ISSN: 0973-7510
E-ISSN: 2581-690X
Microorganisms may cause the almost complete degradation of a material structure, even though symptoms of drastic microbial damage may not be apparent macroscopically. Due to their enzymatic activities and outstanding adaptation abilities to various environmental conditions, actinomycetes constitute a particular threat to a variety of materials. They are able to use, and subsequently degrade any substrate, whether this happens in a matter of days, weeks or years. Given the frequency of occurrence and a number of features that enable Streptomyces to growth on materials, more studies should be undertaken on methods allowing for their proper and rapid identification. Taxonomic studies gain particular importance in the case of damaged valuable historic materials. This paper focuses on identification of actinomycetes isolated from various historic and contemporary materials, including textiles, parchment and ceramics. Strengths and weaknesses of approaches that have been chosen for actinomycetes taxonomic studies are described.
Actinomycetes; Streptomyces, Isolation, Identification, Historic objects
All kinds of materials, exposed temporarily or permanently to moist conditions, are subject to microbial deterioration. A material infection may occur in a production process, arise during use, transportation or storage. Due to their physiology and adaptation abilities, actinomycetes pose a particular threat to diverse materials. These Gram-positive, filamentous bacteria characterised by a high (57–75%) G+C content in their DNA, form a ubiquitous group of prokaryotic organisms. Actinobacteria have an unparalleled ability to produce a vast variety of secondary metabolites, such as: extracellular enzymes, pigments and terpenes. They possess unique abilities to utilize complex and recalcitrant biopolymers such as: cellulose, keratin, chitin, pectin, starch, elastin and lignocellulose.1,2
Actinomycetes taxonomy is very complex. The traditional identification approach includes microscopic techniques and biochemical tests and it has always been considered difficult and time-consuming, requiring a series of costly and specialized tests. Apart from this, classical methods, when used alone, may give ambiguous results and identification is mainly limited to the genus level. To increase the accuracy of the results, a variety of alternative approaches are applied. They include molecular biology techniques, such as 16S rRNA gene sequencing. This tool, apart from being widely used to determine microorganisms’ taxonomic position and to identify new taxa, has also been used to confirm the morphological and biochemical identification results. Nevertheless, despite its specificity, 16S rRNA gene sequencing is still laborious and costly, among other limitations. Recently, Matrix-Assisted Laser Desorption Ionization Time-Of-Flight Mass Spectrometry (MALDI-TOF MS) has emerged as a rapid and cost-effective tool for the identification of microorganisms at the species level.3,4 This technique has been used both for the routine clinical diagnosis of human pathogens5 as well as for the identification of environmental microorganisms.6 It has also been applied to identify fastidious, difficult-to-culture and slow-growing organisms.7 The identification is based on the comparison of species specific protein spectra (molecular fingerprint) to a reference spectra database.
The main objective of the study is to identify actinomycetes isolated from various historic and contemporary materials, including textiles, parchment and ceramics. Strengths and weaknesses of approaches that have been chosen for actinomycetes taxonomic studies are described. Species potentially most destructive to various materials are also indicated.
Isolation of actinomycetes
Actinomycetes were isolated from historic and contemporary materials made of different substrates (organic and inorganic), belonging to various periods and environments. The bacteria isolation was carried out from areas with visible signs of microbial material deterioration such as stains, coatings or other surface alterations (Table 1). A variety of non- invasive methods and microbiological media were applied, taking the material main component (cellulose, protein, etc.), its state of preservation and the group of microorganisms sought into account (Table 1). After 4 weeks of incubation at 28±2oC, colonies of grown microorganisms, apparent after several days or weeks, were picked, deposited on PDA medium and then purified. Pure strains of actinomycetes were maintained on slants at 4oC and in MicrobankTM.
