ISSN: 0973-7510
E-ISSN: 2581-690X
Betaine is a trimethylglycine, serves as osmoregulator to prevent dehydration and plasmolysis under adverse hyperosmotic environments. Choline oxidase gene from Arthrobacter sp. catalyzes two step oxidation reaction of choline to betaine followed by betaine accumulation in cells which in turn help them to survive and thrive in harsh environmental condition. To identify potential choline oxidase gene source, a gram stain positive, rod shaped, catalase and oxidase positive, motile, aerobic bacterial strains designated as HYJE003 and HYJE005 was isolated based on the colony morphology, biochemical and molecular characterization from the industrially polluted soil samples of Hyderabad, India. Optimum growth of the isolated strains was observed at 32°C on nutrient agar media and was found that both the strains were capable of utilizing variety of sugars as carbon source. The 16S rRNA gene sequence analysis revealed that strain HYJE003 was closely related to Arthrobacter globiformis with pairwise sequence similarities of 99.85%, 99.63%, 98.76% and 98.12% respectively. The strain HYJE005 was closely related to Arthrobacter phenanthrenivorans with pairwise sequence similarities of 99.93%, 99.47%, 99.25% and 98.11% respectively. Choline oxidase gene potential of the isolates was studied by feeding the cultures with choline chloride and converted betaine was assessed by the formation of betaine reineckate. Findings revealed that the isolated strain Arthrobacter globiformis-HYJE003 has four times higher conversion rate of choline chloride to betaine than the strain Arthrobacter phenanthrenivorans HYJE005.
Arthrobacter, BLAST, Pleomorphic, Reineckate, Betaine, Snapping division.
Soil bacteria are considered as economical source of useful biocatalysts in numerous industrial processes. Arthrobacter are, a typical soil bacterium widely distributed in diverse environmental conditions. This community gained special interest when compared to other soil microorganisms due to their ability to perform key metabolic functions such as: recycling of elements, recycling of wastes, detoxification of hazardous chemicals1, including heavy metal and organic pollutants2. All Arthrobacter species are pleomorphic, since they are rod shaped during exponential growth phase and cocci during their stationary phase3. Choline oxidase of Arthrobacter family is the only enzyme that catalyzes the two-step oxidation reaction of choline to betaine4. Betaine acts as osmoprotectant and plays an instrumental role in the growth of Arthrobacter in very harsh environmental conditions. The enzyme choline oxidase has high importance because, betaine is one of the limited numbers of compatible solutes that can accumulate in cell cytoplasm at very high concentration in adverse hyperosmotic environments5,6 to prevent dehydration and plasmolysis.
Betaine is an organic base present in fruits, vegetables, grains and seafood, thus betaine rich diet potentially favors human health. Betaine is a product well known to improve meat characteristics, weight gain, growth, carcass characteristics and feed conversion ratio in domestic animals7. Betaine allows a good water retention in meat and poultry, it increases breast meat yield, reduces abdominal fat and reduction of backfat thickness in swine8,9. As betaine is a methyl donor, it reduces the amount of methionine/cysteine from deamination and allows higher protein synthesis9. It modulates the histone and DNA methylation for epigenetic gene expression10. Various studies have demonstrated that betaine play critical role in embryonic and fetal development11. Considering several prospective and retrospective studies on betaine, in 2011 European Food Safety Authority has authorized a health claim of betaine on homocysteine metabolism.
Natural betaine is currently extracted and purified from beetroot, sugar beet and its derivatives12. Due to high demand, betaine is chemically produced in the form of betaine hydrochloride. Recent comparative studies between natural betaine and betaine hydrochloride clearly illustrates that the chemically synthesized betaine hydrochloride is not effective as the natural betaine13. In view from the above findings, the current study is aimed at isolation, identification and characterization of Arthrobacter strains from bacterial consortium of industrially polluted soil to evaluate their choline oxidase gene potential of choline to betaine oxidation.
