ISSN: 0973-7510
E-ISSN: 2581-690X
COVID-19 pandemic due to SARS-CoV-2 has been one of the major global health issues of this aeon. The aim of this study was to evaluate the association of SARS-CoV-2 cycle threshold (Ct) values with multiple factors among COVID-19 patients visiting a tertiary care hospital in Sudurpashchim province of Nepal. A retrospective analysis was performed on the data of randomly selected COVID-19 cases among the total RT-qPCR tested patients from March 2020 to April 2022. The Ct values at the time of patient admission and their clinical outcomes (discharge or death) were compared. Among the COVID-19 patients, survivor group had significantly higher initial Ct value compared to non-survivors [median Ct values 23.21 and 24.39 (P < 0.0001)]. Selected haematological parameters; white blood cells (P<001), neutrophils (P<001), and monocytes (P<0.0001), and all the biochemical parameters were significantly different between these two groups (p < 0.005). Furthermore, significantly increased CRP (61.54±63.00, P<0.0017), D-dimer levels (0.8979± 1.480, P<0.0001), creatinine (0.7931±0.2551, P<0.0001), monocytes (0.6782±0.7981, P<0.0001), and random blood sugar (152.4±34.32, P<0.0001) were observed among non-survivors indicating as cause of disease severity in COVID-19. The findings of this study imply that the Ct value, CRP and D-dimer levels could be a crucial marker for the early detection of severe COVID-19 patients or those at higher risk of developing severe disease. This will eventually help to identify cases requiring immediate and critical medical care and reduce mortality.
SARS-CoV-2, COVID-19, RT-qPCR, Cycle Threshold-Ct, Disease Severity, Severity Marker, Nepal
Human Coronavirus was first confirmed in 1960, 23 years after its description in birds. More than 80% infections caused by this virus are zoonotic in nature. Taxonomically, Coronavirus belongs to the family coronaviridae and subfamily beta coronavirus.1 Coronavirus is an enveloped single-stranded RNA virus with a genome length of approximately 27-30 kb.1 Different coronaviruses evolved with time and at least seven of these viruses have been linked to human infections including the most recent Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2) which caused the COVID-19 pandemic.2-4 SARS-CoV-2 infected more than 769 million people with 6,955,141 deaths globally as of 12 August 2023.1,2
SARS-CoV-2 infection in humans is believed to have originated from wild animals although the intermediate host is not known yet.3 This speculation was based on: 1- it was first reported from a seafood market in Wuhan, China, and 2-coronaviruses spilled over from bat, pivot cat and other wild animals to humans showed similar nucleotide sequence as SARS-COV-2 suggesting the evidence of zoonotic transmission.4
Nepal, a landlocked country, borders with India and China. Due to an open border between India and Nepal, there have been massive cross-border movement of people for various social and economic activities. By the time China controlled the first wave of COVID-19 pandemic, it was rapidly growing in India.2 Despite the countrywide lockdown to contain virus transmission in Nepal and India, the existing porous borders allowed an uncontrollable infusion of people across borders that led to a surge in cases and subsequent bigger clusters in Nepal. Therefore, the number of COVID-19 suspected/confirmed cases and people under quarantine outnumbered in the bordering areas of Nepal compared to other parts. Diagnostic facilities in Nepal had been upgraded and expanded but were not adequate to meet the increased demand indicating the underreporting of the actual community transmission during the early pandemic phase. Subsequently, the situation became worse with the evolution of newer variants of SARS-CoV-2 that caused different waves of pandemic similar to the global trends. Since the first report of COVID-19 on January 13, 2020,4-6 a total of 1,003,424 cases have been confirmed by RT-PCR with 12,031 confirmed deaths as of September 13, 2023.1-7 Simultaneously, SARS-CoV-2 RNA was detected by qPCR in 60% (50/84) in tested waste, river water and hospital centers in Nepal conducted by Tandukar et al.8 In Sudurpaschchim province (eight districts), the first case of COVID-19 was gulf returnee confirmed on March 27, 2020, at Seti Provinical Hospital, Dhangadhi, a total of 6,70,765 COVID-19 cases were detected by RT-qPCR and antigen of 637 reported deaths as 30/09/2023 according to Sudurpaschchim Province of Health Directorate, Nepal.9
