ISSN: 0973-7510
E-ISSN: 2581-690X
This study was conducted to isolate and identify bacteria from the wastewater of the textile printing industry inoculated with a mixture of cow dung, jaggery, and urea to assess the bioremediation potential of isolated bacterial species for reducing color and other polluting parameters of the wastewater. Based on colony characteristics, we were able to isolate nine types of bacteria (Pri 1 to 9), capable of thriving in textile printing wastewater. Out of the nine isolates examined, four (Pri 3, Pri 4, Pri 8, and Pri 9) significantly reduced the color and values of other physicochemical parameters of the wastewater. Isolated bacterial cultures were identified using various biochemical tests, antibiotic sensitivity tests, and rRNA sequence analysis. At the end of a 24-h incubation period at room temperature under shaking conditions at 100 rpm on an orbital shaker, isolate Pri 3 was identified as Alcaligenes aquatilis LMG 22996 (T), capable of reducing color by 86.13%, biological oxygen demand (BOD) by 70.44%, chemical oxygen demand (COD) by 80.65%, total dissolved solids (TDS) by 47.31%, total suspended solids (TSS) by 56.56%, and ammoniacal nitrogen by 75.95%; isolate Pri 4 as Priestia aryabhattai B8W22(T), capable of reducing color by 78.35%, BOD by 66.35%, COD by 74.92%, TDS by 34.94%, TSS by 31.66%, and ammoniacal nitrogen by 71.14%; isolate Pri 8 as Citrobacter werkmanii NBRC 105721(T), capable of reducing color by 90.37%, BOD by 82.13%, COD by 85.06%, TDS by 54.83%, TSS by 61.97%, and ammoniacal nitrogen by 80.76%; and isolate Pri 9 as Shewanella chilikensis JC5(T), capable of reducing color by 90.17%, BOD by 84.68%, COD by 83.46%, TDS by 59.13%, TSS by 62.45%, and ammoniacal nitrogen by 90.37%.
Textile Printing Wastewater, Cow Dung, Bacterial Isolates, Bioremediation, Physicochemical Parameters, Antibiotic Sensitivity, 16S rRNA Sequencing
Since the existence of mankind, human beings have used colors in their routine lifestyles, which were traditionally derived from natural materials.1 However, with the increase in population and industrial development, these natural materials are being replaced by synthetic colors and dyes. Currently, more than 100,000 types of synthetic dyes are being used across various industrial sectors.2 Among these sectors, the textile industry stands out as the largest consumer of synthetic dyes and chemicals in its manufacturing process. Textile manufacturing depends on the freshwater requirements, and approximately 20–25% of the used dyes and chemicals return as wastewater from these industries.3 Many of the textile dyes inhibit oxygen transfer in the generated wastewater, thereby increasing both biological and chemical oxygen demands.4 This may lead to toxicity for aquatic life, and continuous exposure to such dyes might pose health risks to humans.5,6 Consequently, proper treatment of industrial wastewater before discharge into the natural environment becomes crucial. Previously, treatment processes relied on chemical and physical methods,7 which helped in color removal of the dyes to some extent. However, these methods often generate undesirable by-products, such as aromatic amines, which are challenging to maintain,8 Instead, the biological procedure for the removal of such compounds from the wastewater is cheaper due to the availability of the vast array of microbes in the natural environment and their diverse metabolic pathways that remove or convert complex forms of pollutants to simpler forms.9,10 This can be achieved through biosorption, biodegradation, bioaccumulation, and biostimulation.11 As the bacterial system has diverse metabolic pathways, numerous innovations have been conducted in textile wastewater treatment. Various bacterial genera, including Bacillus sp. ADR, ETL-2012, SF, VUS, UN2,12–22 Caulobacter subvibrioides;23 Enterobacter aerogenes and its spp. SXCR, CV–S1;24-26 Klebsiella (ST16.16/034), (facultative) strain VN-31, strain Y3, DA26;11,27,28 Moraxella osloensis;29,30 Paenibacillus alvei MTCC 10625 and P. azoreducens;7,31 Pseudomonas spp. SU-EBT, LBC1, SUK1;32-34 Pseudomonas luteola;35-37 Rhodobacter erythropholis MTCC 4688;38 and Stenotrophomonas spp. BHUSSp X2,12,39 have been utilized in the treatment process.
The success of microbial wastewater treatment highly depends on the isolation of microorganisms from the polluted sample, such as textile wastewater6,26,40-44 textile effluent-contaminated soil,19,43,45 and activated sludge.46 In this study, we isolated bacteria from textile printing wastewater inoculated with a mixture of cow dung, jaggery, and urea. The efficiency of different types of bacteria naturally present in manmade polluted samples such as textile printing wastewater, and those present in natural samples such as cow dung that survived in textile printing wastewater was evaluated for their capability to reduce color and values of other physicochemical parameters of the wastewater.
Collection of textile printing wastewater
Wastewater was collected from the effluent treatment plant of the textile printing industry, Western Overseas, situated at 21°43’25.20″ N and longitude 70°34’10.27″ E, in the village of Jetalsar, Jetpur, Rajkot, Gujarat, India. The sample was collected in a 10-L sterile plastic container and preserved at 4°C in the refrigerator until examination.
Enrichment and isolation of bacteria
Bacteria are present in all ecosystems and are part of all living systems on the Earth.47 Typically, bacteria capable of dye decolorization and degradation are isolated from textile wastewater, textile effluent-contaminated soil, and activated sludge. In this study, we prepared a cocktail of textile printing wastewater, cow dung, jaggery, and urea for isolating bacteria suitable for the bioremediation of pollution caused by the colored effluent.
To obtain a diverse range of bacterial systems, enrichment of bacteria is essential before the isolation process. In 500 L of fresh water, a mixture of 2 kg cow dung, 250 g jaggery, and 100 g urea was prepared and aerated for 48 h using a Nanobubble pump (Century Pump, Udhyog Nagar, Rajkot, India), producing bubbles that can survive in water for up to 60 min or more.48 Subsequently, 250 L of this mixture was combined with 250 L of textile printing wastewater, supplemented with 2 kg cow dung, 250 g jaggery, and 100 g urea. Aeration with the Nanobubble pump was continued for 24 h, and this cycle was repeated for 7 days. This method enriched only those bacteria capable of surviving in textile printing wastewater. The microbially enriched textile printing wastewater was diluted 100 times in sterile distilled water, streaked aseptically onto nutrient agar plates (HiMedia, Mumbai, India) prepared in sterile textile printing wastewater, and incubated at 37°C for 24 h. After incubation, numerous morphologically distinct bacterial colonies were observed on the nutrient agar plates. Nine morphologically different colonies were selected, maintained by subculturing onto nutrient agar slants, and stored in a refrigerator at 4°C.
Study of bacterial growth in textile printing wastewater
For the examination of bacterial growth, nutrient broth (HiMedia, Mumbai, India) was prepared directly in sterile textile printing wastewater. In a 250 mL Erlenmeyer flask, 100 mL of this nutrient broth was added and sterilized in an autoclave. A loopful of the pure culture of all nine selected bacterial isolates was individually inoculated into separate flasks and agitated at 100 rpm in an orbital shaker (Remi, Bharat) at room temperature for 10 h. Aseptically, 10 mL samples were harvested from each flask and collected in 12 mL sterile centrifuge tubes. The tubes containing isolates were centrifuged at 5000 rpm for 10 min using a laboratory centrifuge (Remi, India). The cell-free supernatants were discarded, and pellets of bacterial cells were resuspended in 10 mL distilled water. The absorbance was measured at l660 nm (Systronics 106, India) to estimate the growth.