Strain isolation and purifications
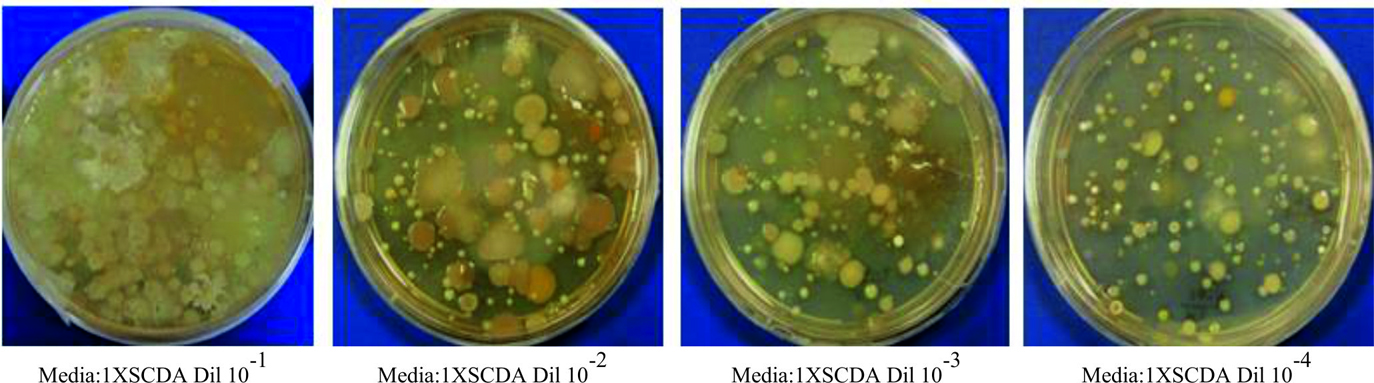
Five soil samples were collected from Jeedimetla industrial area, a well-known heavy metal polluted site in Hyderabad, India14. Samples were aseptically sampled and grinded in lab by using a sterile mortar. From the homogenized soil samples, one gram each weighed and distributed in five respective sterile flasks with 99 ml of sterile saline solution (0.85% of NaCl). After incubation at 25°C for 15 minutes at 200rpm, each sample was diluted to tenfold and from fourth to fifth dilution series 0.1ml of inoculum was added to the surface of 1X, 0.5X, 0.25X and 0.1X strength SCDA (Soya bean Casein Digest Agar) culture medium and later incubated at 30°C for 2days. Based on the cultural condition, colony morphology and microscopic characteristics two Arthrobacter strains were isolated from the culture plate of 0.1X SCDA with 10-4 dilution plating. Further pure cultures were established with the isolates by two level streak plating on nutrient agar media.
Biochemical and molecular analysis
Isolated pure cultures were subjected to biochemical analysis such as catalase, oxidase, urease, nitrate reduction and indole test15. To evaluate the sugar utilization profile of the isolated Arthrobacter strains, Hi-Carbohydrate kit from Hi-Media was used as per manufacture protocol. Later mobility test, gram staining and endospore staining were performed16. The genomic DNA from the isolated strains was extracted from CATB DNA extraction method17. Spectrophotometer and agarose gel electrophoresis was used to quantify the extracted DNA quality18. By using 27F-AGAGTTTGATCCTGGCTCAG and 1492R-GGTTACCTTGTTACGACTT18,19 primers nearly full length 16S rRNA gene was amplified from isolated strains. The PCR solution consist of 2X Go Taq Green mix 10µl, primers of 0.5µl each from 10µM stock, genomic DNA 1µl (10ng) and ddH2O was added to make up the final volume to 20µl. PCR condition of 94°C for 5 minutes and then 94°C for 20 seconds, 60°C for 30 second, 72°C for 1 minutes for 35 cycles followed by final extension of 72°C for 5 minutes. The amplified PCR products was verified by agarose gel electrophoresis, followed by PCR clean up sent for sequencing. Sequencing was performed using Applied Biosystems DNA sequencer, the 16S rRNA sequences were BLAST analyzed (National Centre for Biotechnology Information) to determine the closest available database match20.