Despite the rapid development of different platforms for COVID-19 diagnosis, real-time quantitative polymerase chain reaction (RT-qPCR) has been the gold standard method because of reasonably high sensitivity and specificity.7,10 Apart from the SARS-CoV-2 diagnostic purpose, the potential utility of RT-qPCR Ct values in COVID-19 severity identification/ prediction has not been studied adequately in Nepal, although there are reports coming from other parts of the world.7,10,11 This is very important during the pandemic specially in the health care resource limited countries like Nepal to enable rationale and timely use of intensive facilities for severe patients.6-8 Viral dynamics and Ct values could predict the disease severity in COVID-19 when combined together with other factors such as symptoms, underlying health conditions (including comorbidities), routine laboratory parameters (haematological, immunological and biochemical profiles), chest x-ray, and chest CT scan, etc. Ct values can be influenced by several factors such as specimen collection technique, specimen type, sampling time, viral kinetics, transport and storage conditions, nucleic acid extraction, viral load, primer designing and real-time PCR efficiency and platforms used.7,11 Nevertheless, Ct value could still be a simple yet useful marker for disease severity in COVID-19. Therefore, the main objective of this study was to evaluate the association of Ct values with multiple factors among COVID-19 patients visiting a tertiary care hospital in Sudurpashchim province of Nepal and explore its potential utility in severity prediction.
Study site and its justification
The site, Seti Provinical Hospital (SPH) in Kailali districts was selected because it is the major tertiary-care public hospital in Sudurpashchim province of Nepal. Moreover, this hospital was one of the first to establish COVID-19 PCR laboratory in the Sudurpashchim province. The SPH catchment population area includes one of the major entry and exit portals of Nepalese people visiting India for different purposes including job, treatment and tourism.
Study design
This was a retrospective study conducted on randomly selected COVID-19 patients who were admitted and suspected COVID-19 patients at SPH. Purposive sampling was performed to select cases confirmed with RT-qPCR Ct values and multiple parameters related to COVID-19 disease severity. From March 2020 to April 2022, a total of 127,196 suspected COVID-19 patients were tested by RT-qPCR at SPH and 23,966 of them were confirmed as COVID-19 cases (Ct < 40). Of these confirmed COVID-19 cases, 313 positive cases with a history of discharge and 131 death cases were randomly selected from the SPH database. The main criteria for this selection were based on the availability of complete laboratory parameters obtained within one-week of onset of infection to allow the inclusion of eligible cases.
Specimen collection and RNA extraction
Nasopharyngeal (NP) and oropharyngeal (OP) swab samples were collected from admitted and suspected patients at SPH. The collected swabs were placed in viral transport medium and immediately transported to the COVID-19 laboratory for further analysis maintaining cold chain as per the WHO guidelines.12 Manual and automated, viral RNA extractions were performed following the manufacturer’s instructions (General Biologicals, Hsinchu, Taiwan13 and Nanjing Zhongkebio Medical Technology, Nanjing, China).14
SARS-CoV-2 detection by RT-qPCR
