the final electron acceptor. These qualities have piqued the researcher’s interest in using laccases.
Laccase is typically a highly stable, industrially applicable, heat-resistant enzyme. This enzyme can eliminate xenobiotics and generate polymeric compounds employed in bioremediation procedures. Textile industry-produced phenols and dyes can be eliminated and detoxified using laccase.27 Laccase is a promising method for treating organic micropollutants in wastewater. It effectively degrades and detoxifies various compounds, including pharmaceuticals, steroid hormones, personal care products, pesticides, and industrial chemicals. Laccases have been successfully utilized to remove pharmaceuticals, personal care products, biocides, endocrine-disrupting agents, steroid hormones, and microplastics. Although no commercially available laccase-based wastewater treatment preparations exist, studies demonstrate its effectiveness.28
Agricultural wastes, including sawdust, banana peels, and rice bran, contain lignin and phenolic substances that increase laccase production.29 There are numerous biotechnological uses for laccases in food, textile, paper, cosmetics, and other industries. This is because of their high oxidative capability. Laccases are also employed to break down agricultural items like pesticides and herbicides, protecting the environment from dangerous chemicals. Laccases remediate soil contaminated by oil hydrocarbons, treat wastewater containing textile industry dyes, and perform bio-bleaching procedures.30,31 Laccases, known to have a huge substrate range, can oxidize a wide variety of aromatic chemicals, especially phenolic substrates.32 According to Guan et al.33 the majority of laccases from different microorganisms are identified, described, and studied, especially the Streptomyces laccase from actinomycetes. Simulated textile effluents (STE) can be discolored by E. coli recombinant CotA laccase. Pure and unpurified CotA laccase decolorized dyes more rapidly when simulated textile effluents were buffered at pH 7.34 In roughly four hours, the pure recombinant laccase eliminates over 93% of the tested colors at a basic pH of 9.0. Laccase-mediated dye degradation is an environmentally acceptable substitute for conventional dye degradation methods, which are sometimes costly and have negative effects.35 Recombinant laccase, which is utilized in aquaculture wastewater bioremediation, is produced by B. vallismortis strain fmb103.36 Five of the seven dyes that were tested-Evans Blue, Brom Cresol Purple, Remazol Brilliant Blue, Reactive Black 5, and Amido Black 10B-were destroyed by it.37 Titanium dioxide, iron oxides, aluminum, mica flakes, cadmium, mercury, and lead are among the heavy metals known to be present in the many colored pigments used in the paint industry. According to reports, the laccase enzyme can break down heavy metals utilized in several industries. The laccase enzyme belongs to a broad class of enzymes known as polyphenol oxidases, which are helpful in the printing and tannery, baking, wine and beverage, textile, and pharmaceutical sectors. Laccase is one of the enzymes produced by microorganisms for bioremediation.
The purpose of this study is to separate bacteria from paint industry wastewater, screen for possible laccase-producing bacteria, and produce the laccase enzyme.38
Laccases can be produced from filamentous fungi such as white rot fungi.39 Strong Laccase producers include Pleurotus florida, P. ostreatus, P. pulmonarius,40 P. tailandia.41 and Trichoderma.42 The fungi Trametes sp., Coriolopsis sp., Grifola sp., and many others also create laccases. The Rhus vernicifera tree served as the first source of laccase isolation.22 Bacteria such as E. coli, insects’ example Bombyx, Drosophila, Papilio, Schistocerca, etc., and plants that produce laccases include mango, peaches, and pines.39
Dehydrogenase (EC1.1.1.1)
Dehydrogenases belong to the oxidoreductase family and are found in bacteria, yeast, plants, animals, & human beings. The bacteria’s cell-free extracts that break down xenobiotics manufactured industrially showed signs of polyethylene glycol dehydrogenase activity. Xenobiotic polyvinyl alcohol which is water soluble is broken down by recombinant polyvinyl alcohol dehydrogenase.40,43 It has been discovered that aldehyde dehydrogenase is involved in the anabolic and catabolic processes of aromatic compounds and according to a protein expression analysis of Amycolatopsis tucumanensis DSM 45259 used in the biodegradation of phenanthrene, it is extensively and specifically expressed.44 Table 1 lists a few dehydrogenases together with information about each one’s characteristics and function in bioremediation.
Lipases (EC 3.1.1.3)
Lipases are triacylglycerol ester hydrolyses that facilitate the conversion of triglycerides into glycerol and fatty acids. Lipases catalyze the reaction in which bonds are broken due to acid (acidolysis), alcohol (alcoholysis), and amino acids (aminolysis) in addition to hydrolysis. Lipases are the most adaptable biocatalysts. Plants, animals, and microbes all have lipases in varying concentrations. The most common source of lipases are microorganisms. Bacillus sp. is one of the most common bacteria that produces lipase. Bacillus alcalophilus, Staphylococcus caseolyticus, B. licheniformis, Serratia rubidaea, B. stearothermophilus, Pseudomonas aeruginosa, and Acinetobacter radioresistens are the most common producers of lipase.45
Microbial lipases are used in industry to create biosensors, which are diagnostic instruments used to identify a range of illnesses. In addition to the pharmaceutical, polymerization, pulp and paper, and cosmetic industries, microbial lipases are commercially useful in the bioremediation of oil residues, petroleum pollutants, and effluents.46,47 Lipases can accelerate the bioremediation of oily effluents that are generated from a range of sources that contain fats, proteins, and oils. Lipase from the bacteria Acinetobacter sp., Rhodococcus sp., and Mycobacterium sp., has been used to manage oil spills containing n-alkanes, aromatic hydrocarbons, and PAHs.48,49
Pseudomonas aeruginosa lipase has been observed to degrade castor oil50 and to remediate soil having waste oil released from industries. Bacteria that are isolated from soil polluted with motor oil produce lipase that helps in the remediation of hydrocarbon.51 In household laundry, lipases are used to reduce environmental contaminants and improve the effectiveness of detergent to get rid of stubborn grease or oil stains. Crude lipase isolated from Bacillus subtilis strain is used to lessen the impact of phosphate byproducts in laundry detergents.52 It was discovered that L. plantarum displayed the highest polycaprolactone (PCL) degradation efficiency in comparison with other lipases when it came to the breakdown of artificial polyester polycaprolactone (PCL) by co-cultures of L. brevis and L. plantarum lipases.53 In fungus, lipase makers are Rhizopus (Rhizopus arrhizus, R. niveus), Penicillium, Aspergillus sp., etc. Lipases are expected to expand at the fastest rate among enzymes due to their extensive use in numerous industrial domains. Table 1 and 2 lists a few microbial lipases together with information about each one’s characteristics and bioremediation function.
