ISSN: 0973-7510
E-ISSN: 2581-690X
Diseases caused by multidrug-resistant (MDR) bacteria continue to challenge physicians and endanger their patients’ lives. Polymyxins, including colistin, are the last resort antibiotics to treat serious infections caused by carbapenem-resistant bacteria. The aim of this study is to explore the resistance of Gram negative isolates recovered from 200 clinical specimens to carbapenem and colistin antibiotics, and the prevalence of plasmid-mediated mcr-1 gene in the resistant isolates. Clinical specimens were collected from two teaching hospitals and two private clinical laboratories in Cairo, Egypt. We identified one hundred and thirty isolates as Gram negative. These isolates were screened for their susceptibility to b–lactams antibiotics, carbapenems, colistin, polymyxin B, levofloxacin and amikacin. Thirty isolates were found to be resistant to the tested carbapenems. Of these, five isolates were found to be resistant to both carbapenem and colistin. They were tested for the presence of mcr-1, pmrB and pmrA genes; known to be among the reasons for colistin resistance. One isolate showed the presence of pmrA while three isolates showed the presence of pmrA and pmrB. Only one isolate showed the presence of mcr-1, pmrA and pmrB. This was tested by real time PCR to ensure the activity of this plasmid-mediated gene. Using 16S rRNA sequencing, the isolate showed 100% similarity to Escherichia coli strain K12 (MG1655). Here, we report a carbapenem-resistant and colistin-resistant Escherichia coli strain producing mcr-1gene that is the first to be reported in Egypt between human.
carbapenem resistance, colistin resistance, mcr-1, pmrB, pmrA.
Carbapenem-resistant parts of the family Enterobacteriaceae have disseminated all over the world1,2. Carbapenem-resistant Enterobacteriaceae are coresistant for many classes of antimicrobial agents3. This means poor treatment outcomes can be a considerable threat to public health. Colistin has been recently considered clinical interest, due to being frequently successful in treating infections caused by multidrug resistant (MDR) Pseudomonas, Escherichia and Klebsiella pathogens4. It should be administered intravenously and by inhalation in combination with other drugs to manage infections with Pseudomonas aeruginosa in children and adult patients with cystic fibrosis5. Colistin is a polypeptide anti-microbial which acts through electrostatic interactions. Its cationic moieties disturb the bacterial external membrane, leading to leakage of cell contents and cellular death6. Resistance for Colistin generally due to inefficient binding of polymyxins to the lipid A group of lipopolysaccharide. This can be basically due to the 4’-phosphoethanolamine (PEA) modification of the lipid A on the LPS7,8. This type of chemical modification on the bacterial lipid A can be ascribed to either the chromosome as in Klebsiella pneumonia [9], or the plasmid-transferred mobilized colistin resistance (mcr-1) gene in Enterobacteriaceae like Escherichia coli 10. In previous years, scientists have recorded infections that showed resistance to both carbapenems and colistin-resistant globally as in, Greece 11 and Israel 12. Plasmid-mediated colistin resistance encoded by mcr-1 was discovered in China 10. Reports followed in several nations, among which France13, Denmark14, Venezuela15 and USA16. Recently, Wang and his coworkers at 2018 assembled the largest data set of mcr-1– positive sequenced isolates17. They found that the nations having the largest numbers of mcr-1– positive samples were China, Vietnam and Germany17. Interestingly, more mcr genes have been recently reported to be included in colistin resistance such as mcr-2 and mcr-4 18,19.
In this study, the author screened two hundred isolates, recovered from clinical specimens in Egypt, for susceptibility to selected carbapenems and colistin. Further, the mechanism of resistance was explored.
Isolation and biochemical identification of bacteria
Two hundred clinical specimens were collected from two teaching hospitals (Sayed Jalal and Al Hussein hospitals) and two private clinical laboratories in Cairo, Egypt. These were isolated from urine samples, sputum, pleural fluid, endotracheal secretion and swaps. The samples were collected at November 2016- October 2017. Isolates were collected from patients admitted in ICU,PICU as well as outpatients. Their ages ranged from 6 – 50 years. They suffered from respiratory infection, UTI or Meningitis. Gram negative isolates were identified using standard microbiological tests; Gram stain, motility test, catalase test, triple sugar iron, citrate agar test, oxidase test and witek test.