Viral detection was performed by using the RT-qPCR kit (Uni-Medica, Shenzhen, China)15 according to the manufacturer’s instructions. Briefly, RT-qPCR were performed in a 40 µl reaction mixture containing 18.5 µl reaction buffer, 1.5 µl enzyme mix and 20 µl of RNA template. The reaction was executed in a thermo-cycler (D-Lab, Beijing, China) with the following conditions: reverse transcription at 55°C for 10 min, Taq activation with pre-denaturation at 95°C for 3 min, denaturation at 95°C for 10 s, followed by 45 cycles of amplification with a final annealing, extension and fluorescence acquisition at 60°C for 1 min. For interpretation analysis, samples with a Ct value < 40 of all Orf1ab/N/E genes were considered as positive and Ct value > 40 of Orf1ab/N/E genes as negative for SARS-CoV-2.15
Data collection
Information on RT-qPCR-positive patients’ demographic details including age, gender, Ct value, hospitalization status, health status of patients, laboratory test results (haematological, immunological and biochemical markers), and chest x-ray of COVID-19 patients and clinical outcomes were retrieved from the electronic medical records and database of respective hospital departments. According to Shah et al,16 Ct values were categorically grouped into high (Ct 31–40), moderate (21–30), and low (11–20). Among COVID-19 patients (survivors and non-survivors), Ct values were compared based on clinical outcomes (severe, moderate, and mild disease). The following criteria used to classify as mild, moderate and severe cases if COVID-19 patients had: (i) mild clinical symptoms without evidence of breathlessness or hypoxia (normal saturation), dyspnea or abnormal chest imaging, ii) fever, cough and evidence of lower respiratory disease during clinical assessment or imaging, including oxygen saturation (SpO2) <94% (range 90–94%) on room air, respiratory rate ≥24 per minute and iii) clinical signs of pneumonia plus one of the following; respiratory rate >30 breaths/min, severe respiratory distress with oxygen saturation (SpO2) < 90% at room air, requirement for mechanical ventilation, and/or admission to intensive care unit (ICU).17,18
Data analysis
RT-qPCR Ct values and all available clinical and laboratory parameters were retrieved/ transcribed into the Excel spread sheet followed by data cleaning and verification. Ct values of confirmed COVID-19 cases were further analysed to ascertain the correlation of Ct with clinical manifestations, haematological and biochemical markers, chest x-ray findings and COVID-19 severity levels. GraphPad Prism 9.4 software was used for data analysis. For continuous variables, Student’s t-test and Mann-Whitney U test were used for parametric and non-parametric data, respectively. Chi-square was used for categorical variables. A P-value < 0.05 was considered statistically significant.
Ethics
Ethical approval was obtained from the Institutional Review Committee – Institute of Science and Technology (IRC-IoST), Tribhuvan University, Kathmandu, Nepal (IRCIOST-23-0090). We obtained completely de-identified COVID-19 data of patients. As this was a retrospective analysis conducted in the de-identified data, informed consent was not required.
Ct values and clinical outcomes
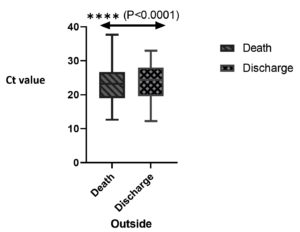
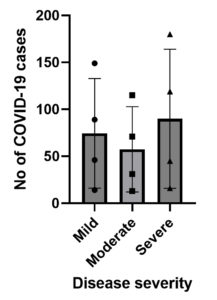
Among 127,196 RT-qPCR-positive cases, 444 cases (131 deaths and 313 survivors) were randomly selected for the analysis. More than half (66.56%, 300/444) were male patients. The median age was 45 years [IQR (63-29)] and the median Ct value was 24.01 (IQR, 19.35-27.750). Comparison of the initial Ct value with clinical outcomes (discharge or death) showed that the survivors had a significantly higher Ct than non-survivors [median Ct (IQR), 23.39 (19.55-28) and 23.21 (19.08-26.68)] (p < 0.0001; Figure 1). The frequency and average Ct values were also analysed according to disease severity; mild (33.55%, median (IQR): 23.16 (18.15-27.70); moderate (25.90%, 24.88 (19.8-28.23) and Severe (40.76%, 24.01 (20.38-27.65) by among the 444 patients included (p> 0.05). Of the death cases, majority had severe pneumonia followed by cardio-respiratory failure, acute respiratory distress syndrome and hypertension (data not shown) which could be the cause of death in COVID-19 patients.