Screening of bacterial isolates for their decolorization potential
The ability of all bacterial isolates to remove color from the effluent was assessed. To prepare an inoculum, a loopful of the pure culture of bacteria was transferred from a nutrient agar slant to a 250 mL sterile Erlenmeyer flask containing 100 mL nutrient broth (pH 7.0) prepared in sterile distilled water. These flasks were placed on an orbital shaker (Remi, Ahmedabad, India) at 100 rpm and room temperature for 8 h. Then, 1.0 mL of actively growing inoculum was transferred to another 250 mL sterile Erlenmeyer flask containing 100 mL nutrient broth prepared in the textile printing wastewater itself (pH 7.0). Similar flasks for all nine bacterial isolates were prepared and kept on an orbital shaker at 100 rpm and room temperature. Visual observations of reduction or change in color were recorded at 24 h and 48 h. At the end of this process, four out of nine isolates were identified as the most effective decolorizers of textile printing wastewater and were selected for further testing.
Reduction in color and values of other polluting parameters of textile printing wastewater by selected bacterial isolates
Four bacterial isolates were subjected to further testing to assess their efficacy in reducing color and the values of other polluting parameters of wastewater, including BOD, COD, TDS, TSS, and ammoniacal nitrogen. To conduct the experiment, 100 mL of textile printing wastewater was introduced into a 250 mL Erlenmeyer flask, and 0.1 g jaggery was added as a nutrient source. The final pH was adjusted to 7.0, and the flasks were sterilized in an autoclave. A control flask without inoculation served as a reference, while test flasks were inoculated with 1 mL of an actively growing bacterial isolate aseptically. Each bacterial isolate had a dedicated test flask, placed on an orbital shaker at 100 rpm and room temperature. Samples were collected from these flasks at 8 h, 16 h, 24 h, and 48 h and analyzed for color reduction (decolorization), BOD, COD, TDS, TSS, and ammoniacal nitrogen. Decolorization was measured using a UV-visible spectrophotometer (Elico, Model No. SL 159) at λ660 nm, while other parameters were measured following the standard methods for the examination of water and wastewater (APHA).49
Decolorization percent was calculated using equation (1):4
- % Decolorization = [ Initial absorbance – Final absorbance) / (Initial absorbance ] x100
Reduction in all other parameters, i.e., BOD, COD, TDS, TSS, and ammoniacal nitrogen, was calculated using the following equation (2):50
- % Reduction in the parameters = [ Initial result-Final result)/(Initial result] x100
Identification of selected bacteria
Morphological examinations
Morphological examination of bacteria involves observing colony characteristics and conducting microscopic examinations of cells. The colony characteristics of bacterial isolates were visually recorded, while cellular characteristics were documented through Gram and spore staining. Additionally, motility was observed through a hanging drop preparation of pure culture suspension.
Biochemical tests
By performing various biochemical tests, bacterial systems can be identified up to the genus level, and sometimes even to the species level.44 These tests were performed following the procedures outlined in Bergey’s Manual of Systematic Bacteriology. The identification process was conducted in VITEK-2 (BioMerieux USA), a fully automated microbial identification system.
Antibiotic sensitivity test
All four selected bacterial isolates underwent testing for their sensitivity to various antibiotics using the disk diffusion method.51 Mueller–Hinton agar plates (HiMedia, Mumbai, India) served as a nutrient medium, with 47 different antibiotic discs, each with a suggested dose for the tests. Antibiotics included amikacin, tobramycin, gentamicin, netilmicin, aztreonam, cefazolin, cefuroxime, ceftizoxime, cefpodoxime, cefixime, ceftazidime, cefepime, piperacillin + tazobactam, cefoperazone + tazobactam, cefoperazone + sulbactam, amoxicillin + clavulanic acid, trimethoprim/sulfamethoxazole, colistin, imipenem, meropenem, doripenem, tetracycline, doxycycline, minocycline, ciprofloxacin, levofloxacin, moxifloxacin, penicillin G, erythromycin, azithromycin, rifampicin, linezolid, vancomycin, and clindamycin. All plates were incubated at 37°C for 24 h to measure sensitivity.
16S rRNA sequencing
For the identification of bacterial species, 16S rRNA sequencing of the selected bacterial isolates was conducted at the National Center for Microbial Resources (NCMR), Pune, India.
Colony morphology of bacterial isolates
Colony morphology refers to the visible characteristics and appearance of bacterial colonies grown on solidified agar media. This information is valuable for understanding the bacterial species present and their growth patterns.52 The Detailed colony characteristics are mentioned in Table 1. The colonies of isolates Pri 1 to 8 exhibited a circular form, whereas those of isolate Pri 9 displayed an irregular form. Colony sizes ranged from 0.5 mm to 3.0 mm among all isolates. The elevation of colonies varied: isolates Pri 1, Pri 3, Pri 4, Pri 7, Pri 8, and Pri 9 had convex elevations; isolates Pri 2 and Pri 6 were umbonate; and isolate Pri 5 had a flat elevation. The margin of colonies showed filamentous characteristics for isolates Pri 5 and Pri 8, whereas that of other isolates had entire margins. The color of colonies differed: isolates Pri 1, Pri 2, Pri 3, and Pri 7 were white; Pri 9 was milky white; Pri 5 and Pri 8 were grey; Pri 4 was yellow; and Pri 6 was bluish white. The surface of the colonies also varied: Pri 1, Pri 2, Pri 3, and Pri 7 had smooth surfaces; Pri 4 and Pri 9 had smooth and shiny surfaces; Pri 5 and Pri 6 had rough and white surfaces; and Pri 8 had rough and shiny surfaces. Most colonies appeared opaque or slightly opaque, with the exception of isolate Pri 8, which produced transparent colonies. Based on these observed colony characteristics, all nine microbial isolates were primarily identified as bacteria.
Table (1):
Colony morphology of bacterial Isolates
Isolates |
Form |
Size (mm) |
Elevation |
Margin |
Surface |
Opacity |
Color |
|---|---|---|---|---|---|---|---|
Pri 1 |
Circular |
>0.5 |
Convex |
Entire |
Smooth |
Opaque |
White |
Pri 2 |
Circular |
<2.0 |
Umbonate |
Entire |
Smooth |
Opaque |
White |
Pri 3 |
Circular |
1.0–1.5 |
Convex |
Entire |
Smoot |
Slightly opaque |
White |
Pri 4 |
Circular |
1.5–1.8 |
Convex |
Entire |
Smooth and shiny |
Opaque |
Yellow |
Pri 5 |
Irregular |
<2.0 |
Flat |
Filamentous |
Rough |
Opaque |
Grey |
Pri 6 |
Circular |
1.5–1.7 |
Umbonate |
Entire |
Rough |
Opaque |
Bluish white |
Pri 7 |
Circular |
1.0–1.5 |
Convex |
Entire |
Smooth |
Slightly opaque |
White |
Pri 8 |
Circular |
2.0–3.0 |
Convex |
Filamentous |
Rough and shiny |
Transparent |
Grey |
Pri 9 |
Irregular |
1.2–1.8 |
Convex |
Entire |
Smooth and shiny |
Opaque |
Milky white |
Microscopic examination of bacterial isolates
Bacterial cells were examined under a compound light microscope to determine their size, shape, organization, motility, and other structural traits. Gram staining was employed to determine the composition of cell walls, and spore staining was employed to assess whether the bacteria possessed a survival mechanism under unfavorable environmental conditions. Additionally, motility observation was conducted to determine the motility of bacteria in the given medium. The Details of the staining and Motility test are mentioned in Table 2. In our study, cells of isolates Pri 1 and Pri 2 were observed as gram-positive cocci, Pri 4 as gram-positive bacilli, Pri 5 and Pri 6 as gram-positive rods, Pri 3 and Pri 7 as gram-negative coccobacilli; and Pri 8 and Pri 9 as gram-negative bacilli. Except for isolate Pri 5, cells of all isolates were found to be non-sporulating. Furthermore, cells of isolates Pri 4, Pri 8, and Pri 9 were motile, whereas those of the other isolates were non-motile.