Antimicrobial susceptibility testing
The antibiotic susceptibility testing was performed by Kirby Bauer disk diffusion method7. Gram negative isolates were screened for susceptibility to six b-lactam antibiotics; meropenem, imipenem, amoxicillin-clavulinic, cefotaxime, amoxicillin and ceftazidime. Resistant isolates were screened for susceptibility to polymyxinB, colistin, amikacin and levofloxacin.
DNA extraction and manipulation
DNA extraction for double resistant (Colistin/Carbapenem) isolates consisted of boiling lysates prepared from the strains; a loopful of culture was suspended in 1 mL of sterile PBS buffer, centrifuged 5 min at 20000 g and the supernatant rejected. Then, the pellet was re-suspended in 100 µL TE buffer (10:1) and boiled for 10 min at 100÷C. For use as template in the polymerase chain reaction, this DNA was further diluted at 1:10 in TE buffer. The DNA extract was checked by running on 0.8 % agarose gel ,stained with ethidium bromide (Thermo Fischer Scientific, USA) using gel electrophoresis (Labnet, USA). The gel was illuminated under Whitman U.V Trans-illuminator (Biometra, Germany) according to Sambrook et al. 20.
PCR amplification of specific sequences of some genes encoding for colistin and polymyxin resistance
Three genes contributing to colistin and polymyxin resistance were selected in this study and amplified by PCR; mcr-1 primers {( CLRr/CLRf (5’CGGTCAGTCCGTTTGTTC’3) CLR/r (5’CTTGGTCGGTCTGTAGGG’3) 94÷C 3min+ 25X(94÷C 30 sec+ 58÷C 90 sec+ 72÷C 60 sec) + 72÷C 10min}10, pmrA primers LT2PMRA/REV{( F (5’CAT TTC CGC GCA CTG TCT GC’3)R (5’CAG CTT TCA GTT GCA AAC AG’3) 95°C for 10s,52°Cfor 5s, 72°C for 5s} and pmrB primers 1519/1520{( F (5’ACC TAC GCG AAA AGA TTG GC’3)R (5’GAT GAG GAT AGC GCC CAT GC’3) 95°C for 30s ,62°C for 30s,72°C for 180s)} 21. Amplification was performed using thermocycler in total volume of 25µL containing 1 µL DNA extract, 1µL of forward primer (10 µM), 1µL reverse primer (10 µM), 12.5µL tag master mix (Qiagen, Germany) and 9.5µL of nuclease-free water.
Amplification using Gene-AMP-PCR-system-9700 thermocycler (Applied bio system). After amplification, 10µL of the PCR mixture was analyzed by gel electrophoresis.(1.5% agarose in tris-EDTA stained with ethidium bromide). . As mcr-1 mostly carried on plasmid and pmr-A, pmr-B are chromosomal genes so mcr-1 dissemination is highly predicted that lead to increase occurrence of colistin resistance globally, so SYBR Green-based real-time PCR assay was designed for specific detection of the mcr-1 gene. Cell lysates (3 µL), prepared as described above, were used as DNA template, MCR -F: 5’-ACGGCGTATTCTGTGCCGTGTAT-3’ and MCR -R 5’- GCTGTTCTTTTGGTGCAAAGGCATTT-3’ were used as primers for PCR analysis of mcr-1real-time PCR were performed using QuantiNova SYBR Green mixture and Qiagen Rotor Gene system (Qiagen, Hilden, Germany). A typical 20 µL of PCR reaction includes 0.7 µM of each primer, 3 µL of lysate and SYBR Green mixture (1x). The PCR conditions used are: 95°C, 10 minute; 30 cycles including 95°C, 15 seconds, 60°C, 1 min; and 72°C, 30 seconds.