Protease (E.C 3.4.21.12)
It is an enzyme that catalyzes the formation of peptide bonds in proteins and is a member of the hydrolase family. Proteases are mainly secluded from Bacillus and Aspergillus species. Microorganism-derived proteases are extremely important because of their low cost, high yield, and practical usage. They are widely used in industries like the food, leather, and wastewater treatment sectors.54 Protease is used in the removal of polymers because it aids in the breakdown of -ester bonds created by polyhydroxyl butyrate (PHB) and c-linkages.55 Because of the presence of keratin protein, which is insoluble in nature, animal horns, nails, poultry feces, and the shedding and molting of appendages are resistant to breakdown. Aside from their disagreeable smell, they are the cause of environmental pollution. By dissolving and recycling keratinous wastes into useful by-products, the protease enzyme keratinase aids in the breakdown of keratin proteins and can be applied to the bioremediation of bird excrement. Stenotrophomonas maltophilia KB13’s keratinase enzyme has shown its ability to use resistant keratinous waste in the biological deprivation of chicken feathers.56 Protease producing bacteria has been discovered and identified from soil of tanneries.57 These are used for silk degumming, leather, waste management, and in silver recovery.58 Bacillus sp. FPF-1 has shown its ability to use resistant keratinous waste biomass from the agricultural industry (accession number MG214993) by degrading chicken feathers at an 82% rate.59 By-products of feather degradation can be employed as ferric ion reducers, fertilisers for plant development, feed additives, free radical scavengers,60 and hazardous hexavalent chromium-reducing agents. Keratinase is utilized in place of the conventional chemicals CaO and Na2S in environmentally friendly enzymatic de-hairing operations in tanneries, which reduce pollution by preventing the release of harmful waste into water bodies.61 The bioremediation of marine crustacean wastes employed in the de-proteinization phase of chitin extraction uses protease enzymes. By producing alkaline protease, Bacillus licheniformis strain MP1 reduces the protein content of shrimp waste by 75%.62 At a temperature of 30 °C and a predominant pH of 7 to 8, Pseudomonas chlororaphis broke down the Impranil substrate while displaying protease activity (beta clearing zone) and esterase activity (alpha clearing zone).63 By breaking down and transforming marine crustacean debris and keratinous waste products into beneficial molecules, proteases lessen environmental pollution.
Microorganisms are a natural source of commercial enzyme preparations as they be cultured in simple conditions, easy cell manipulation etc. About 40%-60% of all enzyme sales from microbial sources is of proteases.64
Peroxidases (E.C. 1.11.1.7)
They are oxidoreductases that utilizes free radical mechanism for transforming a variety of chemical substances into oxidized or polymerized products.65 Peroxidase activity transforms ferricyanides and ascorbic acid into innocuous components. They lessen water pollution by helping in the bioremediation of wastewater contaminated with chlorinated phenolic chemicals, cresol, and phenol. Peroxidases are suitable enzymes that help in the development of enzyme-linked immunosorbent assay (ELISA) kits, as they can produce chromogenic products at very low concentrations.66 These enzymes are used in several industrial and analytical bioprocesses because of their strong reduction potential. They degrade artificial dyes such as brilliant blue, azo dyes, Cibacron red, remazol blue etc.67,68 Peroxidases are also used in the paper industry to hydrolyze cellulose and lignin into carbon dioxide and water, respectively, which breaks down wood components.
Microbes, plants, and animals are the main sources of peroxidases. There have been reports of peroxidase production from a variety of plant sources, including papaya (Carica papaya), banana (Musa paradisiaca), horseradish (Armoracia rusticana),69 and so on. Pseudomonas sp., Escherichia coli, and Bacillus sp. are the main bacterial strains for production of peroxidase. However, Pleurotus ostreatus., Umbelopsis isabelline, Auricularia sp., and, Thanatephorus sp. have been reported to be efficient peroxidase producers in the case of fungal strains.70,71
Cellulases (E.C. 3.2.1.4)
The hydrolysis of cellulosic substrates into monomeric products is facilitated by the cellulase enzymes. They are created when microbial strains hydrolyze the α-1,4-glycosidic linkages of cellulose while growing on cellulosic substrates. Several strains of bacteria, yeast, and fungi have been found to contain cellulases. Because they can use additional pathways to develop higher cellulase activity, fungi are the primary microbiological species that manufacture cellulase enzymes. Aspergillus and Trichoderma are well-known fungal genera that produce cellulase. Strong cellulase production has been reported for A. niger, T. asperellum,72 Trichoderma viride,73 A. fumigatus,74 A. ellipticus,75 and Aspergillus protuberus. Myceliophthora thermophile, Penicillium echinulatum, and Rhizopus oryzae are among the other fungal strains that may have cellulase activity. Thermomonospora sp., Cellulomonas sp., Microbispora sp., Clostridium sp., Cellvibrio sp., and Ruminococcus sp. are among the bacteria that are known to be powerful cellulose-producing genera.76
Cellulase enzyme is mostly utilized in textile industry for bio-polishing, to modify the structure of fibres and are also used in the last phase of textile manufacturing to soften and lessen the tendency of fabric to pill as well as to remove the starch sizing.77
Microbial enzymes mechanism for bioremediation
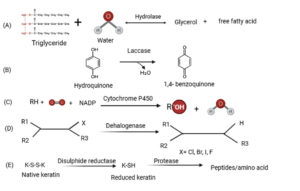
In nature, several genes are known to express different types of enzymes and that regulate how these enzymes function and what kind of structure they should adopt to perform a certain function. The numerous protein folds of the enzyme are regulated by highly distinct sets of genes, which not only adapt the structure of the enzyme for a variety of uses but also determine the catalytic mechanism, which establishes the function of enzymes at a given site. Figure 3 shows the mechanism of action for some of the most important microbial enzymes utilized in bioremediation.