Choline to betaine conversion of isolates
Identified strains were further evaluated for their choline oxidase gene potential to convert betaine from choline chloride by feeding the cultures with 100 mM of choline chloride in the nutrient broth. Cells were harvested from the overnight grown culture, lysed by sonication and centrifuged at high speed of 18000 rpm to remove any cell debris. To the 10 ml of supernatant, 1 ml of 25% trisodium phosphate solution and 10 ml of ammonium reineckate (2%) reagent was added and incubated in ice bath for one hour. Centrifuged at 18000rpm, carefully supernatant was transferred into fresh 15ml centrifuge tube set aside in an ice bath for one hour after addition of 1ml hydrochloric acid (3N). Then centrifuged for twenty minutes at 18000rpm and the clear supernatant was discarded, betaine reineckate pellets were air dried and dissolved in 10ml of acetone water (75:25) mixture. Optical density of the solution was recorded at 525 nm in a spectrophotometer and the amount of the betaine conversion was calculated by a similarly treated betaine standard curve21 (6.25, 12.5, 25, 50, and 100 mg/L).
Table (1):
Colony morphology of isolated strains
| Strain Name | Colonial Morphology | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Shape | Margins | Elevation | Colour | Consistency | Form | Opacity | Structure | Size in mm | |
| HYJE003 | Circular | Smooth | Convex | white | Membranous | Spreading | Opaque | Filamentous | 2 mm |
| HYJE005 | Circular | Smooth | Convex | white | Membranous | Spreading | Opaque | Filamentous | 3 mm |
The PCR was performed to isolate and confirm the choline oxidase gene from the isolated Arthrobacter globiformis HYJE003 strain. The choline oxidase gene was amplified by using oligonucleotide primers of COX-F_AATTGAATTCGATGCACATCGACAACATCGAG and COX-R_ AGCTAAGCTTAGGCGAGGGCCGCGCTCA. The