Confirmation of the identity of isolate harboring mcr-1 by 16S rRNA gene sequence analysis
Identification was confirmed by sequencing of partially amplified 16S rRNA gene of the bacterial isolate that harbored mcr-1 gene. Molecular characterization of the selected strain was carried out according to modified Sambrook et al. 20. The 16S rRNA gene was amplified using universal primers pair: the forward primer sequence was 5-AGAGTTTGATCATGGCTCAG-3 and the reverse primer sequence was 5-ACGAGCTGACGACAGCCATG-3 (IDT, USA). PCR was carried out in a total volume of 50 µL using 25 µL of My Taq PCR Master Mix (2X) (Bio line, Germany), containing 1 µL (10 µM) of each forward and reverse primers together with equivalent micro liters containing 50 µg of the genomic DNA and finally completed to 50 µL by nuclease free water. Thermo cycling was carried out using thermo cycler (Primus 25 Advanced, Germany). It was carried out as follow: first step was denaturation for 3 minutes at 94°C, followed by 40 cycles of denaturation at 94°C for 30 sec, annealing at 50°C C for 30 sec and extension at 72°C for 2 min. Finally, extension was done at 72°C for 10 min. The amplified sequence was verified by gel electrophoresis. PCR products were purified by Gene Jet Gel Extraction Kit (Thermo Fisher Scientific, USA). The purified products were subjected to sequencing. Nucleotide sequence similarities were determined using other known sequences found in the GenBank database using BLAST program of National Center for Biotechnology Information (NCBI) databases.
Isolation and identification of bacteria
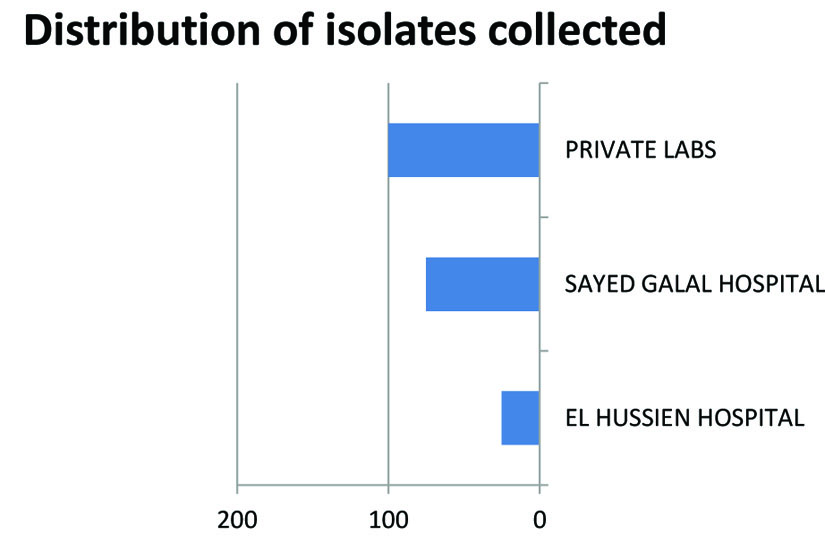
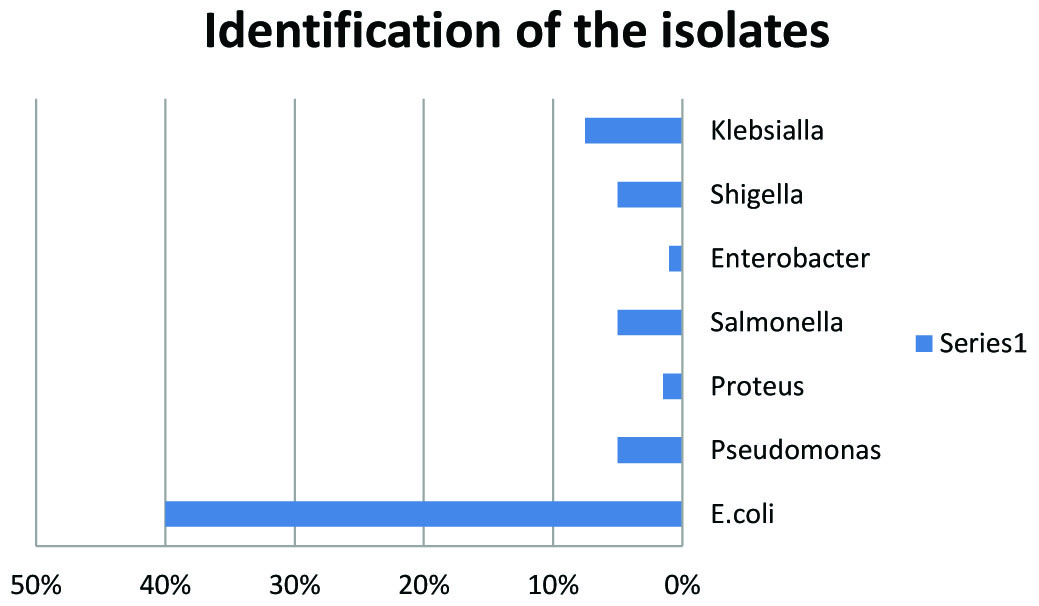
Of the two hundred isolates collected from clinical specimens of various sources distributed (Fig.1). One hundred and thirty isolates were identified as Gram negative bacteria, as presented in Fig.2. The other isolates were excluded from this study.
