ISSN: 0973-7510
E-ISSN: 2581-690X
Trichophyton rubrum is the most common cause of dermatophyte skin infections in humans worldwide. The aim of the current study is to search for effective and safe antifungal agents by investigating the bioactivity of Pimpinella anisum and Moringa oleifera extracts, including the impact on the keratinase gene expression of T. rubrum. Astragalin and cryptochlorogenic acid were detected in M. oleifera leaf extracts by Gas chromatography-mass spectrometry (GC-MS), whereas anethole was extracted from P. anisum oil. The expression of the keratinase gene was assessed in two strains of T. rubrum (KP979791 and KP979787) in Sabouraud Dextrose Broth (SDB) medium (containing 10% keratin) separately supplemented with plant extracts fractions. The results showed that cryptochlorogenic acid and astragalin were active compounds (molecular weights of 353.26 and 449.35 g/mol, respectively that considerably down-regulated the keratinase genes in both strains of T. rubrum. M. oleifera crude extract, astragalin, cryptochlorogenic acid, anise oil, and anethole resulted in the down-regulation of the keratinase genes of T. rubrum strain KP979791 by 0.048, 0.291, 0.001, 0.007, and 0.009 (fold-change), respectively, and that of strain KP979787 by 0.151, 0.42, 0.16, 0.079, and 0.092 as compared to the control (1.00). Nearly all of these outcomes are novel findings that may aid in the identification of novel effective drug targets and antifungal agents.
Trichophyton rubrum, Moringa oleifera, Pimpinella anisum, Chromatography, keratinase gene.
Dermatophytes are a group of parasitizing filamentous fungi that infect the keratinized tissues of the nails, skin, and hair as keratin is utilized as the only carbon energy source, which normally results in universal cutaneous mycoses1. The prevalence of superficial fungal infection under different living circumstances in Babylon Province, Iraq, was investigated2. T. rubrum is a major etiological agent of dermatophytosis and other recurring fungal infections in humans. T. rubrum and T. mentagrophytes account for 27%–76% and 4%–41% of chronic mycotic infections of humans, respectively3. In Iraq, five strains of T. rubrum have been identified to date, which have fairly similar cultural features, but considerably different growth requirements and proteolytic activities; their virulence were also wholly studied4.
T. rubrum is currently a serious epidemic and, at times, an endemic human pathogen2,5. The major mechanism of the pathogenicity of dermatophytes is the production of proteases which makes it possible to penetrate the human skin via keratinized tissues. The proteolytic enzymes produced by dermatophytes are activated under different environmental conditions (i.e., neutral, alkali, and acidic), which have significant roles in the proteolytic activity, pathogenesis, and particularly, the invasion of T. rubrum4. The proteolytic enzymes produced and secreted by this pathogen are major virulence factors6. However, the uneven expression of the protease genes is responsible for the variances in the capacity to cause infections7,8. Keratinases are considered the most prominent virulence factors of dermatophytes9. Trichophyton species have exceptionally high keratinase activities10, and the level of keratinase activity can be used in indirectly estimating the strength of fungal pathogenicity11. It is therefore essential to investigate the expression profiles of genes involved in the virulence of T. rubrum.
Previous gene expression analysis demonstrated that trans-chalcone in commercial antifungals suppresses the expression of virulence-related genes of T. rubrum12. There are three classes of commercially available drugs that are known to interfere with the plasma membrane: azoles and triazoles, which inhibit the enzyme sterol 14 α-demethylase, and allylamines, which inhibit squalene epoxidase and ergosterol biosynthesis. Allylamines also lead to the accumulation of lanosterol, a toxic intermediary compound of the ergosterol biosynthesis pathway13. Sharma et al.14 mentioned that of six antifungal agents, dermatophytes (T. mentagrophytes and T. rubrum) were most resistant to fluconazole and clotrimazole, and voriconazole was the most effective. The failure of any treatment lies in the complexity of the host–fungus interactions and the virulence of the species and the ability to avoid the host immune response15. Several recent studies have improved our understanding of the molecular basis of antifungal resistance. However, the identification of new antifungal targets and the development of novel antifungal compounds from natural as well as synthetic products are needed to promote the development of innovative strategies for antifungal therapies16.
Medicinal plants are valuable sources of antifungal compounds. Moringa oleifera is a precious source of bioactive compounds with different pharmacological activities17. Extracts of M. oleifera reportedly have potential antifungal activities against dermatophytic fungi18 with good remedial properties and few side effects as compared with synthetic antibiotics19. In addition, cell lysis was recognized in fungal tissues of T. rubrum grown on Sabouraud’s dextrose agar (SDA) and treated with anise (Pimpinella anisum) essential oil (EO) and anethole, suggesting that both extracts possessed antimicrobial activities against T. rubrum20,21. The use of natural products, such as plant-derived EOs, as an alternative to synthetic chemical agents is regarded as an environmentally safe alternative for disease control22.