Table (2):
Microscopic examination of bacterial isolates
Isolates |
Grams Stain |
Spore Staining |
Motility |
|---|---|---|---|
Pri 1 |
Gram-positive cocci |
Non-sporulating |
Non-motile |
Pri 2 |
Gram-positive cocci |
Non-sporulating |
Non-motile |
Pri 3 |
Gram-negative coccobacilli |
Non-sporulating |
Motile |
Pri 4 |
Gram-positive rods |
Sporulating |
Motile |
Pri 5 |
Gram-positive rods |
Sporulating |
Non-motile |
Pri 6 |
Gram-positive rods |
Non-sporulating |
Non-motile |
Pri 7 |
Gram-negative coccobacilli |
Non-sporulating |
Non-motile |
Pri 8 |
Gram-negative bacilli |
Non-sporulating |
Motile |
Pri 9 |
Gram-negative bacilli |
Non-sporulating |
Motile |
Growth curve of bacterial isolates
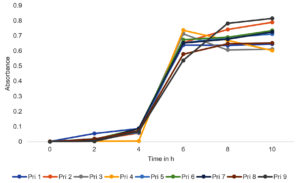
Bacterial growth curve patterns (Figure 1) revealed that all bacterial isolates were capable of surviving and growing in the challenging environment of textile printing wastewater and exhibited normal growth patterns characterized by lag, log, and stationary phases. Consequently, all bacterial isolates demonstrated their potential as viable candidates for textile printing wastewater treatment. However, to comprehensively analyze the reduction in color and values of other polluting parameters of textile printing wastewater by bacterial isolates, additional assays are currently being conducted.
Screening of bacterial isolates for their decolorization potential
Through visual observation for color changes after 24 h and 48 h of incubation, four isolates, namely, Pri 3, Pri 4, Pri 8, and Pri 9, were identified as the most effective candidates for decolorizing textile printing wastewater and the details of the same are mentioned in Table 3. These four bacterial isolates successfully transformed the color of the textile printing wastewater from dark brown to light yellow after 48 h of incubation. In contrast, the remaining bacterial isolates showed minimal alteration in the color of the wastewater.
Table (3):
Screening of bacterial isolates for their decolorization potential
No. |
Isolates |
Visual observation for color change |
Color after 24 h |
Color after 48 h |
|---|---|---|---|---|
1 |
Reference (without bacterial inoculation) |
No color change |
Dark brown |
Dark brown |
2 |
Pri 1 |
Minor decolorization was observed compared with a reference |
Brown |
Light brown |
3 |
Pri 2 |
Minor decolorization was observed compared with a reference |
Brown |
Brown |
4 |
Pri 3 |
A light yellowish-colored layer formed in more than half of the flask |
Light green |
Light yellow |
5 |
Pri 4 |
Complete decolorization was observed compared with a reference |
Light green |
Light yellow |
6 |
Pri 5 |
No decolorization was observed compared with a reference |
Brown |
Brown |
7 |
Pri 6 |
No decolorization was observed compared with a reference |
Brown |
Brown |
8 |
Pri 7 |
A light yellowish-colored layer formed in more than half of the flask |
Yellowish green |
Yellow |
9 |
Pri 8 |
Complete decolorization was observed compared with a reference |
Light yellow |
Light yellow |
10 |
Pri 9 |
Complete decolorization was observed compared with a reference |
Light yellow |
Light yellow |
Bioremediation of textile printing wastewater by selected bacterial isolates
Reduction in wastewater color, BOD, COD, TDS, TSS, and ammoniacal nitrogen was assessed to evaluate the bioremediation potential of selected bacterial isolates Pri 3, Pri 4, Pri 8, and Pri 9.
Color reduction
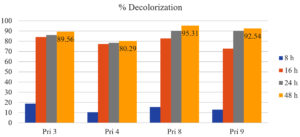
All four bacterial isolates achieved >75% decolorization of textile printing wastewater within 16 h. After 24 h, the decolorization further increased, ranging from 85% to 90%, except for isolate Pri 4, which exhibited <80% decolorization. Subsequently, after 48 h, the decolorization percentages reached 89%, 80%, 95%, and 92% for isolates Pri 3, Pri 4, Pri 8, and Pri 9, respectively. Notably, isolate Pri 8 demonstrated the highest color reduction within 48 h. The results of percent decolorization are illustrated in Figure 2.
BOD reduction
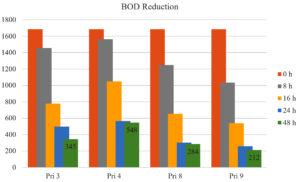
All bacterial isolates exhibited >50% reduction in BOD of textile printing wastewater within 16 h, except for isolate Pri 4 (<36%). After 24 h and 48 h, BOD reduction increased to 70.44% and 79.52% for Pri 3, 66.35% and 67.47% for Pri 4, 82.13% and 83.14% for Pri 8, and 84.68% and 87.41% for Pri 9, respectively. Notably, isolate Pri 9 achieved the maximum BOD reduction within 48 h. The data on BOD reduction are presented in Figure 3.
COD reduction
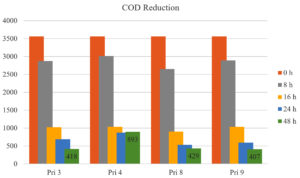
All bacterial isolates demonstrated COD reduction of >70% in textile printing wastewater within 16 h. After 24 h and 48 h, COD reduction increased to 80.65% and 88.26% for Pri 3, 74.92% and 75.46% for Pri 4, 85.06% and 87.95% for Pri 8, and 83.46% and 88.57% for Pri 9, respectively. Notably, isolates Pri 3 and Pri 9 achieved the maximum COD reduction within 48 h. The data on COD reduction are presented in Figure 4.
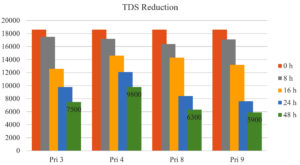
TDS reduction
All bacterial isolates exhibited approximately 50% reduction in TDS after 24 h, except for Pri 4 (<33%). After 48 h, TDS reduction further increased to 59.67% for Pri 3, 47.31% for Pri 4, 66.12% for Pri 8, and 68.27% for Pri 9. Notably, isolate Pri 9 achieved the maximum reduction in TDS within 48 h. The data on TDS reduction are presented in Figure 5.
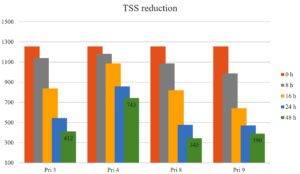
TSS reduction
Approximately 60% reduction in TSS was observed after 24 h by all bacterial isolates, except for isolate Pri 4 (<32%). After 48 h, the reduction increased to 67.22% for Pri 3, 40.89% for Pri 4, 72.55% for Pri 8, and 68.97% for Pri 9. Notably, isolate Pri 8 achieved the maximum reduction in TSS within 48 h. The data on TSS reduction are presented in Figure 6.