PCR was carried with 5% DMSO, with the cycling condition of initial denaturation of one minute at 94°C, followed by 35 cycles of one of minute of cyclic denaturation at 94°C, 30 second at 50°C annealing, cyclic extension of 2 minute at 72°C and a final extension at 72°C for 10 minutes. 10 ng of template DNA, 2U of Pfu Taq DNA polymerase was used and the amplified product was agarose gel purified by QIAGEN kit following manufacture protocol. The amplified choline oxidase gene was then sequenced to confirm the gene sequence.
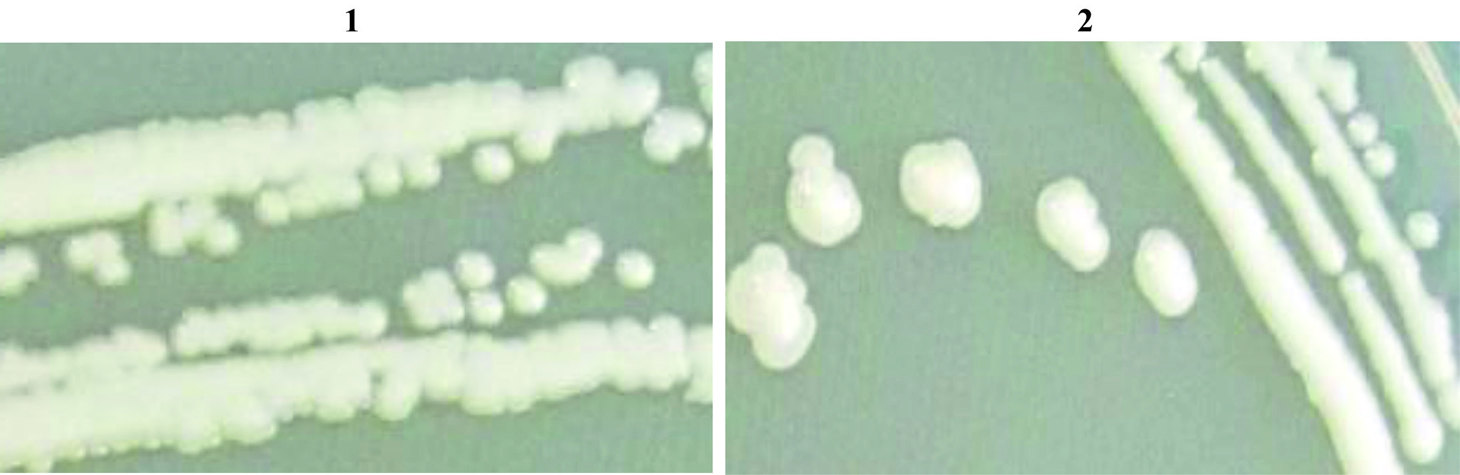
Form the 0.1X strength SCDA media plating with 10-4 dilution (Fig. 1 and Fig. 2) two strains (HYJE003 and HYJE005) were isolated based on colony morphology and snapping cell division3,22 (Fig. 3). Arthrobacter colonies on nutrient agar media have no distinctive pigmentation with undulate colony morphology appeared as smooth, circular with convex elevation, entire margin and nonsporulating (Tabel 1 and Fig. 2). In the early stage, cultures had rod shaped bacteria where as in older culture the bacteria were coccoid23. Microscopic observation reviled the size of the rod cells ranged between 0.5 to 0.8 by 1.5 to 2.0µ meter and the late stage coccoid cells were 0.5 to 0.8µ meter. The isolates showed growth in neutral to slightly alkaline pH and optimal growth was observed at 32°C temperature.
Fig. 3. Grams staining of the isolate to demonstrate snipping cell division (1-Strain HYJE003 and 2-Starin HYJE005)
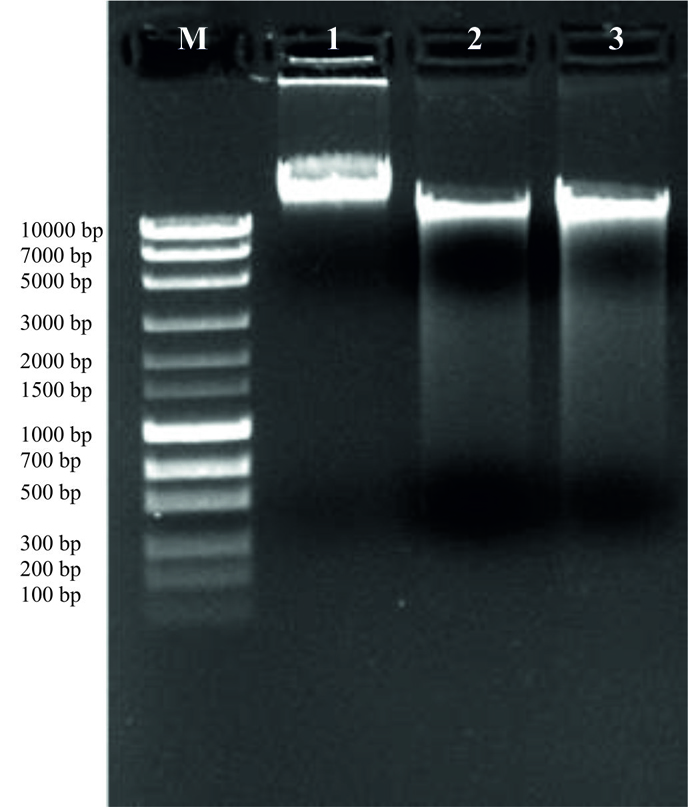
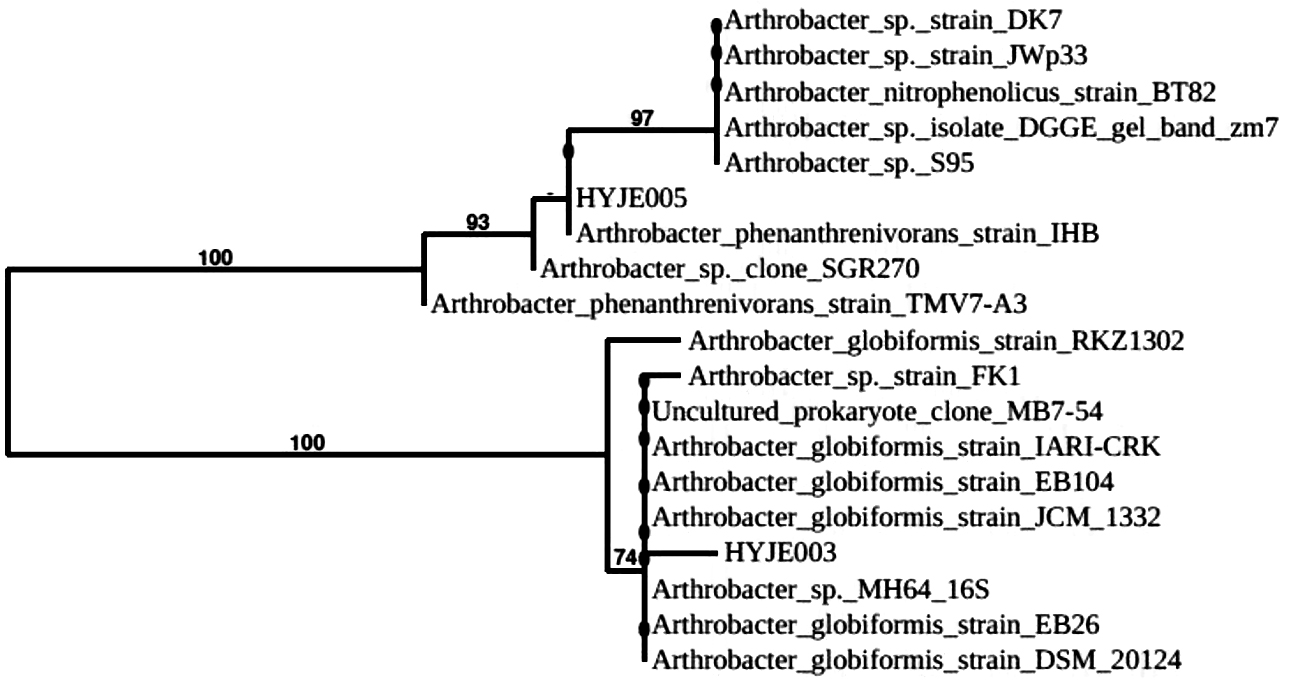
Both the isolated strains were recorded positive for grams staining, catalase test, indole test, urease test, nitrate reduction and gelatin hydrolysis assays (Table 2). On the other hand, both culture types produced negative result for crystal blue stain, motility test and bile esculin assays4 (Table 