Identification of actinomycetes
Classical method
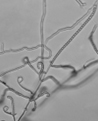
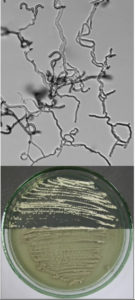
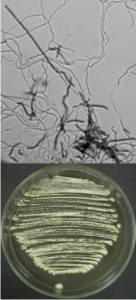
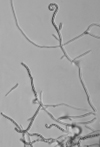
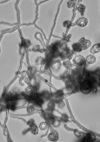
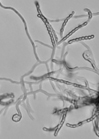
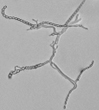
According to the identification key by Nonomura, 19748 and Holt et al., 2000,9 the colony morphology, production of melanoid pigments, soluble pigments other than melanin, spore (chain) morphology and spore-bearing structures were determined. All descriptions were made on 14-day old cultures grown on PDA at 28±2oC. Gram staining was performed for cell morphology. Slide cultures were prepared to study substrate and aerial filaments as well as spore chain morphology. Actinomycetes were examined under a stereoscopic and a transmitted light microscope equipped with a digital camera.
Slide culture method
A small block of agar was placed in the centre of a sterile microscope slide and inoculated with a given, well sporulating actinomycetes strain. A sterile coverslip was placed on top of the block and the slide was kept in a moist chamber. The chamber consisted of a sterile Petri dish lined with a piece of moistened filter paper and a sterile U-shaped glass rod on which the slide was placed. The cultures were incubated at 28±2oC for 14 days and examined periodically. Preparations were made from the actinomycetes which grew on the coverslip and on the sides of the agar block. Details about the spore arrangements were gathered by studying the filaments adhering to coverslips placed at an angle in the growing culture plates.
Molecular techniques
Identification by 16S rRNA gene sequencing
DNA extraction
Prior to DNA extraction the isolates were cultured on PDA medium for 8 days, after which approximately 100 mg of spores and filaments were scraped with a sterile loop into Eppendorf tubes, ground with 2 mm wolfram beads using Retsch MM 400 Mixer Mill (30 Hz, 3 min). Genomic DNA was extracted with Genomic Mini DNA extraction kit (A&A Biotechnology, Poland) and cleaned up with Anty-Inhibitor Kit (A&A Biotechnology, Poland), according to the manufacturer’s instructions. The quality and quantity of the extracted DNA was assessed using BioPhotometer Plus (Eppendorf, Germany). The obtained DNA was stored in a freezer for further analyses.
Identification of isolates
In order to verify the systematic position of the examined isolates, the partial fragments of 16S rRNA gene were amplified using the primers fD1 (5’-AGAGTTTGATCCTGGCTCAG-3’) and rP2 (5’-AGAGTTTGATCCTGGCTCAG-3’). Almost the entire sequence of 16S rRNA gene was amplified using the primers 9F (5’-GAGTTTGATCCTGGCTCAG-3’) and 1541R (5’-AAGGAGGTGATCCAGCC-3’)10. Polymerase chain reaction contained 100 ng of DNA template, 25 pM of each primer, 5 mM of dNTP, 1× PCR buffer and 2 U of DreamTaq DNA polymerase in a total volume of 50 µl. The following temperature profile was used for DNA amplification: initial denaturation at 94°C for 5 min., followed by 25 touchdown cycles of denaturation at 94°C for 30s, annealing starting from 67.5°C with temperature decreasing by 0.5°C in each cycle until 55°C for 30s and elongation at 72°C for 1 min and then 20 cycles of denaturation at 94°C for 30s, annealing at 55°C for 30s and elongation at 72°C for 1 min, final elongation for 10 min. and then stored at 4°C. PCR amplifications were performed in T100TM Thermal Cycler (Bio-Rad, USA). The PCR products were electrophoresed for 60 minutes in 1×TBE Simply Safe (EurX, Poland)-stained (0.5 mg/ml), 1% agarose gel, visualized in UV light and documented by the GelDoc system (BioRad, USA). Subsequently, the amplicons were purified using CleanUp kit (A&A Biotechnology, Poland) and sequenced using primers fD1, rP2 and 9F, 1541R.
Data analysis
The obtained sequences were analyzed using MEGA 6 software.11 The generated 16S rRNA sequences were queried against the NCBI database using BLAST search to determine the most closely related genus. Additionally, in order to compare the phylogenetic position of the studied isolates within the Streptomyces spp. the Bayesian tree was constructed encompassing all examined isolates and 16S sequences of the 36 most closely related Streptomyces spp. reference strains obtained from the GenBank. Mycobacterium tuberculosis H37Rv was used as an outgroup. The calculation was performed with MrBayes 3.1.2.12,13 The best fitted evolutionary model for the analyzed sequences was identified with jModelTest 214 according to the Akaike criterion as HKY with gamma-shaped rates variation among sites and with estimation of the proportion of invariable sites. MCMC simulation was performed in 4 chains per both independent analyzes (three of each heated). The analysis was continued until average standard deviation of split frequencies diagnostic value have reached 0.01 threshold sampling every 100th tree. The initial 25% of trees were discarded as burn-in fraction. 50% majority rule consensus trees were summarized and exported, showing posterior probabilities and average branch lengths and further processed with MEGA 6.