Figure 3. Enzymatic reactions of microbial enzymes employed in bioremediation. (a) Hydrolase, (b) Laccase, (c) Cytochrome P450, (d) Dehalogenase, (e) Protease
Free radicals are created when laccase oxidizes the substrate. Laccase-mediated catalysis can be expanded by utilizing mediators. Laccase oxidizes mediators, which are organic molecules with a low molecular weight. The glycerol backbone of a lipid substrate serves as the typical site of lipase activity in the case of the lipase enzyme.
Electrons are transferred from the substrate to an electron carrier by dehydrogenase enzyme. NAD+, FAD, and NADP+ are common electron acceptors utilized by this subclass. In this process, electron carriers are reduced and are regarded as oxidizers of the substrate. Co-enzymes such as flavin groups, nicotinamide adenine dinucleotide (NAD), flavin mononucleotide (FMN), or nicotinamide adenine dinucleotide phosphate (NADP) catalyze the reaction with dehydrogenase enzymes.83 The dehydrogenase enzyme, which is present in all living organisms, transports hydrogen atoms from organic transporters to electron-acceptor substances.84 Hydroxylase enzymes have active sites containing metal that can catalyze the reactions.
Dioxygen molecules can serve as an electron acceptor for oxidases to use in catalyzing processes.83 Oxidases transfer electrons using a variety of substances, including metals and cofactors. These substances consist of amine oxidases, metals based on alcohol or flavin, or both.85 Utilizing molecular oxygen and NADH or NADPH as a cofactor, cytochrome P450 produces oxidized products and carbon substrate.24 As a source of electrons, cytochrome P450 uses ferredoxin reductase and ferredoxin for catalytic function.
Enzymatic biodegradation potential for sustainability
The enzymatic activity of microorganisms that convert harmful substances into less toxic or non-hazardous compounds is the primary focus of biological remediation of toxic contaminants.96,97 Using microbial enzymes over microbial cell majorly depends on the Standardizable activity, ease of handling and storage, more selectivity, improved mobility because of their smaller size, ability to function in the face of high concentrations of harmful substances, and biodegradability.98 Microbial enzymes enhance the remediation process by yielding non-flammable, non-corrosive byproducts that can be easily disposed of Perpetuini et al.99 It has been seen that immobilization increases the stability and lifetime of enzymes by reducing enzymatic activity.100,101 Recombinant DNA technology has helped to generate more effective enzymes in significant amounts.102 Because enzymes are sustainable, naturally occurring catalysts derived from renewable resources, microbially produced enzyme remediation is a safe and cost-effective bioremediation method. They use as many natural products as possible and are biodegradable.103 Enzyme-mediated degradation of toxic pollutants provides an environmentally friendly technique by reducing post-treatment environmental hazards, making it more socially acceptable.54
Recombinant microbial enzymes produced by different expression systems
Studies have demonstrated effective methods to create various microbial enzymes by diverse microorganisms (Table 3) using a variety of study approaches that have been improved in recent years. Recombinant laccase has been investigated for the treatment of dyes, phenolic compounds, insecticides, and polycyclic aromatic hydrocarbons due to its capacity to oxidize a variety of substrates.104 Laccase-mediated degradation has been investigated for the treatment of a number of pollutants, including dyes, phenolic compounds, insecticides, and polycyclic aromatic hydrocarbons. Endo-1,4-glucanase-encoding Aspergillus fumigatus gene (Afu6g01800) was cloned in the pET-28a (+) vector and was expressed in the Rosetta TM (DE3) strain of E. coli. The findings of this study demonstrated that the afegl7 enzyme belongs to the GH7 family. The afegl7 gene encodes a protein of 460 amino acids, a CBM1 domain at residues 424-460, and a molecular weight of 52kDa.105
Table (3):
Recombinant DNA technology produced microbial enzymes for enhanced bioremediation
No. |
Recombinant enzyme |
Host |
Origin |
Expression vector |
Application |
Ref. |
|---|---|---|---|---|---|---|
1. |
Laccase cotA |
Escherichia coli DH5α |
Bacillus subtilis 168 |
pMD18-T |
Degradation of artificially produced dye |
33 |
2. |
Laccase CueO |
Pichia pastoris GS115 |
Escherichia coli K12 |
pHBM905BDM |
Decolorization of effluent from the textile industry |
105 |
3. |
Cellobiohydrolase (CBH1) |
Pichia pastoris KM71H |
Aspergillus niger-NL-1 |
pPICZαC |
Degradation of pulp, cellulose etc. |
106 |
4. |
Endoglucanase (ReEG I) |
Pichia pastoris GS115, X-33 and KM71H |
Trichoderma reesei Rut C-30 |
pPICZαA |
Degradation of pulp, cellulose, oat xylan, birch xylan, corn straw |
107 |
5. |
Laccase (Fmb- rL 103) |
E. coli BL 21 |
Bacillus vallismortis fmb-103 |
pMD19-Tlac103 |
Degradation of triphenyl methane dye |
36 |
6. |
Cytochrome P450 (CYP105D1) |
Acinetobacter calcoaceticus strain BD413 |
Streptomyces griseus |
pSP19g10L |
Deterioration of pollutants herbicides |
108 |
7. |
Dehydrogenase (17β-HSDx) |
E. coli |
Rhodococcus sp. P14 |
pET-32a |
Bioremediation of steroids contaminated environment |
109 |
8. |
Dehalogenase |
E. coli BL21 (DE3) |
Ochrobactrum sp. T. |
pET30a-a6 |
Degradation of tetra bromo bisphenol A (TBBPA) |
110 |
Laccase cotA produced from Bacillus subtilis, was expressed in E. coli DH5 using the pMD18-T vector, is well known for its synthetic dye efficiency.30 Another laccase, CueO, derived from E. coli K12, was expressed in Pichia pastoris GS115 using the pHBM905BDM vector, demonstrating efficacy in the textile effluents dye decolorization. Laccase Fmb-rL103, from similarly Bacillus vallismortis fmb-103 and expressed in E. coli BL21 using the pMD19-Tlac103 vector, is effective against triphenyl methane dyes. Cellobiohydrolase CBH1 from Aspergillus niger, expressed in Pichia pastoris, is effective in the degradation of pulp and cellulose. Similarly, endoglucanase ReEG I, derived from Trichoderma reesei and expressed in Pichia shepherds, can degrade various substrates including pulp, cellulose, oat xylan, birch xylan, and corn straw.