2). But strain HYJE003 produced positive result for oxidase test and on the other hand strain HYJE005 produced negative result and the nitrate reduction test was also scored positive in the current study4. The sugar utilization result (Table 3, Fig. 4 and 5) reveals isolated strains were able to grow in a vast variety of sugar/carbohydrate source and this ability of Arthrobacter genus make them widely distributed and abundant in soils of harsh environmental conditions. Almost full length of 16S rRNA gene was amplified from the extracted DNA (Fig. 6 and 7) and read length of 1350 bp of high-quality sequences was obtained. The obtained consensus sequences were analyzed and the 16S rRNA sequence were further used to investigate the identity of the isolates. Based on the concept of sequences similarity24 and similarity concept of 16S rRNA analysis25, the isolated strains had 16s rRNA sequence of 1350bp and HYJE003 shows the 99.85% similarity with the Arthrobacter globiformis and the second strain HYJE005 shows the 99.93% similarity with Arthrobacter phenanthrenivorans. The alignment results indicate a significant similarity of the putative strain HYJE003 DNA sequences (identified in the current study) to the 16S rRNA regions of genius Arthrobacter globiformis strains EB104, EB26, FB24, JCM 1332 and DCM 2012420. The identified strain HYJE005 16S rRNA analysis indicate a significant similarity with Arthrobacter phenanthrenivorans strains IHB, TMV7-A3, SGR270 and S9520. 16S rRNA sequence similarity and phylogenetic tree was constructed with MEGA X using Neighbor joining method, showed isolated strains was closely related to the genus Arthrobacter18 (Fig. 8). The 16S rRNA sequence information was deposited in NCBI GenBank with the consecutive accession numbers of MK968148 and MK968149 for the isolated strain HYJE003 and HYJE005 respectively.