Identification by MALDI TOF MS
The MALDI-TOF MS method was applied to identify those actinomycetes strains which, characterised by the above described methods, did not bring definite results. The tests were performed on MALDI Biotyper (Bruker) using a standard procedure recommended by Bruker. Two culture media were applied, i.e., PDA solid medium and Gauss liquid medium. In the latter case, the isolates were grown in Erlenmeyer flasks and were agitated on a rotary shaker at 150 rpm for 5h a day. Agitation and aeration of liquid cultures was necessary to avoid the formation of surface pellicles or cottony sediments and instead obtain even, suspended growth. The isolates were cultured at 28±2°C for 14 days.
All measurements were preceded by a calibration with a standard bacterial test
(E. coli) as a positive control. A non-inoculated matrix solution was used as a negative control. The resulting spectra were analysed using MALDI Biotyper, version 3.0. According to the criteria recommended by the manufacturer, a result was considered valid at the species level whenever the score value attributed by Biotyper was x e” 2.0, valid at the genus level when the score was 1.7 d” x < 2 and as no reliable identification when the score was x < 1.7.
Nucleotide sequence accession numbers
The isolates’ 16S rRNA sequences obtained in this study were submitted to the GenBank. The assigned accession numbers are listed in Table 2.
Actinomycetes isolation
The list of the obtained strains’ IDs together with the description of substrates from which they were isolated, techniques and media used, are presented in Table 1. Other habitats where the species have been found by other authors, the ability to utilize different substrates and enzymes produced are listed in Table 2.
Table (1):
Isolation of studied actinomycetes strains
Sample no. |
Strain ID in author’s1 collection |
Origin / Material from which isolated |
Medium on which isolated; technique applied |
|---|---|---|---|
1 |
BBK13a |
Historic book collection / parchment bookbinding |
CPYA1 solid medium, pH 7.0, swab test, samples collected on sterile swabs from areas with black and brown stains |
2 |
BBO5 |
Sixteenth century textile / cotton lining |
The textile direct imprint on CPYA solid medium, pH 7.0 from the parts of the textile with rusty stains |
3 |
BBK13 |
Historic book collection / parchment bookbinding |
The same as in BBK13a |
4 |
BBA15 |
Peruvian ceramic vessel, Chimu culture |
Samples collected from areas with white coating and transferred to PDA2 medium, pH 5.6, swab test |
5 |
BBT59 |
Sixteenth century tapestry / woolen parts |
Imprint of the textile parts with brown stains on sterile balls of wool (carbon source) placed in sterile Petri dishes and poured with WiC-3 liquid medium, pH 7.5 |
6 |
BBO14 |
Sixteenth century textile / decorative silk textile |
The textile direct imprint on WiC+4 solid medium, pH 7.5 from the parts of the textile with light brown stains |
7 |
BBK11 |
Historic book collection / parchment bookbinding |
The same as in BBK13a |
8 |
BBW92 |
Contemporary woolen textile subjected to disinfection in ethyl alcohol for 2 hours |
Textile placed on CZA5 modified, pH 6.5 |
9 |
BBP12 |
Plastic made of starch composite with ethylene-acrylic acid co-polymer and glycerine and low density polyethylene subjected to 6 months of soil environment. Starch composite content in the plastic: 30%, which corresponds to 20% of pure starch content. |
After the incubation period in soil, samples were removed and subjected to careful observation under a stereoscopic microscope; existing microorganisms were transferred to PDA medium, pH 5.6 |
1CPYA = Czapek Peptone Yeast Agar (recommended for the cultivation of various actinomycetes, favourable for actinomycetes growth)
2PDA = potato dextrose agar
3WiC- = Weary and Canby mineral medium (free of carbon and nitrogen source) [17]
4WiC+ = Weary and Canby enriched with 2g of glucose and 3g of yeast extract [17]
5CZA modified = Czapek medium without sucrose