Therefore, the aim of the present study is to obtain a secure, efficient, naturally acquired, antifungal agent from P. anisum EO and leaf extracts of M. oleifera. These natural products were screened for antifungal effects against T. rubrum, which is responsible for most human dermatophyte infections23. Furthermore, the effects of the phytoconstituents of selected medicinal plants (i.e., anise oil and M. oleifera), especially anethole, astragalin, and cryptochlorogenic acid, on the expression of the keratinase gene of T. rubrum were investigated, since this gene is known to contribute to the pathogenesis of T. rubrum, and that would aid in the design of effective therapeutics without causing any serious adverse reactions, which may provide future insights into the virulence of this clinically important pathogen.
Source of T. rubrum isolate
T. rubrum strains KP979791 (isolate N05 internal transcribed spacer 1, partial sequence; 5.8S ribosomal RNA gene and KP979787 isolate N01 internal transcribed spacer 1, partial sequence; 5.8S ribosomal RNA gene were obtained from Kadhim et al., 2015b7, Department of Biology, Advanced Mycology Laboratory, University of Babylon (Iraq), both strains were previously identified at the molecular level 7.
Preparation and maintenance of T. rubrum
SDA medium was prepared according to the manufacturer’s instructions by dissolving 64 g of SDA in 1000 mL of distilled water. The medium was sterilized by autoclaving prior to culturing and sub-culturing of the T. rubrum isolates24. Briefly, a colony of T. rubrum (diameter, 0.5 cm) was placed at the center of a Petri plate (diameter, 9 cm) containing sterilized SDA supplemented with cycloheximide (0.5 g/L) and chloramphenicol (0.05 g/L) at pH of 5.6 and incubated at 28°C ± 1°C for 14 days and then stored in a refrigerator at 5°C. The fungus was maintained on SDA slants at 4°C and subcultured monthly throughout the study period.
Plant Materials
The leaves of M. oleifera and the seeds of P. anisum were collected to obtain crude plant extracts according to Obaid et al., 2017a20,
Preparation of protease inhibitors
An extract of protease inhibitor from M. oleifera was prepared in a 500 mL conical flask by homogenizing 25 g of plant materials in 100 mL of 0.1 M phosphate buffer (pH 7). The homogenate was further mixed by incubating the contents at room temperature in a rotary shaker for 30 min at a speed of 150 rpm. The slurry was then filtered through muslin cloth and the filtrate was centrifuged at 6000 rpm for 20 min at 4°C to remove any cell debris that remained in the preparation. The clear supernatant (the amount obtained) solution was represented as the crude extract and was assayed for protease inhibitor activity. Ammonium sulphate was used to precipitate the fractions of the extract 20,25.
Preparation of essential oil
Clevenger-type apparatus (TF-500, TEFIC, China), the procedure consists of immersing the dry seeds of P. anisum L. (60 g) using a miller and then transferring it to a 0.5 litre round bottom flask containing 250 mL of distilled water. This mixture was heated until it boiled for 3 hours, and the produced vapour carrying the volatile substances (essential oil) was then passed through a cooling system where condensation occurred. The essential oils were then collected from the surface of the water. The obtained essential oil was stored at 4°C for further lab analyses22,25. The extracted anethole was used as it was described previously26.
Preparation of active materials from the leaves of M. oleifera
The active material from the crude extract was processed by preparative HPLC. The preparative reversed-phase HPLC was performed using column C18-ODS (25 cm × 4.6 mm × 5μm). The mobile phases were determined based on the method adopted by Porel et al.27 and these phases include (A) Methanol: Acetic acid: Distilled water (10:2:88) and (B) Methanol: Acetic acid: Distilled water (90: 3: 7). The gradients are 40% of A, 60% of B for 4 min, 50% of A, 50% of B for 5-8 min and 60% of A, and 40% of B for 8-10 min. The injection volume was 100μL with a flow rate of 1.0 ml/min, and the detection wavelength was 280 nm27.
Detection of the active compounds of M. oleifera extract by liquid chromatography with tandem mass spectrometry (LC-MS/MS)
LC-MS/MS analyses of the M. oleifera protease inhibitors were performed using a Nexera HPLC system (Shimadzu Corporation, Kyoto, Japan) equipped with a C18 reversed-phase Inertsil ODS-4 analytical column (3μm, 150 × 2.1 mm) coupled to a tandem MS instrument. The column temperature was fixed at 40°C. The elution gradient consisted of a mobile phase (A) of water: formic acid (99:1) and a mobile phase (B) of methanol: formic acid (99:1). The flow rate was 0.1 mL/min. Elution was performed at a starting concentration of B of 10%, which was increased to 70% over a period of 30 min. The solvent flow rate was maintained at 500μL/min, and the injection volume was 4μL.