Table (1):
Demographic characteristics and severity levels of COVID-19 patients, Nepal
| Variable | Category | Mild (n = 149) | Moderate (n = 115) | Severe (n = 180) | Chi-square | P value |
|---|---|---|---|---|---|---|
| Clinical outcome | Survived | 147 (46.96%) | 89 (28.43) | 77 (24.60%) | χ 2 =125.93 | P < 0.00001* |
| Death | 2 (1.52%) | 26 (19.84%) | 103 (78.62%) | |||
| Gender | Male | 104 (69.80%) | 76 (66.09%) | 124 (68.89%) | χ 2 =0.43 | P = 0.802 |
| Female | 45 (30.30%) | 39 (33.91%) | 56 (31.11%) | |||
| Age group | 18-25 | 51 (34.23%) | 22 (19.13%) | 18 (10%) | χ 2 =55.43 24.66±13.63 |
P < 0.00001* |
| 26-33 | 28 (18.79%) | 16 (13.91%) | 20 (11.11%) | |||
| 34-42 | 22 (14.77%) | 13 (11.30%) | 16 (8.89%) | |||
| 43-50 | 17 (11.41%) | 16 (13.91%) | 24 (13.33%) | |||
| 51-60 | 7 (6.09%) | 17 (9.44%) | 34 (18.89%) | |||
| >61 | 24 (16.11%) | 31 (29.96%) | 68 (37.78%) |
* P < 0.05 was considered statistically significant
Table 1 provides detailed information on patient demographics and descriptive statistics. Among death group (n =131), majority of patients had severe conditions (103/131, 78.62%) followed by moderate conditions (26/131, 19.84%) and mild conditions (2/131, 1.52%) based on Ct value and clinical outcomes as per the classification criteria.13-15 In contract, among survivors, most patients had mild (147/313, 46.96%), moderate (89/313, 28.43%) followed by severe conditions (77/313, 24.60%). Male patients had higher frequency of severity (124-68.89%) than females (56-31.11%). Moreover, among the elderly patients above 60 years, majority had severe conditions (68-37.78%) and moderate condition (31-29.96%) compared to other age groups. Similarly, age group 18-25 predominantly had mild condition (51-34.23%) (Table 1).
Biochemical parameters of COVID-19 cases
The biochemical parameters i.e. CRP (P<0.0017), D-dimer (0.0017), creatinine (P<0.0001) and potassium (P<0.0001) showed a statistically significant difference (P<0.05) in COVID-19 cases between survival and death groups (Table 2). Interestingly, CRP (< 5 mg/L, 35.15-61.54, dimer (< 0.5mg /dl, 0.895-1.475), alanine aminotransferase (5-42 IU/L, 67.82-72.94), and aspartate aminotransferase (5-37 IU/L, 40.05-48.13) were found to be significantly higher than the normal reference range in comparison of survival with death group.
Table (2):
Biochemical parameters and clinical outcomes COVID-19 patients, Nepal (n=444)
Parameters |
Reference range |
Survived (n=313) |
Death (n=131) |
P-valueb |
|---|---|---|---|---|
CRP (mg/L) |
<5 |
35.15±31.53 |
61.54±63.00 |
< 0.0017 |
D-Dimer (mg/L) |
<0.5 |
1.475±2.018 |
0.8979±1.480 |
< 0.0001 |
HB1AC (%) |
3.8-5.8 |
10.86±10.14 |
8.376±3.813 |
0.7248 |
Urea (mg/dL) |
15-45 |
27.96±14.78 |
25.46±11.01 |
0.0541 |
Creatinine (mg/dL) |
0.6-1.5 |
0.6986±0.4656 |
0.7931±0.2551 |
<0.0001 |
Albumin (mg/dl) |
3.2-5.5 |
3.077±0.4180 |
3.199±0.7377 |
0.0104 |
Sodium (Na+) (mmo/I) |
135-150 |
138±6.468 |
137.5±3.427 |
0.4333 |
Potassium (K+) (mmo/I) |
3.5-5.3 |
4.667±4.415 |
3.884±0.5583 |
< 0.0001 |
ALT (U/L) |
5-42 |
67.82±23.11 |
72.94±47.19 |
0.4937 |
AST (U/L) |
5-37 |
48.13±29.99 |
40.05±21.44 |
0.0389 |
ALP (U/L) |
45-132 |
84.30±18.72 |
86.28±12.86 |
0.1631 |
aMean ±standard deviation; bMann-Whitney U test; P < 0.05 was considered statistically significant.
CRP: C-Reactive Protein, AST: Aspartate Aminotransferase, ALT: Alanine Aminotransferase, ALP: Alkaline Phosphatase
Haematological parameters of COVID-19 cases
Non-survivors (death group) had significantly lower count of WBC (p<0.0001), neutrophil (p<0.0001), and significantly higher levels of monocytes (p<0.0001), haemoglobin (p<0.0001), platelets (p=0.0022), and random blood sugar(P<0.0001) (p < 0.005), while there was no significant difference in lymphocytes (p=0.2489), and platelets counts (p=0.0022) with (p > 0.0.05) (Table 3). Remarkably, random blood sugar level was found significantly higher than the normal reference range (80-120 mg/dl) in both death (152.4±34.32) and survivor groups (128±39.22) with statistically significant difference (p < 0.0001).