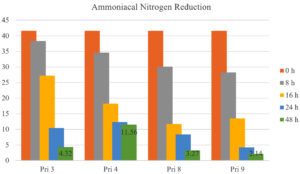
Ammoniacal nitrogen reduction
All bacterial isolates demonstrated >75% reduction in ammoniacal nitrogen after 24 h. After 48 h, the reduction increased to 90.37% for Pri 3, 73.54% for Pri 4, 92.68% for Pri 8, and 95.18% for Pri 9. Notably, isolate Pri 9 achieved the maximum reduction in ammoniacal nitrogen within 48 h. The data on ammoniacal nitrogen reduction are presented in Figure 7.
Identification of the selected bacterial isolates using biochemical tests and VITEK 2
We have tested all the isolates against 47 different Sugars for the identification of Bacteria with a biochemical test, and the results we got are mentioned in Table 4.
Table (4):
Bacterial identification using VITEK-2, the microbial identification system
No. |
Tests |
Mnemonic |
Pri-3 |
Pri-4 |
Pri-8 |
Pri-9 |
|---|---|---|---|---|---|---|
1 |
Ala-Phe-Pro-Arylamidase |
APPA |
– |
– |
– |
– |
2 |
ADONITOL |
ADO |
– |
– |
– |
– |
3 |
L-pyrrolidonyl-Arylamidase |
PyrA |
– |
+ |
+ |
– |
4 |
L-Arabitol |
IARL |
– |
– |
– |
– |
5 |
D-Cellobiose |
dCEL |
– |
+ |
– |
– |
6 |
Beta-Galactosidase |
BGAL |
– |
+ |
+ |
– |
7 |
H2S Production |
H2S |
– |
– |
+ |
– |
8 |
Beta-N-Acetyl-Glucosaminidase |
BNAG |
– |
– |
– |
– |
9 |
Glutamyl Arylamidase pNA |
AGLTp |
– |
– |
– |
– |
10 |
D-Glucose |
dGLU |
– |
+ |
+ |
– |
11 |
Gamma-Glutamyl-Transferase |
GGT |
– |
– |
+ |
– |
12 |
Fermentation/Glucose |
OFF |
– |
– |
+ |
– |
13 |
Beta-Glucosidase |
BGLU |
– |
+ |
– |
– |
14 |
D-Maltose |
dMAL |
– |
+ |
+ |
– |
15 |
D-Mannitol |
dMAN |
– |
+ |
+ |
– |
16 |
D-Mannose |
dMNE |
– |
– |
+ |
– |
17 |
Beta-Xylosidase |
BXYL |
– |
– |
– |
– |
18 |
Beta-Alanine Arylamidase pNA |
BAlap |
– |
– |
– |
– |
19 |
L-Proline Arylamidase |
ProA |
+ |
– |
– |
+ |
20 |
Lipase |
LIP |
– |
– |
– |
– |
21 |
Palatinose |
PLE |
– |
+ |
– |
– |
22 |
Tyrosine Arylamidase |
TyrA |
+ |
+ |
– |
+ |
23 |
Urease |
URE |
– |
– |
+ |
– |
24 |
D-Sorbitol |
dSOR |
– |
– |
+ |
– |
25 |
Saccharose/Sucrose |
SAC |
– |
+ |
– |
– |
26 |
D-Tagatose |
dTAG |
– |
– |
– |
– |
27 |
D-Trehalose |
dTRE |
– |
+ |
+ |
– |
28 |
Citrate |
CIT |
+ |
– |
– |
+ |
29 |
Malonate |
MNT |
– |
– |
+ |
– |
30 |
5-keto-D-Gluconate |
5KG |
– |
– |
+ |
– |
31 |
L-Lactate alkalinization |
ILATk |
+ |
+ |
– |
+ |
32 |
Alpha-Glucosidase |
AGLU |
– |
+ |
– |
– |
33 |
Succinate alkalinization |
SUCT |
+ |
– |
+ |
+ |
34 |
Beta-N-Acetyl-Galactoaminidase |
NAGA |
– |
– |
– |
– |
35 |
Alpha-Galactosidase |
AGAL |
– |
+ |
– |
– |
36 |
Phosphatase |
PHOS |
– |
– |
– |
– |
37 |
Glycine Arylamidase |
GlyA |
– |
– |
– |
– |
38 |
Ornithine Decarboxylase |
ODC |
– |
– |
– |
– |
39 |
Lysine Decarboxylase |
LDC |
– |
– |
– |
– |
40 |
L-Histidine assimilation |
IHISa |
– |
– |
– |
– |
41 |
Coumarate |
CMT |
– |
– |
+ |
– |
42 |
BETA-Glucuronidase |
BGUR |
– |
– |
– |
– |
43 |
O/129 Resistance (Comp-Vibrio) |
O129R |
– |
– |
– |
– |
44 |
Glu-Gly-Arg-Arylamidase |
GGAA |
– |
– |
– |
– |
45 |
L-MALATE assimilation |
IMLTa |
(-) |
– |
– |
(-) |
46 |
Ellman |
ELLM |
+ |
– |
– |
+ |
47 |
L-Lactate assimilation |
ILATa |
+ |
– |
– |
+ |
Antibiotic sensitivity tests of the selected bacterial isolates
We have tested all the isolates against 47 different antibiotics for the selected isolates depending on the biochemical identification test, and the results we got after the incubation period are mentioned in Table 5.
Table (5):
Antibiotic sensitivity tests of the selected bacterial isolates
| Sr. No. | Antibiotics | Pri 3 | Pri 4 | Pri 8 | Pri 9 | ||||
|---|---|---|---|---|---|---|---|---|---|
| Zone | Sensitivity | Zone | Sensitivity | Zone | Sensitivity | Zone | Sensitivity | ||
| 1 | Amikacin | 23 | Sensitive | 23 | Sensitive | 15 | Intermediate | 21 | Sensitive |
| 2 | Tobramycin | 24 | Sensitive | 24 | Sensitive | 14 | Intermediate | 23 | Sensitive |
| 3 | Gentamicin | 21 | Sensitive | 22 | Sensitive | 14 | Intermediate | 21 | Sensitive |
| 4 | Netilmicin | 22 | Sensitive | 21 | Sensitive | 15 | Sensitive | 22 | Sensitive |
| 5 | Aztreonam | 24 | Sensitive | 19 | Sensitive | 22 | Sensitive | 21 | Sensitive |
| 6 | Cefazolin | 18 | Sensitive | 21 | Sensitive | 16 | Resistance | 26 | Sensitive |
| 7 | Cefaclor | — | — | — | — | — | — | — | — |
| 8 | Cefuroxime | 15 | Sensitive | 21 | Sensitive | 18 | Intermediate | 24 | Sensitive |
| 9 | Ceftizoxime | 25 | Sensitive | 23 | Intermediate | 18 | Intermediate | 26 | Sensitive |
| 10 | Cefpodoxime | 22 | Sensitive | 24 | Sensitive | 26 | Sensitive | ||
| 11 | Cefixime | 26 | Sensitive | 26 | Sensitive | — | — | — | — |
| 12 | Ceftazidime | 25 | Sensitive | 23 | Sensitive | 19 | Intermediate | 23 | Sensitive |
| 13 | Cefoperazone | — | — | — | — | 21 | Sensitive | 24 | Sensitive |
| 14 | Cefotaxime | — | — | — | — | 26 | Sensitive | 26 | Sensitive |
| 15 | Ceftriaxone | — | — | — | — | — | — | 23 | Sensitive |
| 16 | Cefepime | 19 | Sensitive | 26 | Sensitive | 23 | Sensitive | 27 | Sensitive |