Antimicrobial susceptibility testing
A hundred and thirty Gram negative isolates were tested for susceptibility to six B-lactam antibiotics. Results were interpreted as resistant (R), intermediate (I) or susceptible (S) based on the criteria listed in CLSI (2016). Thirty isolates were found to be resistant to the tested carpabenems meropenem and imipenem. These were tested for susceptibility to colistin in addition to polymyxin B, levofloxacin and amikacin. As demonstrated in Table (1), five isolates were found to be resistant to both carpabenems and colistin. Of the latter isolates, three were susceptible to levofloxacin, while two isolates were resistant to all antibiotics used in this study, and only one sample was harboring the mcr-1gene. The identity of the mcr-1 harboring double resistant (Colistin/Carbapenem) bacterium was confirmed by 16SRNA genetic analysis.
Table (1):
Susceptibility of selected antimicrobials against carpabenems-resistant Gram negative isolates collected in this study. IMP: Imipenem, MEM: Meropenem ,CT: Colistin, AK: Amikacin, PB: Polymyxin and LEV: Levofloxacin. The antibiotic susceptibility testing was according to Kirby Bauer disk diffusion method.
Isolate No |
IMP |
MEM |
CT |
AK |
PB |
LEV |
|---|---|---|---|---|---|---|
44 |
R |
R |
R |
R |
R |
R |
46 |
R |
R |
S |
S |
S |
S |
50 |
R |
R |
S |
S |
S |
R |
48 |
R |
R |
S |
S |
S |
S |
109 |
R |
R |
S |
S |
R |
S |
77 |
R |
R |
S |
S |
S |
R |
114 |
R |
R |
S |
S |
S |
S |
42 |
R |
R |
S |
R |
S |
R |
35 |
R |
R |
R |
R |
R |
S |
128 |
R |
R |
S |
R |
S |
S |
33 |
R |
R |
R |
R |
R |
S |
2 |
R |
R |
R |
R |
R |
S |
39 |
R |
R |
R |
R |
R |
R |
31 |
I |
R |
S |
S |
I |
S |
121 |
I |
R |
S |
S |
S |
I |
13 |
R |
R |
S |
S |
S |
S |
120 |
S |
R |
S |
I |
S |
I |
41 |
I |
R |
S |
I |
S |
S |
90 |
R |
I |
S |
R |
S |
R |
34 |
R |
S |
S |
R |
S |
S |
101 |
R |
S |
S |
R |
S |
I |
36 |
R |
S |
S |
S |
S |
S |
65 |
I |
R |
S |
R |
S |
I |
21 |
R |
I |
S |
I |
S |
S |
24 |
R |
S |
S |
R |
S |
I |
83 |
R |
I |
S |
R |
S |
S |
11 |
R |
I |
S |
R |
S |
S |
27 |
R |
I |
S |
S |
S |
I |
95 |
I |
R |
S |
R |
S |
R |
97 |
I |
R |
S |
R |
R |
R |
PCR screening of selected genes associated with colistin resistance
The five double-resistant isolates (Carbapenem/ Colistin) showed variable patterns upon analysis using specific primers of mcr-1, pmrA and pmrB genes. One isolate showed the presence of pmrA, while three isolates showed the presence of pmrA and pmrB. Only one isolate (isolate 35) showed the presence of mcr-1(300bp), pmrA(170bp) and pmrB(400bp). Fig. 3. Identification of mcr-1 carrying isolate by 16S rRNA
The PCR product of 16S rRNA of the isolate which has showed mcr-1 was confirmed by sequencing. Using BLAST program of National Center for Biotechnology Information (NCBI) database the sequence showed 100% similarity to Escherichia coli strain K-12 substr. MG1655.