Table (3):
Haematological parameters and clinical outcomes of COVID-19 patients, Nepal (n=444)
Parameters |
Reference range |
Survivala (n=313) |
Deatha (n=131) |
P valueb |
|---|---|---|---|---|
WBC (10×3/ul) |
4.5-11 |
10.47±3.711 |
8.604±3.492 |
<0.0001** |
NEU (10×3/ul) |
1.8-7.7 |
8.85±1.940 |
7.0±2.167 |
<0.0001** |
LYM (10×3/ul) |
1.50-3.0 |
1.221±0.7660 |
1.267±0.8525 |
0.2489 |
MON (10×3/ul) |
0.3-0.5 |
0.4981±0.1600 |
0.6782±0.7981 |
<0.0001** |
HG (g/dl) |
11.5-15.5 |
12.59±1.607 |
13.21±1.553 |
<0.0001** |
PLT (10×3/ul) |
150-400 |
179.2±67.69 |
197.9±55.27 |
0.0022 |
RBS (mg/dl) |
80-120 |
128.4±39.22 |
152.4±34.32 |
<0.0001** |
aMean ±standard deviation; bMann-Whitney U test; P < 0.05 was considered statistically significant.
WBC: White Blood Cell, NEU: Neutrophils, LYM: Lymphocytes, MON: Monocytes, HG: Haemoglobin, PLT: Platelet, RBS: Random Blood Sugar
In the current study, high SARS-CoV-2 viral load (Ct:11-20) was observed more frequently in admitted patients with the consequence of death, which is comparable to the previous reports.15,16,19,20 The reason behind the high viral load could be due to disease severity associated with other endemic, pandemic as well as seasonal respiratory viruses. However, others respiratory viruses, laboratory diagnosis was not performed among the randomly selected COVID-19 patients to draw a conclusion.21-25 Pre-pandemic avian influenza and seasonal influenza cases with high viral loads were associated with more severe form of disease and poorer treatment outcomes similar to SARS-CoV-2 infection. The correlation of viral load with clinical outcome and disease severity has been investigated with different endemic respiratory viruses, but most of the results remained inconclusive.21-25 The association of viral load with clinical consequence and severity of COVID-19 were assessed by employing haematological, biochemical and immunological markers (Table 2 and Table 3).22 Additionally, numerous common laboratory parameters that likely cause severity in COVID-19 infection may play an crucial role during the disease prognosis and forecast for treatment decisions.23-26 Most of the previous studies revealed to be association of disease severity with common parameters include C-reactive protein (CRP), D-dimer, creatinine, potassium, alanine aminotransferase (ALT), alkaline phosphatase (ALP), white blood cell, neutrophil, monocytes and lymphocytes.23-26 Assessment of initial Ct value with clinical consequences exhibited that the survivors had significantly higher levels than non-survivors (P<0.0001; Figure 1). Regarding the Ct values of COVID-19 cases with different clinical outcomes (mild, moderate and severe), current study did not show any significant difference (Figure 2). Ct values could be indirectly associated with the majority of final clinical decisions when combined with routine laboratory profiles, radiological findings and physician’s clinical observation (i.e. clinical signs/scores and disease progression). Therefore, Ct value could be a useful tool for physicians in the management of COVID-19 patients and to make clinical decisions.27
We found, more than half of the patients had severe pneumonia (B/L pneumonia in the lower zone) followed by cardiorespiratory failure, (ARDS) and hypertension among the death group. Similarly, chest x-ray finding in COVID-19 patients was able to detect severe pneumonia as in the previous study where the presence of severe symptoms significantly associated with abnormal chest x-ray findings. Previous findings by Yang et al.28 and Rousan et al.,29 suggesting that the combination of clinical symptoms and radiological findings with Ct value could be helpful as a tool in the diagnosis, follow up of COVID-19 pneumonia and drawing clinical conclusions for prompt treatment of COVID-19 cases in resource-limited countries like Nepal.
Similarly, haematological parameters (WBC, neutrophils, and monocytes) were significantly different between survivor and non-survivor COVID-19 cases which is comparable to previous studies.25,30,31 On the other hand, random blood sugar level was found significantly higher than normal reference range in COVID-19 patients (Table 2). Other studies found that WBC, monocytes, and random blood sugar increased during the viral infection while the neutrophil and lymphocytes counts decreased.27,29 In contrast, a previous study on COVID-19 patients showed an elevated AST, and low leukocyte, neutrophil, lymphocyte, eosinophil, and monocyte counts.32 According to Cheng-Fu et al.,32 decreased CBC was found in immuno-compromised conditions or other inflammatory responses during COVID-19 disease progression. Of note, Wang et al.33 showed that these values were within the normal range in initial stage of COVID-19 disease and subsequently elevated during disease progression with a decreased lymphocyte count.