Cytochrome P105D1, sourced from Streptomyces griseus and expressed in Acinetobacter calcoaceticus using the pSP19g10L vector, plays a key role in the breakdown of various pollutants and herbicides.108 It has been observed that the Dehydrogenase from Rhodococcus sp. P14, expressed in E. coli with pET-32a, significantly contributes to the bioremediation of steroids.106 A study suggests that dehalogenase produced by Ochrobactrum species, and expressed in E. coli BL21 (DE3) using the pET30a-a6 vector, can be used for the degradation of the environmental contaminant Tetra Bromo Bis Phenol A (TBBPA).110 Thus, Several recombinant enzymes have been developed and expressed in various host systems for applications in biodegradation and bioremediation.
Rising contaminants harm biotic components through chronic exposure, even at low concentrations. When optimal conditions of temperature, pH, and concentration are met, microbial enzymes transform pollutant substrates into harmless products. Polyhalogenated aromatics and PAHs interact with cytochrome P450’s active site to produce non-toxic substances. Laccase’s halotolerant capabilities convert antibiotics, synthetic colours, PAHs, and phenolic contaminants through oxidation. Dehalogenase enzymes cleave carbon-halogen bonds, converting halogenated contaminants into substrates that reduce chlorinated environment. Dehydrogenase converts hydroxyl groups in synthetic polymers and alcohols into aldehydes. Protease enzyme breaks down keratinous waste, colours, marine crustacean waste, and biodegradable plastics through hydrolysis. Microbial hydrolases degrade plasticizers, cyanides, and nitrile chemicals into less hazardous byproducts. Microbial lipase decomposes copolymers, synthetic polyester, and parabens into biodegradation products. Therefore, bioremediation by microbial enzymes provides a safe, sustainable method for biodegrading dangerous organic and inorganic materials.
ACKNOWLEDGMENTS
The authors are thankful to the Amity Institute of Microbial Technology, and Amity Institute of Biotechnology, Amity University Rajasthan, for providing support and infrastructure.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
SY, GKA and NJ conceptualized the study. SY collected resources. GKA and NJ performed supervision. SY and NJ performed data curation. NJ performed data validation. SY wrote the original draft. SY, NK, GKA, NJ and AB wrote, reviewed and edited the manuscript. All authors read and approved the final manuscript for publication.
FUNDING
This work was supported by the grant provided by DST-FIST (Project No.: SR/FST/LS-I/2019/502) to the Amity Institute of Microbial Technology and DST-PURSE (Project No.:- SR/PURSE/2021/77) to the Amity University Rajasthan.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
Not applicable.
- Jie H, Khan I, Alharthi M, Zafar MW, Saeed A. Sustainable energy policy, socio-economic development, and ecological footprint: The economic significance of natural resources, population growth, and industrial development. Util Policy. 2023;81:101490.
Crossref - Kesari KK, Soni R, Jamal QMS, et al. Wastewater treatment and reuse: a review of its applications and health implications. Water Air Soil Poll. 2021;232(5):1-28.
Crossref - Elekwachi CO, Andresen J, Hodgman TC. Global use of bioremediation technologies for decontamination of ecosystems. J Bioremed Biodeg. 2014;5(4):1-9.
Crossref - Liu L, Bilal M, Duan X, Iqbal HMN. Mitigation of environmental pollution by genetically engineered bacteria-current challenges and future perspectives. Sci Total Environ. 2019;667:444-454.
Crossref - Prasad B, Kaur PS, Malik T, Rana P, Sharma A. Recent Advances in Nanotechnology and Nanofiltration Methods for Industrial Wastewater Treatment In: Malik T, Shah MP eds. Recent Trends in Industrial Wastewater Treatment. London CRC Press eBooks. 2025:123-145.
Crossref - Prasad B, Malik T, Sarkar O, Gupta S, Negi KS. A comprehensive review on sustainable removal of micropollutants in wastewater by micro-biotechnological approaches with special reference to microbial associated nanoparticles. Bioremediation Journal. 2024:1-27.
Crossref - Usmani Z, Sharma M, Lukk T, et al. Developments in enzyme and microalgae based biotechniques to remediate micropollutants from aqueous systems-A review. Crit Rev Environ Sci Technol. 2022;52(10):1684-1729.