| 17 | Piperacillin + tazobactum | 31 | Sensitive | 22 | Sensitive | 24 | Sensitive | 23 | Sensitive |
| 18 | Ticarcillin + clavulanic acid | 24 | Sensitive | — | — | 25 | Sensitive | — | — |
| 19 | Cefepime + tazobactum | — | — | — | — | 23 | Sensitive | 26 | Sensitive |
| 20 | Cefoperazone + tazobactum | 22 | Sensitive | 26 | Sensitive | — | — | — | — |
| 21 | Cefoperazone + sulbactam | 24 | Sensitive | 24 | Sensitive | 25 | Sensitive | 24 | Sensitive |
| 22 | Amoxicillin + clavulanic acid | 22 | Sensitive | 22 | Sensitive | 18 | Sensitive | 21 | Sensitive |
| 23 | Trimethoprim/sulfamethoxazole | 23 | Sensitive | 24 | Sensitive | 22 | Sensitive | 21 | Sensitive |
| 24 | Colistin | 14 | Sensitive | 13 | Sensitive | 13 | Sensitive | 15 | Sensitive |
| 25 | Imipenem | 25 | Sensitive | 26 | Sensitive | 27 | Sensitive | 25 | Sensitive |
| 26 | Meropenem | 26 | Sensitive | 25 | Sensitive | 28 | Sensitive | 25 | Sensitive |
| 27 | Doripenem | 27 | Sensitive | 27 | Sensitive | 27 | Sensitive | 24 | Sensitive |
| 28 | Tetracycline | 21 | Sensitive | 20 | Sensitive | 21 | Sensitive | ||
| 29 | Doxycycline | 18 | Sensitive | 23 | Sensitive | 17 | Sensitive | 22 | Sensitive |
| 30 | Minocycline | 18 | Sensitive | 19 | Sensitive | 18 | Sensitive | 25 | Sensitive |
| 31 | Tigecycline | 18 | Sensitive | — | — | 19 | Sensitive | 22 | Sensitive |
| 32 | Chloramphenicol | 21 | Sensitive | — | — | 20 | Sensitive | 21 | Sensitive |
| 33 | Nitrofurantoin | 20 | Sensitive | — | — | 19 | Sensitive | 24 | Sensitive |
| 34 | Norfloxacin | 19 | Sensitive | — | — | 21 | Sensitive | 19 | Sensitive |
| 35 | Ciprofloxacin | 23 | Sensitive | 26 | Sensitive | 23 | Sensitive | 27 | Sensitive |
| 36 | Levofloxacin | 24 | Sensitive | 23 | Sensitive | 21 | Sensitive | 25 | Sensitive |
| 37 | Fosfomycin | — | — | — | — | 19 | Sensitive | 23 | Sensitive |
| 38 | Piperacillin | 22 | Sensitive | — | — | 20 | Intermediate | 22 | Sensitive |
| 39 | Ticarcillin | 21 | Sensitive | — | — | 24 | Sensitive | 24 | Sensitive |
| 40 | Moxifloxacin | — | — | 26 | Sensitive | — | — | — | — |
| 41 | Penicillin G | — | — | 28 | Sensitive | — | — | — | — |
| 42 | Erythromycin | — | — | 25 | Sensitive | — | — | — | — |
| 43 | Azithromycin | — | — | 19 | Sensitive | — | — | — | — |
| 44 | Rifampicin | — | — | 23 | Sensitive | — | — | — | — |
| 45 | Linezolid | — | — | 27 | Sensitive | — | — | — | — |
| 46 | Vancomycin | — | — | 19 | Sensitive | — | — | — | — |
| 47 | Clindamycin | — | — | 06 | Resistant | — | — | — | — |
Out of 47 antibiotics tested, isolate Pri 3 exhibited sensitivity to 32; Pri 4 showed sensitivity to 33 and intermediate sensitivity to 1; Pri 8 displayed sensitivity to 26, intermediate sensitivity to 7, and resistance against 1; and isolate Pri 9 demonstrated sensitivity to 35 antibiotics. Thus, the selected bacterial isolates were found to be sensitive to the majority of the antibiotics tested, suggesting vulnerability and positive response to antibiotics in case of infections, even after proper disinfection of the bioremediated textile printing wastewater.
Identification of the selected bacterial isolates by 16S rRNA sequencing
According to the report from NCMR, the results of 16S rRNA sequencing for the selected bacterial isolates are as follows:
The Pri 3 isolate matches with the Alcaligenes aquatillis LMG 22996(T) strain with approximately 99.93% similarities, as per the gene-sequencing database.
TGACGGGCGGTGTGTGCAAGACCCGGGAACGTATTCACCGCGACATTCTGATCCGCGATTACTAGCGATTCCGACTTCACGCAGTCGAGTTGCAGACTGCGATCCGGACTACGATCGGGTTTCTGAGATTGGCTCCCCCTCGCGGGTTGGCGACCCTCTGTCCCGACCATTGTATGACGTGTGAAGCCCTACCCATAAGGGCCATGAGGACTTGACGTCATCCCCACCTTCCTCCGGTTTGTCACCGGCAGTCTCATTAGAGTGCTCTTGCGTAGCAACTAATGACAAGGGTTGCGCTCGTTGCGGGACTTAACCCAACATCTCACGACACGAGCTGACGACAGCCATGCAGCACCTGTGTTCCGGTTCTCTTGCGAGCACTGCCAAATCTCTTCGGCATTCCAGACATGTCAAGGGTAGGTAAGGTTTTTCGCGTTGCATCGAATTAATCCACATCATCCACCGCTTGTGCGGGTCCCCGTCAATTCCTTTGAGTTTTAATCTTGCGACCGTACTCCCCAGGCGGTCAACTTCACGCGTTAGCTGCGCTACTAAGGCCTAACGGCCCCAACAGCTAGTTGACATCGTTTAGGGCGTGGACTACCAGGGTATCTAATCCTGTTTGCTCCCCACGCTTTCGTGTCTGAGCGTCAGTATTATCCCAGGGGGCTGCCTTCGCCATCGGTATTCCTCCACATATCTACGCATTTCACTGCTACACGTGGAATTCTACCCCCCTCTGACATACTCTAGCTCGGCAGTTAAAAATGCAGTTCCAAGGTTGAGCCCTGGGATTTCACATCTTTCTTTCCGAACCGCCTACACACGCTTTACGCCCAGTAATTCCGATTAACGCTTGCACCCTACGTATTACCGCGGCTGCTGGCACGTAGTTAGCCGGTGCTTATTCTGCAGATACCGTCAGCAGTATCTCGTATTAGGAGATACCTTTTCTTCTCTGCCAAAAGTACTTTACAACCCGAAGGCCTTCATCATACACGCGGGATGGCTGGATCAGGGTTTCCCCCATTGTCCAAAATTCCCCACTGCTGCCTCCCGTAGGAGTCTGGGCCGTGTCTCAGTCCCAGTGTGGCTGGTCGTCCTCTCAAACCAGCTACGGATCGTTGCCTTGGTGAGCCTTTACCCCACCAACTAGCTAATCCGATATCGGCCGCTCCAATAGTGAGAGGTTCCGAAGAATCCCCCCCTTTCCCCCGTAGGGCGTATGCGGTATTAGCCACTCTTTCGAGTAGTTATCCCCCGCTACTGGGCACGTTCCGATATATTACTCACCCGTCCGCCACTCGCCACCAAGAGAGCAAGCTCTCTCGTGCTGCCGTTCGACTTGCATGTGTAAAGCATCCCCGCTAGCGTTCAATCTGAGCCAGGAT
The Pri 4 isolate matches with the Priestia aryabhattai B8W22(T) strain with approximately 99.93% similarities, as per the gene-sequencing database.