Detection was performed using an LCMS-8040 Triple Quadrupole system (Shimadzu Corporation) equipped with an electrospray ionization (ESI) source operating in both positive and negative ionization modes. LC-MS/MS data were collected and processed using Lab Solutions software (Shimadzu Corporation). Multiple reaction monitoring was used to quantify the results. Analysis of the investigated compounds was performed following two or three transitions per compound, i.e., the first time for quantitative purposes and the second and/or third time for confirmation to ensure accuracy. The optimum ESI conditions were specified as follows: desolvation line temperature, 250°C; heat block temperature, 400°C; nebulizing gas (N2) flow rate, 3 L/min; and drying gas (N2) flow rate, 15 L/min28. The active material that was detected was Astragalin and cryptochlorogenic acid.
Effect of M. oleifera crude extract, astragalin, cryptochlorogenic acid, anise oil and anethole on the keratinase gene expression of T. rubrum
Cultures of T. rubrum strains KP979791 and KP979787 at turbidities of 0.5 McFarland standard were prepared in SDB containing 10% keratin. The active materials of the crude extracts of M. oleifera, anise oil, astragalin, cryptochlorogenic acid, and anethole were added to the medium at concentrations of 50, 7.8, 20, 20, and 3.9μg/ml respectively, to assess their effects on keratinase gene expression.
RNA extraction
The medium was inoculated with T. rubrum. Following incubation and centrifugation at 5,000×g for 10 min at 4°C, the supernatant was collected for isolation of the fungal cells. Total RNA was extracted using a FAVORGEN® total RNA extraction kit (Fisher Biotec, Wembley, WA, Australia). The extracted RNA was stored at −70°C to keep it stable along the time of experiment until analysis
Real-time polymerase chain reaction (RT-PCR)
RT-PCR primers
Primers for keratinase mRNA expression were designed with reference to the GenBank database (https://www.ncbi.nlm.nih.gov/nuccore/XM_003235234.1). Moreover, the 18S primer pair was designed by Shanghai ShineGene Molecular Biotech, Inc. (Shanghai, China). The primer sequences for the keratinase gene were similar to those in the GenBank database (accession number 003235234): (forward) 5′-CTC TCC TCT ACG AAT GCC AAA G-3′ and (reverse) 5′-GAA TGT GAA CTG GTT GCG TTT C-3′, which produced an amplicon of 159 bp. The following primer sequences were used for amplification of the reference 18S gene (accession number 01000074.1): (forward) 5′- AAA CGG CTA CCA CAT CC A-3′ and (reverse) 5′-CAC CAG ACT TGC CCT CCA-3′, which produced an amplicon of 166 bp. The fungal samples were treated with chemical compounds and plant extracts.
cDNA synthesis
DNase I-treated RNA samples were used to produce the cDNA of the keratinase (target) and 18S genes with the use of FicoScript® Moloney murine leukaemia virus reverse transcriptase (M-MLV RT; Shanghai ShineGene Molecular Biotech, Inc.) in accordance with the manufacturer’s instructions. Each reaction included 5 × RT buffer, 100 ng/μL of primer, M-MLV RTase (200 U), RNase inhibitor, RNA, and DEPC water at volumes of 4, 1, 1, 1, 6, and 7μL, respectively. The conditions for cDNA synthesis were 37°C for 20 s and 95°C for 3 min and the RT-PCR reaction conditions were 95°C for 20 s, 59°C for 20 s, and 72°C for 25 s.
Quantitative reverse transcriptase (qRT)-PCR data analysis
The relative gene expression levels (fold-change) were determined by analyzing the qRT-PCR data of the housekeeping and target genes using the ΔΔCT method as described by Livak and Schmittgen29 as follows:
ΔCT target gene = Ct target gene − Ct reference gene/target …(1)
ΔCT control = Ct control − Ct reference gen/control …(2)
Upon determining the ΔCT values of the target and controls, the following equation was used to determine the ΔΔCT value:
ΔΔCT = Δ Ct target gene − Δ Ct control …(3)
The 2−ΔΔCT method was then used to determine the fold-change in gene expression with the data presented in Table 1.
Table (1):
qRT-PCR data analysis.
Gene Entry |
Test (Treatment group) |
Cal. (Control group) |
|---|---|---|
Target gene |
CT (target, test) |
CT (target, cal) |
Reference gene |
CT (ref, test) |
CT (ref, cal) |
*This table was adopted from reference 29.
Statistical Analysis
SPSS (25) was used for statistical analysis. Variation among the groups was analysed using one-way (ANOVA) test, monitored by LSD’s tests. A minimum range of variance P < 0.05 was reflected significant.