Crossref - Malik A. Metal bioremediation through growing cells. Environ Int. 2004;30(2):261-278.
Crossref - Singh S, Kang SH, Mulchandani A, Chen W. Bioremediation: environmental clean-up through pathway engineering. Curr Opin Biotech. 2008;19(5):437-444.
Crossref - Choi J, Kim H, Ahn YR, et al. Recent advances in microbial and enzymatic engineering for the biodegradation of micro-and nanoplastics. RSC Adv. 2024;14(14):9943-9966.
Crossref - Roth C, Wei R, Oeser T, Then J, Follner C, Zimmermann W, Strater N. Structural and functional studies on a thermostable polyethylene terephthalate degrading hydrolase from Thermobifida fusca. Appl Microbiol Biotechnol. 2014;98(18):7815-7823.
Crossref - Zhang Y, Pedersen JN, Eser BE, Guo Z. Biodegradation of polyethylene and polystyrene: From microbial deterioration to enzyme discovery. Biotechnol Adv. 2022;60:107991.
Crossref - Narayanan M, Ali SS, El-Sheekh M. A comprehensive review on the potential of microbial enzymes in multipollutant bioremediation: Mechanisms, challenges, and future prospects. J Environ Manage. 2023;334:117532.
Crossref - Bhatt P, Ahmad S, Joshi S, Bhatt K. Recent Advancement in Microbial Enzymes and Their Industrial Applications In: Bhatt P eds. Industrial Applications of Microbial Enzymes. London CRC Press eBooks. ; 2022:1-17.
Crossref - Bhandari S, Poudel DK, Marahatha R, et al. Microbial enzymes used in bioremediation. J Chem. 2021;2021(1):8849512.
Crossref - Masood H, Khan S, Khan SM. Enzyme Applications in Textile Industry: A Step Toward Sustainable Development Goals. In: Arshad, M. eds. Enzymes in Textile Processing: A Climate Changes Mitigation Approach. SDGs and Textiles. Springer, Singapore. 2025: 19-33
Crossref - Singh R, Kumar M, Mittal A, Mehta PK. Microbial enzymes:industrial progress in 21st century. 3 Biotech. 2016;6(2):174.
Crossref - Adrio JL, Demain AL. Microbial enzymes: Tools for biotechnological processes. Biomolecules. 2014;4(1):117-139.
Crossref - Sarrouh B, Santos TM, Miyoshi A, Dias R, Azevedo V. Up-to-date insight on industrial enzymes applications and global market. J Bioprocess Biotech. 2012;4:002.
Crossref - Niyonzima FN. Production of microbial industrial enzymes. Acta Sci Microbiol. 2019;2(12):75-89.
Crossref - Zu L, Li G, An T, Wong PK. Biodegradation kinetics and mechanism of 2,4,6-tribromophenol by Bacillus sp. GZT: A phenomenon of xenobiotic methylation during debromination. Bioresource Technol. 2012;110:153-159.
Crossref - Yoshida H. Chemistry of laquer (urushi). J Chem Soc. 1883;43:472-486.
Crossref - Anzenbacher P, Anzenbacherova E. Cytochromes P450 and metabolism of xenobiotics. Cell Mol Life Sci. 2001;58:737-747.
Crossref - Guengerich FP. Mechanisms of cytochrome P450-catalyzed oxidations. ACS Catal. 2018;8(12):10964-10976.
Crossref - Chakraborty J, Das S. Molecular perspectives and recent advances in microbial remediation of persistent organic pollutants. Environ Sci Pollut R. 2016;23(17):16883-16903.
Crossref - Shraddha N, Shekher R, Sehgal S, Kamthania M, Kumar A. Laccase: microbial sources, production, purification, and potential biotechnological applications. Enzyme Res. 2011;2011(1):217861.
Crossref - Sondhi S, Sharma P, George N, Chauhan PS, Puri N, Gupta N. An extracellular thermo-alkali-stable laccase from Bacillus tequilensis SN4, with a potential to biobleach softwood pulp. 3 Biotech. 2015;5(2):175-185.
Crossref - Janusz G, Skwarek E, Pawlik A. Potential of laccase as a tool for biodegradation of wastewater micropollutants. Water. 2023;15(21):3770.
Crossref - Muthukumarasamy NP, Jackson B, Joseph RA, Sevanan M. Production of extracellular laccase from Bacillus subtilis MTCC 2414 using agroresidues as a potential substrate. Biochem Res Int. 2015:2015(1):765190.
Crossref - Sharma A, Thakur VV, Shrivastava A, et al. Xylanase and laccase based enzymatic kraft pulp bleaching reduces adsorbable organic halogen (AOX) in bleach effluents: a pilot scale study. Bioresour Technol. 2014;169:96-102.
Crossref - Martin-Sampedro R, Eugenio ME, Villar JC. Effect of steam explosion and enzymatic pre-treatments on pulping and bleaching of Hesperaloe funifera. Bioresour Technol. 2012;111:460-467.
Crossref - Guan Y, Chen X. Recent applications of flavin-dependent monooxygenases in biosynthesis, pharmaceutical development, and environmental science. Catalysts. 2023;2023;13(12):1495.
Crossref - Guan ZB, Luo Q, Wang HR, Chen Y, Liao XR. Bacterial laccases: Promising biological green tools for industrial applications. Cell Mol Life Sci. 2018;75(19):3569-3592.
Crossref - Wang TN, Zhao M. A Simple strategy for extracellular production of CotA laccase in Escherichia coli and decolorization of simulated textile effluent by recombinant laccase. Appl Microbiol Biotechnol. 2017;101(2):685-696.
Crossref - Vaithyanathan VK, Vaidyanathan VK, Cabana H. Laccase-driven transformation of high-priority pesticides without redox mediators: towards bioremediation of contaminated wastewaters. Front Bioeng Biotechnol. 2022;9:770435.