Fig. 6. Extracted DNA quality check on 1.0% agarose get electrophoresis. M-1Kb DNA ladder, 1- ʎDNA 200ng, 2- HYJE003 and 3-HYJE005.
Fig. 7. 16s rRNA PCR Gel Image of isolated strains showing amplicon size of around 1600 bp.M-1Kb DNA ladder, 1- HYJE003 and 2-HYJE005.
Fig. 8. Phylogenetic tree of Arthrobacter sp. HYJE003 and HYJE05 based on 16S rRNA sequences. Each branch represents bootstrap values (≥ 50%) for 1000 replications from pairwise neighbor joining.
Table (2):
Biochemical result of the isolated strains
Test |
Strain HYJE003 |
Strain HYJE005 |
|---|---|---|
Grams Satin |
Positive |
Positive |
Crystal Blue Satin |
No Spores |
No Spores |
Motility Test |
Negative |
Negative |
Catalase Test |
Positive |
Positive |
Oxidase Test |
Positive |
Negative |
Indole Test |
Positive |
Positive |
Nitrate Reduction Test |
Positive |
Positive |
Urease Test |
Positive |
Positive |
Gelatin Hydrolysis Test |
Positive |
Positive |
Bile Esculin |
Negative |
Negative |
Table (3):
Hi-Carbohydrate kit result for isolated strains
| Name | Sugar Name | HYJE003 | HYJE005 |
|---|---|---|---|
| Part-A | Lactose | Positive | Negative |
| Xylose | Positive | Positive | |
| Maltose | Positive | Positive | |
| Fructose | Positive | Positive | |
| Dextrose | Positive | Positive | |
| Galactose | Negative | Negative | |
| Raffinose | Negative | Positive | |
| Trehalose | Positive | Negative | |
| Sucrose | Positive | Positive | |
| Melibiose | Positive | Positive | |
| L-Arabinose | Positive | Negative | |
| Mannose | Positive | Positive | |
| Part-B | Inulin | Positive | Negative |
| Sodium gluconate | Positive | Positive | |
| Glycerol | Positive | Negative | |
| Salicin | Positive | Negative | |
| Dulcitol | Positive | Negative | |
| Inositol | Negative | Positive | |
| Sorbitol | Positive | Positive | |
| Mannitol | Positive | Positive | |
| Adonitol | Positive | Positive | |
| Arabitol | Positive | Positive | |
| Erythritol | Positive | Positive | |
| Alpha-Methyl-D-glucoside | Negative | Negative | |
| Part-C | Rhamnose | Negative | Negative |
| Cellobiose | Negative | Negative | |
| Melezitose | Positive | Positive | |
| Methyl-D-Mannoside | Positive | Positive | |
| Xylitol | Positive | Negative | |
| ONPG | Positive | Positive | |
| Esculin | Positive | Positive | |
| D-Arabinose | Positive | Negative | |
| Citrate | Positive | Positive | |
| Malonate | Positive | Positive | |
| Sorbose | Positive | Positive |
Table (4):
Spectrophotometer reading for betaine estimation
# |
Sample ID |
Mean O.D. Value |
Standard Deviation |
Standard Error |
|---|---|---|---|---|
1 |
STD-1(100mg/L) |
0.921 |
0.005508 |
0.00318 |
2 |
STD-2(50mg/L) |
0.458 |
0.012055 |
0.00696 |
3 |
STD-3(25mg/L) |
0.234 |
0.006083 |
0.003512 |
4 |
STD-4(12.5mg/L) |
0.101 |
0.004933 |
0.002848 |
5 |
STD-5(6.25mg/L) |
0.056 |
0.003215 |
0.001856 |
6 |
HYJE003 |
0.085 |
0.005568 |
0.003215 |
7 |
HYJE005 |
0.011 |
0.003055 |
0.001764 |
Spectrophotometer observance of the betaine reineckate against standard curve relieved 9.8 mg/L and 1.8 mg/L of betaine in the cultures of HYJE003 and HYJE005 respectively4,21 (Table No-4 and Fig. 9). Choline oxidase gene from the HYJE003 strain was successfully amplified and sequenced. The sequenced choline oxidase gene from the strain HYJE003 was deposited in NCBI GenBank with the accession numbers of MK988621.