HPLC of M. oleifera extract
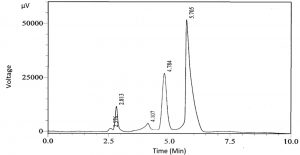
As shown in the chromatograph presented in Fig. 1, there were two main peaks. Two compounds in the extracts of the M. oleifera protease inhibitor sample were obtained by HPLC (Fig. 1), with retention times of 4.784 and 5.705 min for the first and second compounds, respectively. Therefore, LC-MS/MS analysis was performed to further characterize the detectable compounds.
Fig. 1. HPLC chromatogram of the M. oleifera protease inhibitor. Two main peaks with retention times of 4.784 and 5.705 min were detected.
LC-MS/MS analysis of the M. oleifera active compounds
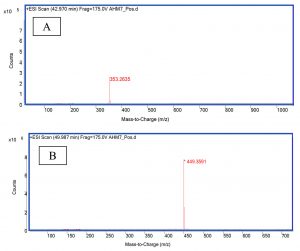
The LC-MS/MS results show that the protease inhibitor (extracted from M. oleifera leaves) after purification and isolation with HPLC had two predominant peaks: cryptochlorogenic acid (C16H18O9; molecular weight, 353.26 g/mol; retention time, 5.61 min; Fig. 2A) and astragalin (C21H20O11; molecular weight, 449.35 g/ mol; retention time, 9.19 min; Fig. 2B). The inhibitory effects on the protease enzyme can be attributed to the presence of these two compounds.
Fig. 2. LC-MS/MS chromatograms of: A. cryptochlorogenic acid (molecular weight, 353.26), B. astragalin (molecular weight, 449.35).
Lin et al.30 reported that cryptochlorogenic acid and astragalin were two of the basic compounds present in M. oleifera leaf extracts with molecular weights of 355 and 447, respectively, which is consistent with the results of the present study. Cryptochlorogenic acid possesses inhibitory effects against proteases and many other enzymes, including hepatic glucose-6-phosphatase31. In addition, Rohn et al.32 found that the inhibitory activities of cryptochlorogenic acid against alpha-amylase, trypsin, and lysozyme were deactivated. Inhibition of this enzyme is dependent on the reactivity of the phenolic and related ingredients tested and on the type of the substrate. The decreased activities were accompanied with reductions in the amounts of free amino and thiol groups as well as tryptophan residues, which resulted from the covalent attachment of phenolic and related compounds to reactive nucleophilic sites of the enzymes.
Flavonoids, such as astragalin, are an important group of natural compounds that can interfere with the activities of various enzymes. Almost all flavonoids inhibit aldehyde oxidase activity, consistent with the results of the present study, which confirmed the inhibitory effect of astragalin33. Braz34 found that a number of flavonoids inhibited the enzymatic activity of glyceraldehyde-3-phosphate dehydrogenase.
Gene expression
The results of the RT-PCR analysis showed that the keratinase mRNA levels of T. rubrum were significantly decreased in the treated samples, as compared with the control group. The down-regulation expression of the keratinase gene in T. rubrum by the plant extracts and chemical compounds was evident. The Livak technique was applied to normalize the gene expression of the target gene (keratinase) with the housekeeping gene (18S) expression as a reference gene when determining the relative expression of the keratinase gene in treated and untreated T. rubrum.
In both plots, the target and reference genes as well as the gene expression in the crude isolates (i.e. the control or calibrator) are presented. The target gene-threshold cycle numbers were first normalized to that of the reference gene in the two tested isolates to obtain the ΔCT for target gene and control, equation number (1) and (2), the ΔΔCT was calculated as shown in the equation number (3). Then, the 2-ΔΔCT can be calculated as the fold change that represented the gene expression.
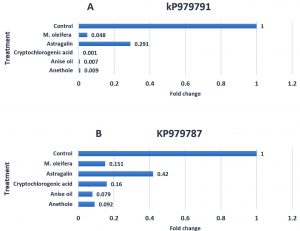
The results of the calculations demonstrated that the relative expression of the keratinase gene in the T. rubrum (KP979791) treated with M. oleifera crude extract, astragalin, cryptochlorogenic acid, anise oil extract and anethole was 0.048, 0.291, 0.001, 0.007, and 0.009, respectively, while in T. rubrum (KP979787) was 0.151, 0.42, 0.16, 0.079, 0.092 respectively compared with a high gene expression value in the control which was 1 (Tables 2A and 2B).
Table (2A):
The analysis of the Keratinase gene’s relative gene expression for the T. rubrum KP979791 (ΔΔCT) Technique.