TCGAGCGAACTGATTAGAAGCTTGCTTCTATGACGTTAGCGGCGGACGGGTGAGTAACACGTGGGCAACCTCCTGTAAGACTGGGATAACTTCGGGAAACCGAAGCTAATACCGGATAGGATCTTCTCCTTCATGGGAGATGATTGAAAGATGGTTTCGGCTATCACTTACAGATGGGCCCGCGGTGCATTAGCTAGTTGGTGAGGTAACGGCTCACCAAGGCAACGATGCATAGCCGACCTGAGAGGGTGATCGGCCACACTGGGACTGAGACACGGCCCAGACTCCTACGGGAGGCAGCAGTAGGGAATCTTCCGCAATGGACGAAAGTCTGACGGAGCAACGCCGCGTGAGTGATGAAGGCTTTCGGGTCGTAAAACTCTGTTGTTAGGGAAGAACAAGTACGAGAGTAACTGCTCGTACCTTGACGGTACCTAACCAGAAAGCCACGGCTAACTACGTGCCAGCAGCCGCGGTAATACGTAGGTGGCAAGCGTTATCCGGAATTATTGGGCGTAAAGCGCGCGCAGGCGGTTTCTTAAGTCTGATGTGAAAGCCCACGGCTCAACCGTGGAGGGTCATTGGAAACTGGGGAACTTGAGTGCAGAAGAGAAAAGCGGAATTCCACGTGTAGCGGTGAAATGCGTAGAGATGTGGAGGAACACCAGTGGCGAAGGCGGCTTTTTGGTCTGTAACTGACGCTGAGGCGGAAAGCGTGGGGAGCAAACAGGATTAGATACCCTGGTAGTCCACGCCGTAAACGATGAGTGCTAAGTGTTAGAGGGTTTCCGCCCTTTAGTGCTGCAGCTAACGCATTAAGCACTCCGCCTGGGGAGTACGGTCGCAAGACTGAAACTCAAAGGAATTGACGGGGGCCCGCACAAGCGGTGGAGCATGTGGTTTAATTCGAAGCAACGCGAAGAACCTTACCAGGTCTTGACATCCTCTGACAACTCTAGAGATAGAGCGTTCCCCTTCGGGGGACAGAGTGACAGGTGGTGCATGGTTGTCGTCAGCTCGTGTCGTGAGATGTTGGGTTAAGTCCCGCAACGAGCGCAACCCTTGATCTTAGTTGCCAGCATTTAGTTGGGCACTCTAAGGTGACTGCCGGTGACAAACCGGAGGAAGGTGGGGATGACGTCAAATCATCATGCCCCTTATGACCTGGGCTACACACGTGCTACAATGGATGGTACAAAGGGCTGCAAGACCGCGAGGTCAAGCCAA
The Pri 8 isolate matches with the Citrobacter werkmanii NBRC 105721(T) strain with approximately 99.78 % similarities, as per the gene-sequencing database.
ACGGGCGGGTGTGTGCAAGGCCCGGGAACGTATTCACCGTGGCATTCTGATCCACGATTACTAGCGATTCCGACTTCATGGAGTCGAGTTGCAGACTCCAATCCGGACTACGACATACTTTATGAGGTCCGCTTGCTCTCGCGAGGTCGCTTCTCTTTGTATATGCCATTGTAGCACGTGTGTAGCCCTACTCGTAAGGGCCATGATGACTTGACGTCATCCCCACCTTCCTCCAGTTTATCACTGGCAGTCTCCTTTGAGTTCCCGGCCTAACCGCTGGCAACAAAGGATAAGGGTTGCGCTCGTTGCGGGACTTAACCCAACATTTCACAACACGAGCTGACGACAGCCATGCAGCACCTGTCTCAGAGTTCCCGAAGGCACCAAAGCATCTCTGCTAAGTTCTCTGGATGTCAAGAGTAGGTAAGGTTCTTCGCGTTGCATCGAATTAAACCACATGCTCCACCGCTTGTGCGGGCCCCCGTCAATTCATTTGAGTTTTAACCTTGCGGCCGTACTCCCCAGGCGGTCGACTTAACGCGTTAGCTCCGGAAGCCACGCCTCAAGGGCACAACCTCCAAGTCGACATCGTTTACGGCGTGGACTACCAGGGTATCTAATCCTGTTTGCTCCCCACGCTTTCGCACCTGAGCGTCAGTCTTTGTCCAGGGGGCCGCCTTCGCCACCGGTATTCCTCCAGATCTCTACGCATTTCACCGCTACACCTGGAATTCTACCCCCCTCTACAAGACTCTAGCCTGCCAGTTTCGGATGCAGTTCCCAGGTTGAGCCCGGGGATTTCACATCCGACTTGACAGACCGCCTGCGTGCGCTTTACGCCCAGTAATTCCGATTAACGCTTGCACCCTCCGTATTACCGCGGCTGCTGGCACGGAGTTAGCCGGTGCTTCTTCTGCGAGTAACGTCAATTGCTGCGGTTATTAACCACAACACCTTCCTCCTCGCTGAAAGTACTTTACAACCCGAAGGCCTTCTTCATACACGCGGCATGGCTGCATCAGGCTTGCGCCCATTGTGCAATATTCCCCACTGCTGCCTCCCGTAGGAGTCTGGACCGTGTCTCAGTTCCAGTGTGGCTGGTCATCCTCTCAGACCAGCTAGGGATCGTCGCCTAGGTGAGCCGTTACCCCACCTACCAGCTAATCCCATCTGGGCACATCCGATGGCAAGAGGCCCGAAGGTCCCCCTCTTTGGTCTTGCGACATTATGCGGTATTAGCTACCGTTTCCAGTAGTTATCCCCCTCCATCGGGCAGTTTCCCAGACATTACTCACCCGTCCGCCACTCGTCACCCGAGAGCAAGCTCTCTGTGCTACCGTTCGACTTGCATGTGTTAGGCCTGCCGCCAGCGTTCAATCTGAGCCA
The Pri 9 isolate has 100% similarities with the Shewanella chilikensis JC5(T) strain, as per the gene-sequencing database.