Crossref - Sun J, Zheng M, Lu Z, Lu F, Zhang C. Heterologous production of a temperature and pH-stable laccase from Bacillus vallismortis fmb-103 in Escherichia coli and its application. Process Biochem. 2017;55:77-84.
Crossref - Mandic M, Djokic L, Nikolaivits E, et al. Identification and characterization of new laccase biocatalysts from Pseudomonas species suitable for degradation of synthetic textile dyes. Catalysts. 2019;9(7):629.
Crossref - Oyedeji BA, Oloye NR, Adebajo SO, Taiwo MO, Folarin BT, Akintokun AK. Optimization and production of laccase enzyme by Bacillus cereus isolated from effluent of paint producing industry. Bull Nat l Res Cent. 2025;49(1):27.
Crossref - Fernandez-Fernandez M, Sanroman MA, Moldes D. Recent developments and applications of immobilized laccase. Biotechnol Adv. 2013;31(8):1808-1825.
Crossref - Tychanowicz GK, Zilly A, de Souza CGM, Peralta RM. Decolourisation of industrial dyes by solid-state cultures of Pleurotus pulmonarius. Process Biochem. 2004;39(7):855-859.
Crossref - Mahmud MA, Anannya FR. Sugarcane bagasse – A source of cellulosic fiber for diverse applications. Heliyon. 2021;7(8):e07771.
Crossref - Kalra K, Chauhan R, Shavez M, Sachdeva S. Isolation of laccase producing Trichoderma spp. and effect of pH and temperature on its activity. Int J Chem Environ Technol. 2013;5(5):2229-2235.
- Hirota-Mamoto R, Nagai R, Tachibana S, et al. Cloning and expression of the gene for periplasmic poly (vinyl alcohol) dehydrogenase from Sphingomonas sp. strain 113P3, a novel-type quinohaemoprotein alcohol dehydrogenase. Microbiol. 2006;152(7):1941-1949.
Crossref - Bourguignon N, Irazusta V, Isaac P, Estevez C, Maizel D, Ferrero MA. Identification of proteins induced by polycyclic aromatic hydrocarbon and proposal of the phenanthrene catabolic pathway in Amycolatopsis tucumanensis DSM 45259. Ecotoxicol Environ Saf. 2019;175:19-28.
Crossref - Thakur S. Lipases, its sources, properties and applications:a review. Int J Sci Eng Res. 2012;3(7):1-29.
- Arora NK, Mishra J, Mishra V, eds. Microbial enzymes: roles and applications in industries. Berlin Heidelberg: Springer. 2020;11:1-110.
Crossref - Gurung N, Ray S, Bose S, Rai V. A broader view: Microbial enzymes and their relevance in industries, medicine, and beyond. BioMed Res Int. 2013;2013(1):329121.
Crossref - Hassan SWM, Abd El Latif HH, Ali SM. Production of cold-active lipase by free and immobilized marine Bacillus cereus HSS: application in wastewater treatment. Front Microbiol. 2018;9:2377.
Crossref - Casas-Godoy L, Duquesne S, Bordes F, Sandoval G., Marty A. Lipases: An Overview. In: Sandoval, G. eds. Lipases and Phospholipases. Methods in Molecular Biology. Totowa, New Jersey Humana Press. 2012;861:3-30.
Crossref - Amara AA, Salem SR. Degradation of castor oil and lipase production by Pseudomonas aeruginosa. Am Eurasian J Agric Environ Sci. 2009;5(4):556-63.
- Mahmood MH, Yang Z, Thanoon RD, Makky EA, Rahim MHA. Lipase production and optimization from bioremediation of disposed engine oil. J Chem Pharm Res. 2017;9(6):26-36.
- Saraswat R, Verma V, Sistla S, Bhushan I. Evaluation of alkali and thermotolerant lipase from an indigenous isolated Bacillus strain for detergent formulation. Electron J Biotechnol. 2017;30:33-38.
Crossref - Wang L, Liu T, Sun H, Zhou Q. Transesterification of para-hydroxybenzoic acid esters (parabens) in the activated sludge. J Hazard Mater. 2018;354:145-152.
Crossref - Kumar A, Gudiukaite R, Gricajeva A, et al. Microbial lipolytic enzymes-promising energy-efficient biocatalysts in bioremediation. Energy. 2020;192:116674.
Crossref - Haider TP, Volker C, Kramm J, Landfester K, Wurm FR. Plastics of the future? The impact of biodegradable polymers on the environment and on society. Angew Chem Int Ed Engl. 2019;58(1):50-62.
Crossref - Bhange K, Chaturvedi V, Bhatt R. Feather degradation potential of Stenotrophomonas maltophilia KB13 and feather protein hydrolysate (FPH) mediated reduction of hexavalent chromium. 3 Biotech. 2016;6:1-9.
Crossref - Maitig AMA, Alhoot MA, Tiwari K. Isolation and screening of extracellular protease enzyme from fungal isolates of soil. J Pure Appl Microbiol. 2018;12(4):2059-2067.
Crossref - Rathinamoorthy R, Bharathi TS, Snehaa M, Swetha, C. Mycelium as sustainable textile material-review on recent research and future prospective. Int J Cloth Sci Tech. 2023;35(3):454-476.
Crossref - Nnolim NE, Okoh AI, Nwodo UU. Bacillus sp. FPF-1 produced keratinase with high potential for chicken feather degradation. Molecules. 25(7):1505.
Crossref - Laba W, Zarowska B, Chorazyk D, et al. New keratinolytic bacteria in valorization of chicken feather waste. AMB Expr. 2018;8(1):1-14.
Crossref - Akhter M, Wal Marzan L, Akter Y, Shimizu, K. Microbial bioremediation of feather waste for keratinase production: an outstanding solution for leather dehairing in tanneries. Microbiol Insights. 2020;13:1178636120913280.