Confirmation of mcr-1 gene expression
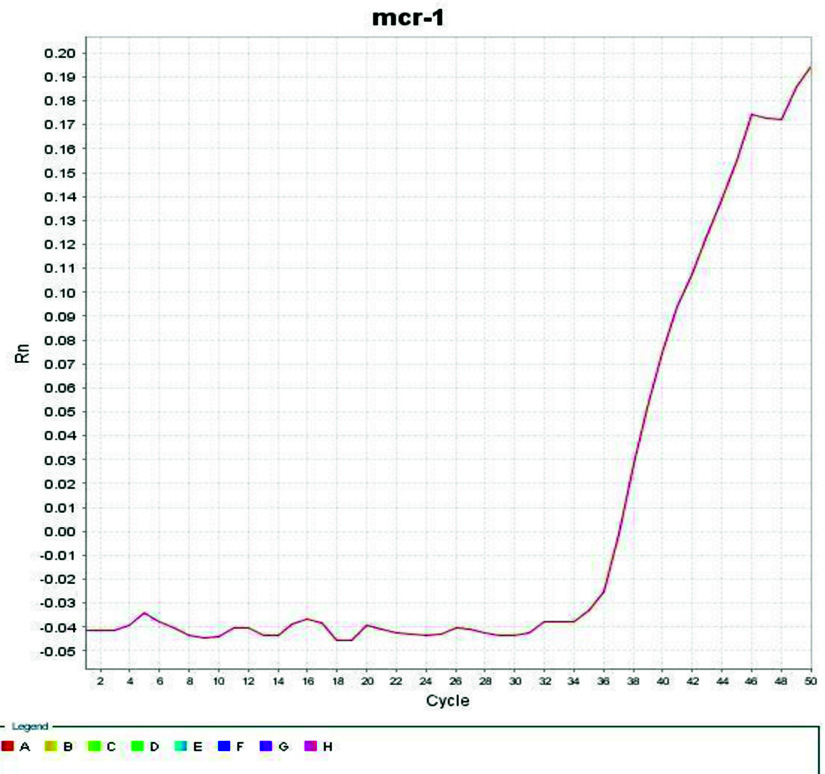
A SYBR Green-based real-time PCR assay was designed for detection of the mcr-1 gene expression in the isolate. The experiment showed positive mcr-1 gene expression and show CT point after 36 cycles as illustrated at Fig. 4.
 Fig. 3. ) Using 1000 pb ladder, isolate no 35 shows the presence of mcr-1(300bp), pmrA(170bp) and pmrB(400bp). In figure 3(a), primer 1519/1520 was used to detect the presence of pmr-B gene. In figure 3(b), primers LT2PMRA/REV and CLRr/CLRf were used to detect the presence of pmrA and mcr-1, respectively.