In conclusion, the data presented in this study extends our knowledge on the sequential isolation, biochemical and molecular characterization of Arthrobacter strains from industrially polluted soil samples and evaluation of their choline oxidase gene potential to convert betaine from choline chloride. Based on conversion result the strain HYJE003- Arthrobacter globiformis showed four-fold higher rate of choline to betaine conversion than the strain HYJE005- Arthrobacter phenanthrenivorans. Thus, the choline oxidase gene from isolated strain HYJE003 has shown higher rate of betaine conversion, further research needed for its purified enzyme activity and its commercial feasibility of betaine production.
Acknowledgements
None.
Conflict of Interest
The authors declares that there is no conflict of interest.
Authors’ Contribution
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Funding
None.
Data Availability
All datasets generated or analyzed during this study are included in the manuscript.
Ethics Statement
This article does not contain any studies with human participants or animals performed by any of the authors.
- Camargo, Flavio, Bento, Fatima, C Okeke, Frankenberger, William. Hexavalent Chromium Reduction by an Actinomycete, Arthrobacter crystallopoietes. Biological Trace Element Research, 2004; 97: 183-94.
Crossref - Tahri Joutey, Nezha, Sayel, Hanane, Bahafid, Wifak, el ghachtouli, Naima. Mechanisms of Hexavalent Chromium Resistance and Removal by Microorganisms. Reviews of environmental contamination and toxicology, 2015; 233: 45-69.
Crossref - Jones D and Keddie. The Genus Arthrobacter, 2006; pp. 945– 960. The prokaryotes, Springer, New York, N.Y, USA.
Crossref - Ikuta S, Imamura S, Misaki H, Horiuti Y. Purification and characterization of choline oxidase from Arthrobacter globiformis. J. Biochem., 1977; 82(6): 1741-1749.
Crossref - Teresa Caldas, Nathalie Demont-Caulet, Alexandre Ghazi, Gilbert Richarme. Thermo protection by glycine betaine and choline. Microbiology, 1999; 145: 2543–2548.
Crossref - Boch, J, Kempf B, Bremer E. Osmoregulation in Bacillus subtilis: Synthesis of the osmoprotectant glycine betaine from exogenously provided choline. Journal of Bacteriology, 1997; 176: 5364–5371.
Crossref - Sayed M, and Downing J. The effects of water replacement by oral rehydration fluids with or without betaine supplementation on performance, acid base balance, and water retention of heatstressed broiler chickens. Poult. Sci., 2011; 90: 157–167.
Crossref - Honarbakhsh SS, Zaghari M, Shivazad M. Can exogenous betaine be an effective osmolyte in broiler chicks under water salinity stress. Asian-Australasian Journal of Animal Sci., 2007; 20(11): 1729-1737.