Crossref - Jellouli K, Ghorbel-Bellaaj O, Ayed HB, Manni L, Agrebi R. Nasri, M. Alkaline-protease from Bacillus licheniformis MP1: purification, characterization and potential application as a detergent additive and for shrimp waste deproteinization. Process Biochem. 2011;46(6):1248-1256.
Crossref - Howard GT, Ruiz C, Hilliard NP. Growth of Pseudomonas chlororaphis on apolyester-polyurethane and the purification and characterization of a polyurethanase-esterase enzyme. Int Biodeter Biodegr. 1999;43(1-2):7-12.
Crossref - Vandelook S, Elsacker E, Van Wylick A, De Laet L, Peeters E. Current state and future prospects of pure mycelium materials. Fungal Biol Biotechnol. 2021;8:1-10.
Crossref - Amaro BG, Vandenberghe LPS, Martínez-Burgos WJ, et al. Emerging contaminants bioremediation by enzyme and nanozyme-based processes – A review. iScience. 2023;26(6):106785.
Crossref - Singh RS, Singh T, Pandey A. Microbial Enzymes—An Overview Author links open overlay panel In: Singh RS, Singhania RR, Larroche C, eds. Advances in Enzyme Technology A volume in Biomass, Biofuels, Biochemicals. Elsevier. 2019;1-40.
Crossref - Thurston CF. The structure and function of fungal laccases. Microbiol. 1994;140(1):19-26.
Crossref - Chivukula M, Spadaro JT, Renganathan V. Lignin peroxidase-catalyzed oxidation of sulfonated azo dyes generates novel sulfophenyl hydroperoxides. Biochem. 1995;34(23):7765-7772.
Crossref - Bhunia A, Durani S, Wangikar PP. Horseradish peroxidase catalyzed degradation of industrially important dyes. Biotechnol Bioeng. 2001;72(5):562-567.
Crossref - Liers C, Bobeth C, Pecyna M, Ullrich R, Hofrichter M. DyP-like peroxidases of the jelly fungus Auricularia auricula-judae oxidize nonphenolic lignin model compounds and high-redox potential dyes. Appl Microbiol Biotechnol. 2010;85(6):1869-1879.
Crossref - Yang Q, Yang M, Pritsch K, et al. Decolorization of synthetic dyes and production of manganese-dependent peroxidase by new fungal isolates. Biotechnol Lett. 2003;25:709-713.
Crossref - Raghuwanshi S, Deswal D, Karp M, Kuhad RC. Bioprocessing of enhanced cellulase production from a mutant of Trichoderma asperellum RCK2011 and its application in hydrolysis of cellulose. Fuel. 2014;124:183-189.
Crossref - Nathan VK, Rani ME, Rathinasamy G, Dhiraviam KN, Jayavel S. Process optimization and production kinetics for cellulase production by Trichoderma viride VKF3. SpringerPlus. 2014;3(92):1-12.
Crossref - Das A, Paul T, Halder SK, et al. Production of cellulolytic enzymes by Aspergillus fumigatus ABK9 in wheat bran-rice straw mixed substrate and use of cocktail enzymes for deinking of waste office paper pulp. Bioresour Technol. 2013;128:290-296.
Crossref - Agrawal D, Matkar K. Enhancement of cellulase production by substrate manipulation in three Aspergillus sp. Indian J Appl Res. 2016;6(3).
- Yadav PS, Shruthi K, Prasad BS, Chandra MS. Enhanced production of β-glucosidase by new strain Aspergillus protuberus on solid state fermentation in rice husk. Int J Curr Microbiol App Sci. 2016;5(12):551-564.
Crossref - Lason-Rydel M, Sieczynska K, Gendaszewska D, Lawinska K, Olejnik TP. Use of enzymatic processes in the tanning of leather materials. Autex Res J. 2024;24(1):20230012.
Crossref - Romero S, Blanquez P, Caminal G, et al. Different approaches to improving the textile dye degradation capacity of Trametes versicolor. Biochem Eng J. 2006;31(1):42-47.
Crossref - Margot J, Bennati-Granier C, Maillard J, Blanquez P, Barry DA, Holliger C. Bacterial versus fungal laccase: potential for micropollutant degradation. AMB Expr. 2013;3(1):63.
Crossref - Sodhi AS, Bhatia S, Batra N. Laccase: Sustainable production strategies, heterologous expression and potential biotechnological applications. Int J Biol Macromol. 2024;280(1):135745.
Crossref - Mazhar H, Abbas N, Ali S, Sohail A, Hussain Z, Ali SS. Optimized production of lipase from Bacillus subtilis PCSIRNL-39. Afr J Biotechnol. 2017;16(19):1106-1115.
Crossref - Saranya P, Selvi PK, Sekaran G. Integrated thermophilic enzyme-immobilized reactor and high-rate biological reactors for treatment of palm oil-containing wastewater without sludge production. Bioprocess Biosyst Eng. 2019;42(6):1053-1064.
Crossref - Phale PS, Sharma A, Gautam K. Microbial degradation of xenobiotics like aromatic pollutants from the terrestrial environments In: Prasad MNV, Vithanage M, Kapley A, eds. Pharmaceuticals and Personal Care Products: Waste Management and Treatment Technology Emerging Contaminants and Micro Pollutants. Oxford (UK), Butterworth-Heinemann, 2019:259-278.
Crossref - Dotaniya ML, Aparna K, Dotaniya CK, Singh M, Regar KL. Role of Soil Enzymes in Sustainable Crop Production. In: Mohammed Kuddus eds. Enzymes in Food Biotechnology Production, Applications, and Future Prospects. Academic Press. 2019:569-589.
Crossref - Razzaq A, Shamsi S, Ali A, et al. Microbial proteases applications. Front Bioeng Biotechnol. 2019;7:110.