Fig. 3. ) Using 1000 pb ladder, isolate no 35 shows the presence of mcr-1(300bp), pmrA(170bp) and pmrB(400bp). In figure 3(a), primer 1519/1520 was used to detect the presence of pmr-B gene. In figure 3(b), primers LT2PMRA/REV and CLRr/CLRf were used to detect the presence of pmrA and mcr-1, respectively. The intense overuse and misuse of antibiotics has led to emergence of multi-drug resistant strains which is difficult to treat. In this study, approximately a quarter of the tested isolates were found to be carbapenems resistant, while five isolates were resistant to both carbapenems and colistin. Only one of them was confirmed to harbor mcr-1 and was identified to be Escherichia coli strain k12 substr MG1655. The results obtained from the real-time PCR showed obvious presence of mcr-1 which was also confirmed by conventional PCR, suggesting a probability that mcr genes may be currently prevalent in EGYPT. This study is the first to detect a multi-resistant Escherichia coli strain which is resistant to both carbapenem and colistin in Egypt. A previous work has reported an mcr-1 mediated Escherichia coli isolate detected in Egypt22. This had phenotypic resistance to colistin, but was susceptible to carbapenems22.Worthy to note, the SENTRY antimicrobial surveillance program carried out a worldwide survey in 2009 and reported that rates of resistance to polymyxins among Gram-negative pathogens ranged between 0.1% to 1.5% 23. However, a trend of rise of resistance followed worldwide in later years to reach 3.2%in 201624.
This study showed that the carbapenem-colistin double resistant Escherichia coli was sensitive to levofloxacin. Therefore, colistin might be combined with levofloxacin in treatment regimen, as it seems to produce a synergistic or additive effect. The use of colistin in combination with other antibiotics that are typically active against Gram-positive bacteria has been also explored by others and confirmed to be effective for highly drug-resistant Gram-negative pathogens expressing mcr-1. Most commonly rifampicin25 and carbapenems26 but also macrolides27, minocycline28, tigecycline29, and glycopeptides30.
The coexistence of mcr-1 resistance in the carbapenem-resistant strains might be explained by the fact that mcr-1 has been observed on plasmids containing other antimicrobial resistance genes such as carbapenemases31,32 and extended-spectrum b-lactamases13,33. This suggests is a significant clinical concern that world is approaching a pan drug- resistant era for which the use of colistin and other antimicrobials might become ineffective.
The spread of carbapenem-resistant Enterobacteriaceae has become a serious problem worldwide that represents a marked threat to public health. To the best of our knowledge, this study constitutes the first to report in Egypt of mcr-1 positive Escherichia coli that is carbapenem-resistant and colistin-resistant. That gives an alarming sign for the dissemination of resistance in the area, which can considerably limits treatment options. Our finding from this study stresses how important to monitor the use of colistin in treatment of both human and animal infections and emphasizes the need for tightened infection control practices to restrict further dissemination of multi-resistant isolates.
Acknowledgments
Thanks to departments of Microbiology and Immunology, Faculty of Pharmacy, Cairo University and Beni-Suef University for their rich educational experience, cooperation and continuous support.
Conflict Of Interest
The authors declare that there is no conflict of interest.
Authors’ Contribution
All authors have made substantial, direct and intellectual contribution to the work and approved it for publication.
Funding
None
Data Availability
All datasets generated or analyzed during this study are included in the manuscript.
Ethics Statement
This article does not contain any studies with human participants or animals performed by any of the authors.
- Antoniadou, A., F. Kontopidou, G. Poulakou, E. Koratzanis, I. Galani, E. Papadomichelakis, H. Giamarellou. Colistin-resistant isolates of Klebsiella pneumoniae emerging in intensive care unit patients: first report of a multiclonal cluster. J Antimicrob Chemother, 2007; 59(4):786-790.
- Kitchel, B., J.K. Rasheed, J.B. Patel, A. Srinivasan, S. Navon-Venezia, Y. Carmeli, C.G. Giske. Molecular epidemiology of KPC-producing Klebsiella pneumoniae isolates in the United States: clonal expansion of multilocus sequence type 258. Am J Clin Pathol, 2009; 53(8):3365-3370.
- Schwaber, M.J., S. Klarfeld-Lidji, S. Navon-Venezia, D. Schwartz, A. Leavitt, and Y. Carmeli. Predictors of carbapenem-resistant Klebsiella pneumoniae acquisition among hospitalized adults and effect of acquisition on mortality. Antimicrob Agents Chemother, 2008; 52(3):1028-1033.
- Tangden, T. and C. Giske. Global dissemination of extensively drug resistant carbapenemase producing Enterobacteriaceae: clinical perspectives on detection, treatment and infection control. J Intern Med, 2015; 277(5):501-512.