Treatments |
CT (Keratinase) |
CT (18S) |
ΔCT |
ΔΔCT |
2-ΔΔCT |
|---|---|---|---|---|---|
M. oleifera crude extracts |
19.80333333 |
16.72666667 |
3.08 |
4.36 |
0.048 |
Astragalin |
16.37111111 |
15.87111111 |
0.5 |
1.78 |
0.291 |
Cryptochlorogenic acid |
20.29444444 |
12.12666667 |
8.17 |
9.45 |
0.001 |
Anise oil |
20.25444444 |
14.49333333 |
5.76 |
7.04 |
0.007 |
Anethole |
19.83888889 |
14.42333333 |
5.41 |
6.69 |
0.009 |
Control |
18.67555556 |
19.95333333 |
-1.28 |
0 |
1 |
Table (2B):
The analysis of the Keratinase gene’s relative gene expression for the T. rubrum KP979787 (ΔΔCT) Technique.
Treatments |
CT (Keratinase) |
CT (18S) |
ΔCT |
ΔΔCT |
2-ΔΔCT |
|---|---|---|---|---|---|
M. oleifera crud extracts |
18.28666667 |
17.13444444 |
1.15 |
2.72 |
0.151 |
Astragalin |
19.48777778 |
19.8 |
-0.32 |
1.25 |
0.420 |
Cryptochlorogenic acid |
19.85666667 |
18.79666667 |
1.07 |
2.64 |
0.160 |
Anise oil |
18.79666667 |
16.70555556 |
2.09 |
3.66 |
0.079 |
Anethole |
19.90111111 |
18.03222222 |
1.87 |
3.44 |
0.092 |
Control |
16.83222222 |
18.37555556 |
-1.57 |
0 |
1 |
From the results, all fractions (M. oleifera extract, astragalin, cryptochlorogenic acid, anethole and anise oil) had significantly down-regulation effects on the keratinase gene expression of T. rubrum KP979791 (Fig. 3A) and KP979787 (Fig. 3B) strains. Gene expression in KP979791 was substantially higher than that in KP979787 compared with that in control (1). There is a significant difference in ability of M. oleifera extract, astragalin, cryptologic acid, anise oil and anethole to decrease gene expression for KP979791 by 95%, 70%,99%,99% and 99% and for KP979787 by 84%. 58%, 84% 92% and 90% respectively compared to that in control (0%). From these results, the anise oil and anethole have the greatest ability to inhibit keratinase compared to M. oleifera extract, Astragalin and cryptochlorogenic acid. In contrast, the astragalin showed the lowest ability (70 %) in KP979791 and 58% in KP979787.
Fig. 3. The effects of plant extracts (Control, M. oleifera, Astragalin, Cryptochlorogenic acid, Anise oil and Anethole) on the keratinase gene expression of T. rubrum KP979791 (A) and KP979787 (B).
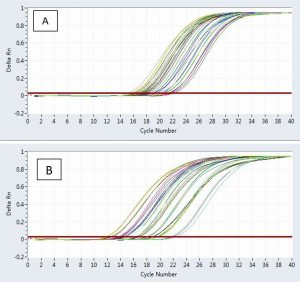
A great deal of variation has been demonstrated using the PCR technique to determine the dermatophyte’s detection rate (Fig. 4). This detection is determined by the target gene employed in the analysis35. Since the experimentation of dermatophytes using traditional laboratory approaches is a difficult problem and in most cases inadequate as a result of fungal phenotypic inconsistency and pleomorphism, the use of established molecular diagnostics provides an efficient method for the rapid analysis of pathogenic dermatophytes with unusual specificity, sensitivity and accuracy36.
Fig. 4. An actual PCR magnification graph (cycle 1-12) of keratinase (A) and the 18S gene (B) in the T. rubrum genotypes treatment group concerning the normalization control set.
The analysis and sequencing of many dermatophyte genomes, as well as developments in the methods of gene therapy of dermal cells, provide tools, which make it much easier for these diffusion species to clarify the direction of virulence. Understanding the basic virulence contributing factors to skin dermatitis can help design effective treatments and help develop new therapeutic strategies37.
The plant extracts and chemical compounds investigated in this study down-regulated the keratinase gene in T. rubrum strains KP979791 and KP979787. The results revealed that all fractions in this experiment had inhibitory impacts on gene expression by down-regulating the keratinase gene expression of T. rubrum strains. Turkoglu et al.38 demonstrated that the analysis of gene expression proved that the Ficus carica leaf extract had a significant down-regulation of SRD5A2, IL-1a, TNF-a, and VEGF compared to the untreated cells. Additionally, a qRT-PCR study was performed using essential oils to distinguish the extent of the gene expression, focusing on the production of ochratoxin A biosynthesis and two regulatory genes: laeA and vea. All six essential oils reduced the expression of the investigated genes, specifically the ackps that was down-regulated by 99.2%, which in this study is the maximum39.