Our findings on biochemical parameters in COVID-19 cases demonstrated an increase in CRP, creatinine, ALT, and ALP (Table 3) and there was a significant increase in death group compared to survivor group. An inflammatory disorder that changes fibrin levels is linked with elevated D-dimer levels.34 This is similar to a report by Tang et al.35 which showed a considerable increase in D-dimers, CRP and liver enzymes in non-survivors. Mardani et al.36 detected increased levels of biochemical parameters i.e. ALT, AST, CRP, bilirubin and albumin so as to forecast COVID-19 progression. Furthermore, study findings also suggest that Ct value, clinical outcome, haematological and biochemical parameters may be useful to predict the progression of disease in the early phase and thereby help forecasting the clinical management of COVID-19 patients. Our finding is in accordance with a previous report by Yang et al.28 which showed a significant increase in the levels of D-dimer, CRP, and creatinine in severe COVID-19 patients compared to non-severe. These findings would be of potential utility to guide treatment, diagnosis and evaluate prognosis. Moreover, CRP and D-dimer can also be considered as an early marker for inflammatory reactions elevated in COVID-19 disease.35-39
In summary, SARS-CoV-2 viral dynamics demonstrated by RT-qPCR Ct values correlated to disease severity in COVID-19 patients at a tertiary care hospital, and this association was influenced by multiple factors including haematological, biochemical, chest x-ray and clinical findings. Our study findings suggest that Ct values could be used as an important tool for the identification of potential severe patients at the early stage COVID-19 thereby supporting in initial diagnosis, and clinical management when combined with haematological, biochemical, radiological and clinical findings.
Thus, viral and human genetics, genomics, and immunology research studies will be necessary to further understand those affecting factors. Further longitudinal studies with larger sample size will also be needed to understand the actual mechanism of clinical illness, COVID-19 severity and their consequences by employing RT-qPCR Ct values.
The results of this study imply that the Ct value could be a crucial tool as an early detection marker of severe COVID-19 disease or in identifying those at higher risk of disease severity. The Ct value also helps to identify cases requiring immediate and critical medical care and can be utilized for initial diagnosis, and prediction of severity and treatment options in COVID-19 when combined with clinical symptoms, haematological, biochemical, and chest x-ray findings. Nevertheless, these findings might be useful for policymakers or planners in improving the strategies to diagnose and treat on time to prevent further transmission of SARS-CoV-2 to healthy populations, as well as in preparedness plans.
ACKNOWLEDGMENTS
The authors express gratitude to Dr. Eddie Samuneti (University Teaching Hospital, Lusaka, Zambia) for critically reviewing the manuscript. The authors also sincerely acknowledge Dr. Pawan Joshi, Dr. Pragya Joshi, Dr. Aaditya Singh, and all the health and laboratory staff for their substantial support in this study conducted at Seti Provincial Hospital, Ministry of Social Development, Sudurpashchim Province, Dhangadhi, Kailali, Nepal.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
JJ, OPJ, SBK, GRA, YS, RS, SK, PRP, SR, SRS, DKP and HRJ conceptualized and designed the study. JJ, YS, HRJ, RPO, CRJ, KP, SPD, RSD, LRB, GRA, KSK and BDP performed methodology. JJ, SPD, YS, KP, RS, GRA and BDP supervised the study. JJ, OPJ, YS, RPO, SK, SR, PRP, KSK and BDP wrote original draft. JJ, OPJ, SBK, GRA, CRJ, KP, SPD, YS, HRJ, RS, LRB, RPO, RSD, GRA, SBS, SK, SR, PRP, SRS, DKP, KSK and BDP wrote and edited the manuscript. All authors have read and approved the final manuscript for publication.
FUNDING
This study was partially funded by Province Health Directorate, Sudurpaschim Province, Rajpur, Doti, Nepal, with grant number 2078/79-841.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
This study was apporved by Institutional Review Committee – Institute of Science and Technology (IRC-IoST), Tribhuvan University, Kathmandu, Nepal (Ref IRCIOST-23-0090).