CAAGGCCCGGGAACGTATTCACCGTGGCATTCTGATCCACGATTACTAGCGATTCCGACTTCATGGAGTCGAGTTGCAGACTCCAATCCGGACTACGACCGGCTTTATGAGATTAGCTCCACCTCGCGGCTTCGCAACCCTCTGTACCGACCATTGTAGCACGTGTGTAGCCCTACTCGTAAGGGCCATGATGACTTGACGTCGTCCCCACCTTCCTCCGGTTTATCACCGGCAGTCTCCCTAAAGTTCCCGGCATTACCCGCTGGCAAGTAAGGATAGGGGTTGCGCTCGTTGCGGGACTTAACCCAACATTTCACAACACGAGCTGACGACAGCCATGCAGCACCTGTCTCTCAGTTCCCGAAGGCACACCTGCGTCTCCGCTGGCTTCTGAGGATGTCAAGAGTAGGTAAGGTTCTTCGCGTTGCATCGAATTAAACCACATGCTCCACCGCTTGTGCGGGCCCCCGTCAATTCATTTGAGTTTTAACCTTGCGGCCGTACTCCCCAGGCGGTCTACTTAATGCGTTAGCTTGAGAGCCCAGTGTTCAAGACACCAAACTCCGAGTAGACATCGTTTACGGCGTGGACTACCAGGGTATCTAATCCTGTTTGCTCCCCACGCTTTCGTGCCTGAGCGTCAGTCTTTGTCCAGGGGGCCGCCTTCGCCACCGGTATTCCTCCAGATCTCTACGCATTTCACCGCTACACCTGGAATTCTACCCCCCTCTACAAGACTCTAGTTTGCCAGTTCGAAATGCGGTTCCCAGGTTGAGCCCGGGGCTTTCACATCTCGCTTAACAAACCGCCTGCGCACGCTTTACGCCCAGTAATTCCGATTAACGCTCGCACCCTCCGTATTACCGCGGCTGCTGGCACGGAGTTAGCCGGTGCTTCTTCTGCGAGTAACGTCACAGCTAGCAGGTATTAACTACTAACCTTTCCTCCTCGCTGAAAGTGCTTTACAACCCGAAGGCCTTCTTCACACACGCGGCATGGCTGCATCAGGGTTTCCCCCATTGTGCAATATTCCCCACTGCTGCCTCCCGTAGGAGTCTGGGCCGTGTCTCAGTCCCAGTGTGGCTGATCATCCTCTCAGACCAGCTAGGGATCGTTGCCTAGGTGAGCCATTACCTCACCTACTAGCTAATCCCACCTGGGCTTATCCATCAGCGCAAGGACCGAAGTTCCCCTGCTTTCCCCCGTAGGGCGTATGCGGTATTAGCAGTCGTTTCCAACTGTTATCCCCCACAAATGGGCAAATTCCCAGGCATTACTCACCCGTCCGCCGCTCGTCATCTTCAAAAGCAAGCTTTTGAAATGTTACCGCTCGACTTGCATGTGTTAGGCCTGCCGCCAGCGTTCAATCTGAGCCA
Addressing environmental restoration amidst the current state of pollution poses a significant challenge. Despite decades of research in colored wastewater treatment, finding an effective approach remains elusive. In light of this, the present study explored the utilization of textile printing wastewater inoculated with cow dung to isolate potential bacterial species for the efficient biological treatment of textile printing wastewater. This bioremediation strategy demonstrated the effectiveness of four enriched bacterial species, namely, A. aquatilis LMG 22996(T), P. aryabhattai B8W22(T), C. werkmanii NBRC 105721(T), and S. chilikensis JC5(T), in substantially reducing color, BOD, COD, and other polluting parameters of textile printing wastewater. However, considering their superior capacity to decolorize a diverse range of dyes in natural settings and achieve the complete mineralization of wastewater, C. werkmanii NBRC 105721(T) and S. chilikensis JC5(T) emerged as the most effective biological candidates among the four isolates. To further understand the depth of dye decolorization by these isolates, we are currently studying the biochemical mechanisms involved. Additionally, we are attempting to isolate the genes responsible for dye degradation. We hope that, in the future, cow dung will be recognized as a reservoir of microbial species with high potential for the bioremediation of pollution.
ACKNOWLEDGMENTS
The authors would like to thank M/s. Western Overseas (Jetalsar Junction) for providing textile printing wastewater samples and Environment Audit laboratory (Research Analytical laboratory) of the School of Engineering, RK University for providing research facilities to carry out analysis of wastewater samples.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
DATA AVAILABILITY
The datasets generated and/or analysed during the current study are available from the corresponding author on reasonable request.
ETHICS STATEMENT
Not applicable.
- Shah MP. Azo Dye Removal Technologies. Austin J Biotechnol Bioeng. 2018;5(1):1090.
- Asad S, Amoozegar MA, Pourbabaee AA, Sarbolouki MN, Dastgheib SMM. Decolorization of textile azo dyes by newly isolated halophilic and halotolerant bacteria. Bioresour Technol. 2007;98(11):2082-2088.
Crossref - Sheth NT, Dave SR. Optimisation for enhanced decolourization and degradation of reactive Red BS C.I. 111 by Pseudomonas aeruginosa NGKCTS. Biodegradation. 2009;20(6):827-836.
Crossref - Najme R, Hussain S, Maqbool Z, et al. Biodecolorization of Reactive Yellow-2 by Serratia sp. RN34 Isolated from Textile Wastewater. Water Environ Res. 2015;87(12):2065-2075.
Crossref - Pandya A, Pandya C, Dave B. Biodegradation Of Toxic Dyes And Textile Dye Effluent – A Review. Int J Adv Eng Res Dev. 2018;5(3):1246-1249.
- Hassan MM, Alam MZ, Anwar MN. Biodegradation of Textile Azo Dyes by Bacteria Isolated from Dyeing Industry Effluent. Int Res J Biol Sci. 2013;2(8):27-31.
- Pokharia A, Ahluwalia SS. Decolorization of Xenobiotic Azo Dye- Black WNN by Immobilized Paenibacillus alvei MTCC 10625. Sci Educ Publ. 2016;4(2):35-46.
Crossref - Simelane LP, Fosso-Kankeu E, Njobeh P, Pandey S. Response of bacterial biosorbents to chemical treatment as influenced by cell membrane structure and impact on the adsorption behaviour of dyes. Curr Sci. 2018;114(4):826-834.
Crossref - H. DB, D. VB, C. MR, et al. Influence of some chemical parameters on decolorization of textile dyes by bacterial strains isolated from waste water treatment plant. African J Microbiol Res. 2014;8(11):1213-1220.
Crossref - Pedrosa Brandão-Costa RM. Fungi of Biotechnological Interest in the Discoloration of Textile Effluents. Trends Text Eng Fash Technol. 2018;4(3):1-4.
Crossref - Jagiasi SR, Patel SN. Microbial Decolorization of Methyl Orange by Klebsiella Spp. DA26. International Journal of Research in BioSciences (IJRBS), 2020; 4(3):27-36.
- Khehra MS, Saini HS, Sharma DK, Chadha BS, Chimni SS. Biodegradation of azo dye C.I. Acid Red 88 by an anoxic – Aerobic sequential bioreactor. Dye Pigment. 2006;70(1):1-7.
Crossref - Thakur JK, Paul S, Dureja P, Annapurna K, Padaria JC, Gopal M. Degradation of sulphonated azo dye red HE7B by Bacillus sp. and elucidation of degradative pathways. Curr Microbiol. 2014;69(2):183-191.
Crossref - Anusiya G, Flora G, Solaieswari P. Isolation and Screening of Azo Dye Decoloizing Bacterial Isolates from Shrimp Shell Contaminated Soil in Thoothukudi Coast. Int J Res Appl Sci Eng Technol. 2017;5(Vii):1351-1354.
- Dubey A, Mishra N, Singh N, Deb A, Verma S. Isolation of dye degrading microorganism. Electron J Environ Agric Food Chem. 2010;9(9):1534-1539.
- Telke AA, Kalyani DC, Dawkar VV, Govindwar SP. Influence of organic and inorganic compounds on oxidoreductive decolorization of sulfonated azo dye C.I. Reactive Orange 16. J Hazard Mater. 2009;172(1):298-309.
Crossref - P Shah M. Microbial degradation of Textile Dye (Remazol Black B) by Bacillus spp. ETL-2012. J Bioremediation Biodegrad. 2012;4(2):6-11.
Crossref - Ramalho PA, Cardoso MH, Ramalho MT, Gübitz GM, Cavaco-Paulo AM. Decolourisation of a synthetic textile effluent using a bacterial consortium. Biotechnol J. 2007;2(3):370-373.
Crossref - Dawkar V V, Jadhav UU, Jadhav SU, Govindwar SP. Biodegradation of disperse textile dye Brown 3REL by newly isolated Bacillus sp. VUS. J Appl Microbiol. 2008;105(1):14-24.
Crossref - Dawkar V V., Jadhav UU, Ghodake GS, Govindwar SP. Effect of inducers on the decolorization and biodegradation of textile azo dye Navy blue 2GL by Bacillus sp. VUS. Biodegradation. 2009;20(6):777-787.