Crossref - Kettunen H, Tiihonen K, Peuranen S, Saarinen MT, Remus JC. Dietary betaine accumulates in the liver and intestinal tissue and stabilizes the intestinal epithelial structure in healthy and coccidia-infected broiler chicks. Comparative Biochemistry and Physiology. Molecular & Integrative Physiology, 2000; 130(4): 759-769.
Crossref - Murdoch Brenda M., Murdoch Gordon K., Greenwood Sabrina, McKay Stephanie. Nutritional Influence on Epigenetic Marks and Effect on Livestock Production. Frontiers in Genetics, 2016; 7: 182.
Crossref - Joselit Y, Nanobashvili K, Jack-Roberts C. Maternal betaine supplementation affects fetal growth and lipid metabolism of high-fat fed mice in a temporal-specific manner. Nutr. Diabetes, 2018; 8(1): 41.
Crossref - Luca Rivoira, Sylwia Studziסska, Malgorzata Szultka, Maria Concetta Bruzzoniti, Bogus³aw Buszewsk. New approaches for extraction and determination of betaine from Beta vulgaris samples by hydrophilic interaction liquid chromatography-tandem mass spectrometry. Anal. Bioanal. Chem., 2017; 409: 5133–5141.
Crossref - Amerah AM. The differences between natural betaine and betaine hydrochloride. International Poultry Production, 2013; 22: 11-13.
- Govil, Pradip, Govil PK. Distribution and characterization of heavy metals in Jeedimetla industrial area, Hyderabad. Journal Pollution Research, 2001; 20(2): 245-255.
- Vashist, H, Sharma D and Gupta A. A review on commonly used biochemical test for bacteria. Innovare Journal of Life Sciences, 2013; 1(1): 1-7.
- Gregersen, T. Rapid method for distinction of gram-negative from gram-positive bacteria. European J. Appl. Microbiol. Biotechnol, 1978; 5: 123.
Crossref - Konstantinos Minas, Neil R McEwan, Charles Jamie Newbold, Karen P Scott. Optimization of a high-throughput CTAB-based protocol for the extraction of qPCR-grade DNA from rumen fluid, plant and bacterial pure cultures, FEMS Microbiology Letters, 2011; 325(2): 162–169.
Crossref - Weisburg WG, Barns SM, Pelletier DA and Lane DJ. 16S ribosomal DNA amplification for phylogenetic study. Journal of Bacteriology, 1991; 173 (2): 697–703.
Crossref - Davis Gislin , Dorairaj Sudarsanam, Gnanaprakasam Antony Raj, Kathirvelu Baskar. Antibacterial activity of soil bacteria isolated from Kochi, India and their molecular identification. Journal of Genetic Engineering and Biotechnology, 2018; 16(2):287–294.
Crossref - Vetrovsky T and Baldrian P. The Variability of the 16S rRNA Gene in Bacterial Genomes and Its Consequences for Bacterial Community Analyses. PLoS ONE, 2013; 8(2).
Crossref - Bandelin and pankratz. The Estimation of Betaine and Choline in Mixtures. J. Am. Pharm. Assoc. Am. Pharm. Assoc., 1953; 42: 442-3.
Crossref - Chan EC, Steveson L. On the biotin requirement of Arthrobacter globiformis. Canadian Journal of Microbiology, 1962; 8: 403-405.
Crossref - Busse HJ. Review of the taxonomy of the genus Arthrobacter. Int. J. Syst. Evol. Microbiol., 2016; 66(1): 9-37.
Crossref - Bosshard P. Abels PS, Zbinden R, Bottger EC, and Altwegg M. Ribosomal DNA sequencing for identification of aerobic gram-positive rods in the clinical laboratory. Journal of Clinical Microbiology, 2003; 41(9): 4134–4140.
Crossref - Janda JM and Abbott SL. 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory pluses, perils, and pitfalls. Journal of Clinical Microbiology, 2007; 5(9): 2761–2764.
Crossref
© The Author(s) 2019. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.