The results of the current study and other related studies found contrasting capabilities of plant extracts (EO or leaf extract) because of the nature of the plant extract, the concentration in the medium, and the active material, which may be related to the virulence and physiology of the microorganism. These factors can explain the discrepancy in the effects of the plant materials on the gene expression profiles of different microorganisms. Liu et al.40 reported a high difference in genes expression of strain of T. rubrum that grow under the same condition.
However, other studies have revealed that treatment with lemongrass EO resulted in the up-regulation of acpP, whereas the increase of the amount of lemongrass EO had a profound impact on the expression levels of acpP, hly, and inlJ. However, M. oleifera extracts considerably down-regulated the mRNA expression levels of PPARα1, PPARγ, and HMG-CoAR41.
The excellent antifungal agent has been assumed as the compound that represses possible virulence factors, affects fungi physiology, and also acts on particular fungal cells. Moreover, these natural compounds may be more effective relative to common antifungal agents, thus synergistically improving their antifungal activity and acting as chemosensitizers to overcome fungal resistance. Therefore, the natural sources may expand the possibilities to develop antifungal therapies16. Our results revealed that kp979791 strain was more sensitive to M. oleifera extract, Astragalin, Cryptochlorogenic acid, Anise oil and Anethole than that in kp979787 by inhibiting a keratinase gene which led to down-regulation of the keratinase genes of kp979791 strain more than kp979787. More recent studies42 suggested that M. oleifera could be a source of metabolites with terbinafine to be used against Micrsporum canis, particularly during combination with a distinguished antifungal agent (terbinafine) which interacts with subtilisin virulence genes in M. canis43. Also, when culture filtrates of Marasmius palmivorus and Pleurotus ostreatus were combined with terbinafine, the mixture exhibited downregulation of citrate and isocitrate genes in T. rubrum43. Moreover, M. palmivorus is a strong antagonist fungus and could be used as bioagent against various phytopathogenic fungi44
We concluded from the aforementioned study that M. oleifera leaf extract, cryptochlorogenic acid, Anise oil and Anethole had pharmaceutical effects against the virulence factors (keratinase gene) of T. rubrum. These findings should prove tremendously helpful in the development of new regimens for the treatment of T. rubrum infections, especially in Babylon Province, Iraq, which has a fairly high rate of such infections. To the best of our knowledge, this is the first report on the impact of astragalin, cryptochlorogenic acid, and anethole on the keratinase gene as a target for T. rubrum infections.
Additional file: Additional Table
ACKNOWLEDGMENTS
Authors thanks Department of Biology, College of Science, University of Babylon, Iraq for providing lab facilities. Sincerely grateful to the Deanship of Al-Mustagbal University College, Hilla, Iraq, for supporting the publication cost.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
This work was financially supported by the deanship of College of Science, University of Babylon, Iraq. (The deanship of College of Science, University of Babylon, provides financial funds for distinguished scientific research projects for Postgraduate students.) Grant number (27856).
ETHICS STATEMENT
This article does not contain any studies with human participants or animals performed by any of the authors.
AVAILABILITY OF DATA
All datasets generated or analyzed during this study are included in the manuscript.
- Achterman R, White TC. Dermatophyte virulence factors: identifying and analyzing genes that may contribute to chronic or acute skin infections. International Journal of Microbiology, 2012; 8 Pages,
Crossref - Abed Ali FH, Al-Janabi JKA, Alhattab MK. Prevalence of dermatophyte fungal infection in Hillah City, Iraq. International Journal of Chem. Tech. Research., 2017; 16(6): 827-837.
- Elewski BE, Rich P, Pollak R, Pariser DM, Watanabe S, Senda H, Ieda C, Smith K, Pillai R, Ramakrishna T9 Olin J T. Efinaconazole 10% solution in the treatment of toenail onychomycosis: Two phase III multicenter, randomized, double-blind studies. J Am Acad Dermatol., 2012; 68(4): 600-608.
Crossref - Kadhim SK, Al-Janabi, JK, Al-Hamadani AH. In vitro, determination of optimal conditions of growth and activity of clinical isolates of Trichophyton rubrum. J. Contemp Med. Sci., 2015a; 1(3): 9-19.
- Bitencourt TA, Macedo C, Franco ME, Assis AF, Komoto TT, Stehling EG, Beleboni RO, Malavazi I, Marins M, Fachin AL. Transcription profile of Trichophyton rubrum conidia grown on keratin reveals the induction of an adhesin-like protein gene with a tandem repeat pattern. Bio. Medical Center Genomics, 2016; 17: 249.
Crossref - Chen J, Yi J, Liu L, Yin S, Chen R, Li M. Substrate adaptation of Trichophyton rubrum. Microb Pathog., 2010; 48(2): 57–61.