- Herrmann, G., L. Yang, H. Wu, Z. Song, H. Wang, N. Hoiby, G. Doring. Colistin-tobramycin combinations are superior to monotherapy concerning the killing of biofilm Pseudomonas aeruginosa. J Infect Dis, 2010; 202(10):1585-1592.
- Davis, S.D., A. Iannetta, and R.J. Wedgwood. Activity of colistin against Pseudomonas aeruginosa: inhibition by calcium. J Infect Dis, 1971; 124(6):610-612.
- Bauer, A., W. Kirby, J.C. Sherris, and M. Turck. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol, 1966; 45(4):493.
- Schwarz, S. and A.P. Johnson. Transferable resistance to colistin: a new but old threat. J Antimicrob Chemother, 2016; 71(8):2066-2070.
- Cannatelli, A., M.M. D’Andrea, T. Giani, V. Di Pilato, F. Arena, S. Ambretti, G.M. Rossolini. In vivo emergence of colistin resistance in Klebsiella pneumoniae producing KPC-type carbapenemases mediated by insertional inactivation of the PhoQ/PhoP mgrB regulator. Antimicrob Agents Chemother, 2013; 57(11):5521-5526.
- Liu, Y.Y., Y. Wang, T.R. Walsh, L.X. Yi, R. Zhang, J. Spencer, X. Huang. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis, 2016; 16(2):161-168.
- Zarkotou, O., S. Pournaras, E. Voulgari, G. Chrysos, A. Prekates, D. Voutsinas, A. Tsakris. Risk factors and outcomes associated with acquisition of colistin-resistant KPC-producing Klebsiella pneumoniae: a matched case-control study. J Clin Microbiol, 2010; 48(6):2271-2274.
- Samra, Z., O. Ofir, Y. Lishtzinsky, L. Madar-Shapiro, and J. Bishara. Outbreak of carbapenem-resistant Klebsiella pneumoniae producing KPC-3 in a tertiary medical centre in Israel. Int J Antimicrob Agents, 2007; 30(6):525-529.
- Haenni, M., L. Poirel, N. Kieffer, P. Chatre, E. Saras, V. Metayer, J.Y. Madec. Co-occurrence of extended spectrum b lactamase and MCR-1 encoding genes on plasmids. Lancet Infect Dis, 2016; 16(3):281-282.
- Hasman, H., A.M. Hammerum, F. Hansen, R.S. Hendriksen, B. Olesen, Y. Agersר, R.S. Kaas. Detection of mcr-1 encoding plasmid-mediated colistin-resistant Escherichia coli isolates from human bloodstream infection and imported chicken meat, Denmark 2015. Euro Surveill, 2015; 20(49).
- Delgado-Blas, J.F., C.M. Ovejero, L. Abadia-Patiסo, and B. Gonzalez-Zorn. Coexistence of mcr-1 and blaNDM-1 in Escherichia coli from Venezuela. Antimicrob Agents Chemother, 2016; 60(10):6356-6358.
- Mediavilla, J.R., A. Patrawalla, L. Chen, K.D. Chavda, B. Mathema, C. Vinnard, B.N. Kreiswirth. Colistin-and carbapenem-resistant Escherichia coli harboring mcr-1 and blaNDM-5, causing a complicated urinary tract infection in a patient from the United States. MBio, 2016; 7(4):e01191-16.
- Wang, R., L. Dorp, L.P. Shaw, P. Bradley, Q. Wang, X. Wang, A. Rieux. The global distribution and spread of the mobilized colistin resistance gene mcr-1. Nat Commun, 2018; 9(1):1179.
- Carattoli, A., L. Villa, C. Feudi, L. Curcio, S. Orsini, A. Luppi, C.F. Magistrali. Novel plasmid-mediated colistin resistance mcr-4 gene in Salmonella and Escherichia coli, Italy 2013, Spain and Belgium, 2015 to 2016. Euro Surveill, 2017; 22(31)
- Zhang, J., L. Chen, J. Wang, A.K. Yassin, P. Butaye, P. Kelly, M. Li. Molecular detection of colistin resistance genes (mcr-1, mcr-2 and mcr-3) in nasal/oropharyngeal and anal/cloacal swabs from pigs and poultry. Scientific reports, 2018; 8(1):3705.