Crossref - Ghosh, P, Ghosh, U. Bioconversion of Agro-waste to Value-Added Product Through Solid-State Fermentation by a Potent Fungal Strain Aspergillus flavus PUF5. In: Ghosh S. eds. Utilization and Management of Bioresources. Springer, Singapore. 2018:291-299.
Crossref - D’Souza-Ticlo D, Sharma D, Raghukumar C. A thermostable metal-tolerant laccase with bioremediation potential from a marine-derived fungus. Mar biotechnol. 2009;11:725-737.
Crossref - Soares GMB, Costa-Ferreira M, de Amorim MTP. Decolorization of an anthraquinone-type dye using a laccase formulation. Bioresour Technol. 2001;79(2):171-177.
Crossref - Elisashvili V, Kachlishvili E. Physiological regulation of laccase and manganese peroxidase production by white-rot Basidiomycetes. J Biotech. 2009;144(1):37-42.
Crossref - Olicon-Hernandez DR, Gonzalez-Lopez J, Aranda E. Overview on the biochemical potential of filamentous fungi to degrade pharmaceutical compounds. Front Microbiol. 2017;8:1792.
Crossref - Wingate KG, Stuthridge T, Mansfield SD. Colour remediation of pulp mill effluent using purified fungal cellobiose dehydrogenase: reaction optimisation and mechanism of degradation. Biotechnol Bioeng. 2005;90(1):95-106.
Crossref - Tilli S, Ciullini I, Scozzafava A, Briganti F. Differential decolorization of textile dyes in mixtures and the joint effect of laccase and cellobiose dehydrogenase activities present in extracellular extracts from Funalia trogii. Enzyme Microb Technol. 2011;49(5):465-471.
Crossref - Takio N, Yadav M, Yadav HS. Catalase-mediated remediation of environmental pollutants and potential application-a review. Biocatalysis and Biotransformation. 2021;39(6):389-407.
Crossref - Sethi S, Gupta S. Optimization of Protease Production from Fungi Isolated from Soil. Int J Appl Biol Pharm Technol. 2015;6(3):149-154.
- Verma A, Singh H, Anwar S, et al. Microbial keratinases: industrial enzymes with waste management potential. Crit Rev Biotechnol. 2017;37(4):476-491.
Crossref - Yamaguchi H, Miyazaki M. Bioremediation of Hazardous Pollutants Using Enzyme-Immobilized Reactors. Molecules. 2024; 29(9):2021.
Crossref - Skendzic S, Zovko M, Zivkovic IP, Lesic V, Lemic D. The impact of climate change on agricultural insect pests. Insects. 2021;12(5):440.
Crossref - Chia XK, Hadibarata T, Kristanti RA, Jusoh MNH, Tan IS, Foo HCY. The function of microbial enzymes in breaking down soil contaminated with pesticides: a review. Bioprocess Biosyst Eng. 2024;47(5):597-620.
Crossref - Perpetuini G, Fossi PAN, Kwak S, et al. Pesticides in foods: Towards bioremediation biocatalysts? Catalysts. 2023;13(7):1055.
Crossref - Kisic I, Hrenovic J, Zgorelec Z, Durn G, Brkic V, Delae D. Bioremediation of agriculture soil contaminated by organic pollutants. Energies. 2022;15(4):1561.
Crossref - Fernandez-Lafuente R. Enzyme immobilization. Molecules. 2023;28(3):1373.
Crossref - Mousavi SM, Hashemi SA, Moezzi SMI, et al. Recent advances in enzymes for the bioremediation of pollutants. Biochem Res Int. 2021;2021(1):5599204.
Crossref - Dave S, Jayashankar Das J. Role of microbial enzymes for biodegradation and bioremediation of environmental pollutants: challenges and future prospects In: Saxena G, Kumar V, Shah MP, eds. Bioremediation for Environmental Sustainability Toxicity, Mechanisms of Contaminants Degradation, Detoxification, and Challenges. Elsevier 2021:325-346.
Crossref - Kesebir AO, Kylyc D, Sisecioglu M, Adyguzel A, Kufrevioglu OI. Recombinant laccase production from Bacillus licheniformis O12: Characterization and its application for dye decolorization. Biologia. 2021;76(11):3429-3438.
Crossref - Ma X, Liu L, Li Q, et al. High-level expression of a bacterial laccase, CueO from Escherichia coli K12 in Pichia pastoris GS115 and its application on the decolorization of synthetic dyes. Enzyme Microb Technol. 2017;103:34-41.
Crossref - Li GQ, Chai CS, Fan S, Zhao LG. Cloning of a cellobiohydrolase gene (cbh1) from Aspergillus niger and heterogenous expression in Pichia pastoris. Adv Mater Res. 2012;347-353:2443-2447.
Crossref - Tao Y, Yang L, Yin L, et al. Novel approach to produce biomass-derived oligosaccharides simultaneously by recombinant endoglucanase from Trichoderma reesei. Enzyme Microb Technol. 2020;134:109481.
Crossref - Lamb DC, Kelly DE, Masaphy S, Jones GL, Kelly SL. Engineering of heterologous cytochrome P450 in Acinetobacter sp.: application for pollutant degradation. Biochem Biophys Res Commun. 2000;276(2):797-802.
Crossref - Ye X, Peng T, Feng J, et al. A novel dehydrogenase 17β-HSDx from Rhodococcus sp. P14 with potential application in bioremediation of steroids contaminated environment. J Hazard Mater. 2019;362:170-177.
Crossref - Liang Z, Li G, Xiong J, Mai B, An T. Purification, molecular characterization and metabolic mechanism of an aerobic tetrabromobisphenol A dehalogenase, a key enzyme of halorespiration in Ochrobactrum sp. T. Chemosphere. 2019;237:124461.
Crossref
© The Author(s) 2025. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.