Crossref - Kadhim SK, Al-Hamadani AH, Al-Janabi JKA. Genotyping and subgenotyping of Trichophyton rubrum isolated from dermatophytosis in Iraqi patients using RFLP-PCR. IOSR Journal of Pharmacy and Biological Sciences (IOSR-JPBS), 2015b; 10(6): 61-67.
- Hay RJ. Tinea Capitis: Current Status. Mycopathologia, 2017; 182: 87-93.
Crossref - Samdani AJ, Dykes PJ, Marks R. The proteolytic activity of strains of T. mentagrophytes and T. rubrum isolated from tinea pedis and tinea unguium infections. J. Med. Vet. Mycol., 1995; 33(3): 167–170.
Crossref - Sharma HK, Vyas N, Mishra RK and Sharma B. In vitro antifungal Susceptibility testing of Dermatophytes isolated from clinical samples in tertiary care hospital. International Journal of Medical and Health Research, 2019; 5(5): 40-45.
- Huang H, Huang M, Lv W, Hu Y, Wang R, Zheng X, Ma Y, Chen C and Tang H. Inhibition of Trichophyton rubrum by 420-nm Intense Pulsed Light: In Vitro Activity and the Role of Nitric Oxide in Fungal Death. Front. Pharmacol., 2019; 10: 1143.
Crossref - Komoto TT, Bitencourt TA, Silva G, Beleboni RO, Marins M, and Fachin AL. Gene expression response of Trichophyton rubrum during coculture on keratinocytes exposed to antifungal agents. Evid. Based Complement. Alternat. Med., 2015; 2015; 180535.
Crossref - Caceres A, Cabrera O, Morales O, Mollinedo P, Mendia P. Pharmacological properties of Moringa oleifera. 1: Preliminary screening for antimicrobial activity. J. Ethnopharmacol., 1991; 33(3): 213-6.
Crossref - Sharma A, Chandra S and Sharma M. Difference in keratinase activity of dermatophytes at different environmental conditions is an attribute of adaptation to parasitism. Mycoses, 2012; 55: 410-415.
Crossref - Sardana K, Kaur R, Arora P, Goyal R, Ghunawat S. Is antifungal resistance a cause for treatment failure in dermatophytosis: A study focused on tinea corporis and cruris from a tertiary center? Indian Dermatol Online J, 2018; 9: 90-5.
Crossref - Martinez-Rossi NM, Bitencourt TA, Peres NTA, Lang EAS, Gomes EV, Quaresemin NR, Martins MP, Lopes L and Rossi A. Dermatophyte Resistance to Antifungal Drugs: Mechanisms and Prospectus. Front. Microbiol., 2018; 9: 1108.
Crossref - Wang L, Chen X, Wu A. Mini Review on Antimicrobial Activity and Bioactive Compounds of Moringa oleifera. Med Chem (Los Angeles), 2016; 6: 578-582.
Crossref - Abd-Rani NZ, Husain K, Kumolosasi E, Moringa Genus: A Review of Phytochemistry and Pharmacology. Front. Pharmacol., 2018; 9: 108. [Google Scholar] [CrossRef] [PubMed]
Crossref - Manikandan PA, Gnanasekaran P, Julikarthika, and Arvind Prasanth, D. Antibacterial Efficacy of Moringa oleifera Leaf against Medically Important Clinical Pathogens. Int. J. Curr.Microbiol. App.Sci. 2016; 5(4): 109-116.
Crossref - Obaid AJ, Al-Janabi JKA and Taj-Aldin WR. Antifungal Activity of Anise Essential Oil Against Growth and Morphological Characteristics of Trichophyton rubrum. Journal of Global Pharma Technology, 2017a; 7(9): 53- 68.
- Obaid AJ, Al-Janabi JKA, and Taj-Aldin WR. Chemical Composition and Bioactivity Characteristics of Pimpinella Anisum Essential Oil against Trichophyton rubrum. Journal of Global Pharma Technology, 2017b; 8(9): 44-56.
- Ibrahim SY and Abd El-Salam MM. Anti-dermatophyte efficacy and environmental safety of some essential oils commercial and in vitro extracted pure and combined against four keratinophilic pathogenic fungi. Environ Health Prev Med., 2015; 20: 279–286.
Crossref - Peres NT, Maranhao FC, Rossi A and Martinez-Rossi NM. Dermatophytes: host–pathogen interaction and antifungal resistance. An Bras Dermatol., 2010; 85(5), 657–667.
Crossref - Odds FC, Sabaraud(s’) agar, J. Med. Vet. Mycol.,.1991; 29: 355-359.
Crossref - Bijina B, Chellappan S, Krishna JG, Basheer SM, Elyas KK, Bahkali AH and Chandrasekaran M. Protease inhibitor from Moringa oleifera with potential for use as a therapeutic drug and as seafood preservative. Saudi Journal of Biological Sciences, 2011; 18: 273–281.