- Lu R, Zhao X, Li J, et al. Genomic characterization and epidemiology 0f 2019 novel coronavirus: implications for virus and receptor binding. Lancet. 2020;395(10224):565-574.
Crossref - Su S, Wong G, Shi W, et al. Epidemiology, genetic recombination and pathogenesis of coronaviruses. Trends Microbiol. 2016;24(6):490-502.
Crossref - Cavanagh D. Cornavirus avian infectious bronchitis virus. Vet Res. 2007;38(2):281-297.
Crossref - Bastola A, Sah R, Rodriguez-Morales AJ, et al. The first 2019 novel coronavirus case in Nepal. Lancet Infect Dis. 2020;20(3):279-280.
Crossref - Sah R, Jha R, Rodriguez-Morales AJ, et al. Complete genome sequence of a 2019 novel coronavirus (SARS-COV-2) strain isolated in Nepal. Microbiol Resour Announc. 2020;9(11):e00169-20.
Crossref - The Ministry of Health and Population, Government of Nepal. https://covid19.mohp.gov.np/. Acessed on 30 September, 2023
- World Health Organization. https://covid19.who.int/ Acessed on 13 September 2023
- Tandukar S, Sthapit N, Thakali O, et al. Detection of SARS-CoV-2 RNA in wastewater, river water, and hospital wastewater of Nepal. Sci Total Environ. 2023;824:153816.
Crossref - Province Health Directorate, Rajpur, Dipayal, Doti. Acessed on30 September 2023 https://hd.sudurpashchim.gov.np/en
- Gupta GP, Shah Y, Pant DK, et al. Preparatory phase for clinical trials of COVID-19 vaccine in Nepal. Hum Vaccin Immunother. 2021;17(2):418-419.
Crossref - Pandey BD, TunM, Pandey K, et al. How an Outbreak of COVID-19 Circulated Widely in Nepal: A Chronological Analysis of the National Response to an Unprecedented Pandemic. Life (Basel). 2022;12(7):1087.
Crossref - World Health Organization. Laboratory testing for 2019 novel coronavirus (2019-nCoV) in suspected human cases Interim guidance. 2020. https://iris.who.int/bitstream/handle/10665/330676/9789240000971-eng.pdf?sequence=1
- General Biological corporation Laprep viral DNA/RNA min kit Taiwan. https://www.biovendor.com/labprep-viral-dnarna-mini-kit-100
- Nanjing ZhongkeBio Medical Technology Co. LTd Nucleic acid extraction and Purification Kit, China. https://www.en.njzkbio.com/product/9/
- Uni-Medica. Real-time PCR kit for Novel Coronavirus 2019-nCoV (ORF1ab, N, E genes).Shenzhen Uni-medica Technology Co. Ltd, China. https://uni-medica.cn/
- Shah S, Singhal T, Davar N, Thakkar P. No correlation betweenCt values and severity of disease or mortality in patients with COVID-19 disease. Indian J Med Microbiol. 2021;39(1):116-117.
Crossref - Clinical management protocol for COVID-19. 2020. https://covid19.india.gov.in/document/clinical-management-protocol-for-covid-19-2. Accessed on 28 January 2021.
- Pocket Book of clinical management of COVID-19 in Healthcare setting Department of Health Services, Epidemiology and disease control divison, Teku, Kathmandu, Nepal. https://edcd.gov.np/uploads/resource/61b8533ddd84d.pdf. Accessed on 14 December 2021
- Yuan XY, Hauang W, Ye B, et al. Changes of hematological and immunological parameters in COVID-19 patients. Int J Hematol. 2020;112(4):553-559.
Crossref - Faico-filho KS, Passarelli VC, Bellei N. Is higher viral load in SARS-C0v2 associated with Death. Amp J Trop Med Hyg. 2020;103(5):2019-2021.
Crossref - Feikin DR, Fu W, Park DE, et al. Is higher viral load in the upper respiratory tract associated with severe pneumonia? Findings from the PERCH study. Clin Infect Dis. 2017;64(S3):S337-S346.