- Maniatis, T., E.F. Fritsch, and J. Sambrook, Molecular cloning: a laboratory manual. Vol. 545. 1982: Cold Spring harbor laboratory Cold Spring Harbor, NY.
- Jayol, A., L. Poirel, A. Brink, M.-V. Villegas, M. Yilmaz, and P. Nordmann. Resistance to colistin associated with a single amino acid change in protein PmrB among Klebsiella pneumoniae isolates of worldwide origin. Antimicrob Agents Chemother, 2014; 58(8):4762-4766.
- Elnahriry, S.S., H.O. Khalifa, A.M. Soliman, A.M. Ahmed, A.M. Hussein, T. Shimamoto, and T. Shimamoto. Emergence of plasmid-mediated colistin resistance gene mcr-1 in a clinical Escherichia coli isolate from Egypt. Antimicrob Agents Chemother, 2016; 60(5):3249-3250.
- Gales, A.C., R.N. Jones, and H.S. Sader. Contemporary activity of colistin and polymyxin B against a worldwide collection of Gram-negative pathogens: results from the SENTRY Antimicrobial Surveillance Program (2006–09). J Antimicrob Chemother, 2011; 66(9):2070-2074.
- Wise, M.G., M.A. Estabrook, D.F. Sahm, G.G. Stone, and K.M. Kazmierczak. Prevalence of mcr-type genes among colistin-resistant Enterobacteriaceae collected in 2014-2016 as part of the INFORM global surveillance program. PloS one, 2018; 13(4):e0195281.
- Hogg, G., J. Barr, and C. Webb. In-vitro activity of the combination of colistin and rifampicin against multidrug-resistant strains of Acinetobacter baumannii. J Antimicrob Chemother, 1998; 41(4):494-495.
- Zusman, O., T. Avni, L. Leibovici, A. Adler, L. Friberg, T. Stergiopoulou, M. Paul. Systematic review and meta-analysis of in vitro synergy of polymyxins and carbapenems. Antimicrob Agents Chemother, 2013; 57(10):5104-5111.
- Timurkaynak, F., F. Can, OK. Azap, M. Demirbilek, H. Arslan, and S. Karaman. In vitro activities of non-traditional antimicrobials alone or in combination against multidrug-resistant strains of Pseudomonas aeruginosa and Acinetobacter baumannii isolated from intensive care units. Int J Antimicrob Agents, 2006; 27(3):224-228.
- Chopra, I. and K. Hacker. Uptake of minocycline by Escherichia coli. J Antimicrob Chemother, 1992; 29(1):19-25.
- Pournaras, S., G. Vrioni, E. Neou, J. Dendrinos, E. Dimitroulia, A. Poulou, and A. Tsakris. Activity of tigecycline alone and in combination with colistin and meropenem against Klebsiella pneumoniae carbapenemase (KPC)-producing Enterobacteriaceae strains by time–kill assay. Int J Antimicrob Agents, 2011; 37(3):244-247.
- Gordon, N., K. Png, and D. Wareham. Potent synergy and sustained bactericidal activity of a vancomycin-colistin combination versus multidrug-resistant strains of Acinetobacter baumannii. Antimicrob Agents Chemother, 2010; 54(12):5316-5322.
- Poirel, L., N. Kieffer, N. Liassine, D. Thanh, and P. Nordmann. Plasmid-mediated carbapenem and colistin resistance in a clinical isolate of Escherichia coli. Lancet Infect Dis, 2016; 16(281):00006-2.
- Yao, X., Y. Doi, L. Zeng, L. Lv, and J.-H. Liu. Carbapenem-resistant and colistin-resistant Escherichia coli co-producing NDM-9 and MCR-1. Lancet Infect Dis, 2016; 16(3):288-289.
- Zhang, H., C.H. Seward, Z. Wu, H. Ye, and Y. Feng. Genomic insights into the ESBL and MCR-1-producing ST648 Escherichia coli with multi-drug resistance. Sci Bull, 2016; 61(11):875-878.
© The Author(s) 2019. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.