Crossref - Ullah H, Mahmood A, Iqbal Awan M and Honermeier B. Effect of Row Spacing and Seed Rate on Fruit Yield, Essential Oil and Composition of Anise (Pimpinella anisum L.). Pakistan Journal of Agricultural Sciences, 2015; 52(2): 349-357.
- Porel A, Sanyal Y and Kundu A. Simultaneous HPLC Determination of 22 Components of Essential Oils; Method Robustness with Experimental Design. Journal of Indian of Pharmacy and Science, 2014; 76(1): 19–30.
- Tohma H, Koksal E, Kilic O A, Abdullah M, Gulcin l, Bursal E and Alwasel SH. RP-HPLC/MS/MS Analysis of the Phenolic Compounds, Antioxidant and Antimicrobial Activities of Salvia L. Species. Journal of Antioxidants, 2016; 5: 1-15.
Crossref - Livak KJ and Schmittgen TD. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 22DDCT Method. Methods Journal, 2001; 25: 402–408.
Crossref - Lin Y, Xu W, Huang M, Xu W, Li H, Ye M, Zhang X, and Chu K. Qualitative and quantitative analysis of phenolic acids, flavonoids and iridoid glycosides in yinhua kanggan tablet by UPLC-QqQ-MS/MS. Journal of Molecules, 2015; 3: 20 (7): 9-28.
Crossref - Galvez, JS, Zevallos LC and Velazquez DAJ. Chlorogenic Acid: Recent Advances on Its Dual Role as a Food Additive and a Nutraceutical against Metabolic Syndrome. Journal of molecules, 2015; 20: 12209-12228.
- Rohn S, Rawel HM and Kroll J. Inhibitory effects of plant phenols on the activity of selected enzymes. Journal of Agriculture Food and Chemistry, 2007; 50(12):71.
Crossref - Pirouzpanah S, Hanaee J, Razavieh SV and Rashidi M R. Inhibitory effects of flavonoids on aldehyde oxidase activity. Journal of Enzyme Inhibition and Medicinal Chemistry, 2009; 24(1):1-7.
Crossref - Braz J. Enzymatic inhibition studies of selected flavonoids and chemosystematic significance of polymethoxylated flavonoids and quinoline alkaloids in Neoraputia (Rutaceae). Journal of the Brazilian Chemical Society, 2007; 14(3):1-6.
Crossref - Kabir S. Detection of dermatophytes DNA by polymerase chain reaction: A review Dermatophytosis Journal, 2004; 9: 115-123.
Crossref - Behzadi P, Behzadi E and Ranjbar R. Dermatophyte fungi: infections, diagnosis, and treatment. SMU Medical Journal, 2014; 1: 50-62.
- Wang H and Ng TB. Ganodermin, an antifungal protein from fruiting bodies of the medicinal mushroom Ganoderma lucidum. Peptides, 2006; 27: 27–30.
Crossref - Turkoglu M, Pekmezci E, Kilic S, Dundar C and Sevinc H. Effect of Ficus carica leaf extract on the gene expression of selected factors in HaCaT cells. Journal of cosmetic dermatology, 2017; 16(4): 54-58.
Crossref - El Khoury R, Atoui A, Verheecke C, Maroun R, El Khoury A and Mathieu F. Essential Oils Modulate Gene Expression and Ochratoxin A Production in Aspergillus carbonarius. Journal of Toxins, 2016; 8(8): 1-19.
Crossref - Liu T, Xu X, Leng W, Xue Y, Dong J, and Jin Q. Analysis of gene expression changes in Trichophyton rubrum after skin interaction. J Med Microbiol., 2014; 63(5): 642–648.
Crossref - Sangkitikomol W, Rocejanasaroj A and Tencomnao T. Effect of Moringa oleifera on advanced glycation end-product formation and lipid metabolism gene expression in HepG2 cells. Genetics and Molecular Research, 2014; 13(1): 723-735.
Crossref - Khazia MQ and Al-Janabi JKA. Antifungal activity of combination of medicinal plant extracts with terbinafine through regulating subtilisin virulence genes in Microsporum canis, Drug Invention Today, 2019; 12(11): 2580-2588.
- Abdullah MH and Al-Janabi JKA. Efficiency of antifungal agents with culture filtrates of two basidiomycetes fungi on growth and on citrate and isocitrate genes of Trichophyton rubrum. Drug Invention Today, 2019; 11(11): 3085-3096.
- Tamur AH, Al-Janabi HJ, Al-Janabi JKA, Mohsin LY and Al-Yassiry ZAN. Characterization and Antagonistic Activity of New Causal Agent of Wilt Disease in Imperata cylindrica (Marasmius palmivorus), J Pure Appl Microbiol., 2019; 13(3): 1525-1536.
Crossref
© The Author(s) 2020. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.