Crossref - Kadji FM, Okamoto M, Furuse Y, et al. Differences in viral load among human respiratory syncytial virus genotypes in hospitalized children with severe acute respiratory infections in the Philippines. Virol J. 2016;13:113.
Crossref - Granados A, Peci A, McGeer A, Gubbay JB. Influenza and rhinovirus viral load and disease severity in upper respiratory tract infections. J Clin Virol. 2017;86:14-19.
Crossref - Lalueza A, Folgeira D, Munoz-Gallego I, et al. Influence of viral load in the outcome of hospitalized patients with influenza virus infection. Eur J Clin Microbiol Infect Dis. 2019;38(4):667-673.
Crossref - Ghazanfari T, Salehi MR, Namaki S, et al. Interpretion of haematological, biochemical and immunological findings of COVID-19 Disease: Biomarkers associated with severity and mortality. Iran J Allergy Asthma Immunol. 2021;20(1):46-66.
Crossref - Al Badi E, Al Shukri I, Al Mahruqi S. Correlation of Viral Load with the Biochemical and Hematological Profiles of COVID-19 Patients in Al Buraimi Hospital, Sultanate of Oman: A Cross-Sectional Study. Cureus. 2023;15(2):e35228.
Crossref - Rao SN, Manissero D, Steele V R, Pareja J. A Systematic Review of the Clinical Utility of Cycle Threshold Values in the Context of COVID-19. Infect Dis Ther. 2020;9(3):573-586.
Crossref - Yang W, Cao Q, Qin Le, et al. Clinical characteristics and imaging manifestations of the 2019 novel coronavirus disease (COVID-19):A multi-center study in Wenzhou city, Zhejiang, China. J Infect. 2020;80(4):388-393.
Crossref - Rousan LA, Elobeid E, Karrar M, Khader Y. Chest x-ray findings and temporal lung changes in patients with COVID-19 pneumonia. BMC Pulm Med. 2020;20(1):245.
Crossref - Bairwa M, Kumar R, Beniwal K, Kalita DJ, Bhaurupi Y. Hematological profile and biochemical markers of COVID-19 non-survivors: A retrospective analysis. Clin Epidemiol Glob Health. 2021;11;100770.
Crossref - Huang D, Yang H, Yu H, et al. Diagnostic Value of Hematological and Biochemical Parameters Combinations for Predicting Coronavirus Disease 2019 (COVID-19) in Suspected Patients. Am J Med Sci. 2021;362(4):387-395.
Crossref - Fu Y, Cheng Y, Wu Y. Understanding SARS-CoV-2-Mediated Inflammatory Responses: From Mechanisms to Potential Therapeutic Tools. Virol Sin. 2020;35(3):266-271.
Crossref - Wang Q, Zhao H, Liu L, et al. Characteristics and change patterns of liver function in 105 hosputalised adults patients with COVID-19 in Beijing, China. Research Square. 2020;10:21203.
Crossref - Eljilany I, Elzouki AN. D-Dimer, Fibrinogen, and IL-6 in COVID-19 Patients with Suspected Venous Thromboembolism: A Narrative Review. Vasc Health Risk Manag. 2020;16:455-462.
Crossref - Tang N, Li D, Wang X, Sun Z. Abnormal coagulation parameters are associated with poor prognosis in patients with novel coronavirus pneumonia. Journal of Thromb Haemost. 2020;18(4):844-847.
Crossref - Mardani R, Vasmehjani AA, Zali F, et al. Laboratory parameters in detection of COVID-19 patients with positive Rt-PCR; a diagnostic accuracy study. Arch Acad Emerg Med. 2020;8(1):e43.
- Yagci AK, Sarinoglua RC, Bilgin H, et al. Relationship of the cycle threshold values of SARS-CoV2 polymerase chain reaction and total severity score of computerized tomography in patients with COVID 19. Int J Inf Dis. 2020;101:160-166.
Crossref - Khalid A, Jaffar MJ, Khan T, et al. Hematological and biochemical parameters as diagnostic and prognostic markers in SARS-CoV2 infected patients of Pakistan: a retrospective comparative analysis. Hematology. 2021;26(1):529-542.
Crossref - Case study:CBC& CRP results of a critically ill COVID-19 patient. 2020. https://www.mindray.com/en/covid-19-response/case-study-cbc-crp-results-of-a-critically-ill-covid-19-patient.
© The Author(s) 2024. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.