Crossref - Dawkar V V., Jadhav UU, Tamboli DP, Govindwar SP. Efficient industrial dye decolorization by Bacillus sp. VUS with its enzyme system. Ecotoxicol Environ Saf. 2010;73(7):1696-1703.
Crossref - Zhao M, Sun PF, Du LN, Wang G, Jia XM, Zhao YH. Biodegradation of methyl red by Bacillus sp. strain UN2: Decolorization capacity, metabolites characterization, and enzyme analysis. Environ Sci Pollut Res. 2014;21(9):6136-6145.
Crossref - Saranraj P, Manigandan M. Enzymes Involved In Bacterial Decolourization And Degradation Of Textile Azo Dyes. Indo – Asian J Multidiscip Res. 2018;4(1):1369-1376.
Crossref - Shyamala Gowri R, Meenambigai P, Vijayaraghavan R, Raja Rajeswari P, Vinitha R. Enhanced Biodegradation of Red Rr By Adapted Bacterial Isolates. Int J Adv Res. 2007;5(4):2320-5407.
Crossref - Prasad SS, Aikat K. Study of bio-degradation and bio-decolourization of azo dye by Enterobacter sp. SXCR. Environ Technol (United Kingdom). 2014;35(8):956-965.
Crossref - Sikdar B, Roy AK, Biswas SK, et al. Biodegradation of Crystal Violet dye by bacteria isolated from textile industry effluents. PeerJ. 2018;6:e5015.
Crossref - Meerbergen K, Willems KA, Dewil R, Van Impe J, Appels L, Lievens B. Isolation and screening of bacterial isolates from wastewater treatment plants to decolorize azo dyes. J Biosci Bioeng. 2018;125(4):448-456.
Crossref - Cui D, Li G, Zhao M, Han S. Decolourization of azo dyes by a newly isolated Klebsiella sp. strain Y3, and effects of various factors on biodegradation. Biotechnol Biotechnol Equip. 2014;28(3):478-486.
Crossref - Nachiyar CV, Sunkar S, Kumar GN, et al. Biodegradation of Acid Blue 113 Containing Textile Effluent by Constructed Aerobic Bacterial Consortia: Optimization and Mechanism. J Bioremediation Biodegrad. 2012;3(9).
Crossref - Karunya A, Rose C, Valli Nachiyar C. Biodegradation of the textile dye Mordant Black 17 (Calcon) by Moraxella osloensis isolated from textile effluent-contaminated site. World J Microbiol Biotechnol. 2014;30(3):915-924.
Crossref - Forgacs E, Cserháti T, Oros G. Removal of synthetic dyes from wastewaters: a review. Environ Int. 2004;30(7):953-971.
Crossref - Telke AA, Joshi SM, Jadhav SU, Tamboli DP, Govindwar SP. Decolorization and detoxification of Congo red and textile industry effluent by an isolated bacterium Pseudomonas sp. SU-EBT. Biodegradation. 2010;21(2):283-296.
Crossref - Telke AA, Kim SW, Govindwar SP. Significant reduction in toxicity, BOD, and COD of textile dyes and textile industry effluent by a novel bacterium Pseudomonas sp. LBC1. Folia Microbiol (Praha). 2012;57(2):115-122.
Crossref - Kalyani DC, Telke AA, Dhanve RS, Jadhav JP. Ecofriendly biodegradation and detoxification of Reactive Red 2 textile dye by newly isolated Pseudomonas sp. SUK1. J Hazard Mater. 2009;163(2-3):735-742.
Crossref - Srinivasan A, Viraraghavan T. Decolorization of dye wastewaters by biosorbents: A review. J Environ Manage. 2010;91(10):1915-1929.
Crossref - Mishra S, Maiti A. The efficacy of bacterial species to decolourise reactive azo, anthroquinone and triphenylmethane dyes from wastewater: a review. Environ Sci Pollut Res. 2018;25(9):8286-8314.
Crossref - Lade HS, Waghmode TR, Kadam AA, Govindwar SP. Enhanced biodegradation and detoxification of disperse azo dye Rubine GFL and textile industry effluent by defined fungal-bacterial consortium. Int Biodeterior Biodegrad. 2012;72:94-107.
Crossref - Kadam AA, Kulkarni AN, Lade HS, Govindwar SP. Exploiting the potential of plant growth promoting bacteria in decolorization of dye Disperse Red 73 adsorbed on milled sugarcane bagasse under solid state fermentation. Int Biodeterior Biodegrad. 2014;86:364-371.
Crossref - Kumari L, Tiwary D, Mishra PK. Biodegradation of C.I. Acid Red 1 by indigenous bacteria Stenotrophomonas sp. BHUSSp X2 isolated from dye contaminated soil. Environ Sci Pollut Res. 2016;23(5):4054-4062.
Crossref - Pandey A, Singh P, Iyengar L. Bacterial decolorization and degradation of azo dyes. Int Biodeterior Biodegrad. 2007;59(2):73-84.
Crossref - Mahmood S, Arshad M, Khalid A, Nazli ZH, Mahmood T. Isolation and screening of azo dye decolorizing bacterial isolates from dye-contaminated textile wastewater. Soil Environ. 2011;30(1):7-12.
- Parvin F, Rahman MM, Islam MM, et al. Isolation of mixed bacterial culture from Rajshahi silk industrial zone and their efficiency in azo dye decolorization. Indian J Sci Technol. 2015;8(10):950-957.
Crossref - Sharma M, Bagewadi ZK, Vernekar AG, et al. Biological Degradation of Reactive Dyes by Using Bacteria Isolated from Dye Effluent Contaminated Soil. Middle-East J Sci Res. 2013;17(12):1695-1700.
Crossref - Saranraj P, Sivasakthivelan P. Prevalence of Bacterial Isolates in Textile Dye Effluent and Analysis of its Dye Degrading Efficiency. Middle-East J Sci Res. 2014;21(5):721-725.
Crossref - Agarry SE, Owabor CN. Evaluation of microbial systems for biotreatment of cassava mill waste water in Nigeria: biodegradation of cyanide. Int J Environ Eng. 2012;4(3-4):315.
Crossref - Buthelezi SP, Olaniran AO, Pillay B. Textile Dye Removal from Wastewater Effluents Using Bioflocculants Produced by Indigenous Bacterial Isolates. Molecules. 2012;17(12):14260-14274.
Crossref - Zwietering MH, Jongenburger I, Rombouts FM, Van’t Riet K. Modeling of the bacterial growth curve. Appl Environ Microbiol. 1990;56(6):1875-1881.
Crossref - Sakr M, Mohamed MM, Maraqa MA, et al. A critical review of the recent developments in micro–nano bubbles applications for domestic and industrial wastewater treatment. Alexandria Eng J. 2022;61(8):6591-6612.
Crossref - Brandi J, Wilson-Wilde L. Standard Methods. Standard_Methods_For_the_Examination_of_water_and_wastewater. 2013:522-527.
Crossref - Boguniewicz-Zablocka J, Klosok-Bazan I, Naddeo V, Mozejko CA. Cost-effective removal of COD in the pre-treatment of wastewater from the paper industry. Water Sci Technol. 2020;81(7):1345-1353.
Crossref - Jain RM, Mody KH, Keshri J, Jha B. Biological neutralization and biosorption of dyes of alkaline textile industry wastewater. Mar Pollut Bull. 2014;84(1-2):83-89.
Crossref - Nasution MY, Ahmad Shafwan S, Pulungan, Prasetya E, Rezeqi S. Isolation and Characterization of Bacteria from River Origin in Mandailing Natal North Sumatera. J Phys Conf Ser. 2021;1819(1):012044.
Crossref
© The Author(s) 2024. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.