The emerging colonies showed typical actinomycetes characteristics. They appeared dry, leathery, with or without colour, adhering to the medium with vegetative filaments and spore forming. As apparent from Table 1, most of the strains were obtained from historic objects and only two out of nine (no. 8 and 9) from modern materials. The majority of strains, i.e., BBK13a, BBK13, BBT59, BBO14, BBK11, BBW92, were isolated from proteinaceous substrates of which three were derived from parchment, two from wool and one from silk. Strain BBO5 was isolated from cotton, BBA15 from pottery and BBP12 from modern composite material. A common feature of all these substrates is that they are hardly available sources of nutrients for microorganisms. To utilise them and grow, microbes must possess a number of specific features, such as a capability to produce substrate-specific enzymes. Actinomycetes are known to be microorganisms of extraordinary physiological potential with a wide range of adaptabilities. They are able to use, and subsequently degrade any substrate, whether this happens in a matter of days, weeks or years. Even in the case of inorganic materials, such as ceramics, the organic residues present in the material can provide sufficient nutrients. Also, environmental conditions contribute to the bacterial development through variations of humidity, temperature etc. Colonna-Preti and Eeckhout, 201315 suggest that the presence of hygroscopic salts inside the ceramics also participates, as it favours the availability of water, so crucial for microbial growth.
Identification of actinomycetes
Classical method
The description of colony morphology, macro- and microscopic features of the isolated actinomycetes grown on PDA medium for 14 days are presented in Table 2.
Table (2):
Characteristics of studied Streptomyces strains
Strain no. |
Strain name / microscopic and macroscopic features,14-day cultures |
GenBank accession number of obtained nucleotide sequences |
Characteristics of Streptomyces isolates on PDA medium, 14-day cultures |
Microscopic features |
Literature based species characteristics |
|---|---|---|---|---|---|
1 |
KT274748 |
Aerial filaments (AF): colourless Colonies (C): circular, smooth Reverse (R): colourless Diffusible pigment (DP): nd1 Earthy smell (ES)3: slight |
Open loops and open spiral chains of spores on top of long aerial filaments, oval spores |
Isolated from: antarctic soil samples [18], soil, Michigan [19], desert sand [20]; amylolythic activity2 |
|
2 |
KT274749 |
AF: grayish-pink C: circular, smooth to powdery R: pink DP: pink (carotenoid pigment) ES: strong |
Long, branched chains of spores, straight, flexous to open loops, oval spores |
Isolated from: El Mellah lake, Algieria [4], mangrove [21], soil, China [22]. Produces: acarviostatins [23], glutaminase [24]; amylolythic activity |
|
3 |
KT274750 |
AF: green C: irregular, rough R: colourless to greenish DP: nd ES: slight |
Flexous to open spiral chains of spores, oval spores |
Strong amylolythic activity |
|
4 |
KT274751 |
AF: dark beige to brownish C: circular, powdery R: brown DP: brown (melanoid pigment) ES: moderate |
Straight, flexous to hook shape chains of spores, oval spores |
Isolated from: a mixture of fresh horse and pig manure [19], mouldy hay, mushroom compost [20]. Produses: chitinase [25], azocaseinase, granaticin4 [19], xylanase [26], α-amylase, protease, cellulase [27]; strong amylolythic activity. Carbon sources utilization: dextrose, fructose, galactose, lactose, maltose, sucrose, starch, mannitol, xylose, inositol, rhamnose [27] Growth temperature: 25-57oC |
|
5 |
KT274752 |
AF: willow-green C: circular, powdery R: brown DP: greenish brown (melanoid pigment) ES: strong |
Long, straight chains of spores, oval spores |
Isolated from: limestone of Tell Basta tomb, Zagazig City, Egypt [28], SS John and Paul crypt, Roma [2], soil, muddy soil from riverbank, potato scab [20], hen house soil [29], endophytic strains isolated from mangrove plants from tropical China; terrestrial lichens and marine macroalgae; deep-sea ecosystems; gorgonian corals and other invertebrates living up to 4700 m depth, at 2–4 °C; atmospheric precipitations (rain drops, snow, hailstone). Halotolerant5, psychrotolerant, barotolerant, pH tolerance 4 to 11 [30]. Can sporulate in liquid cultures, conditions in which Streptomyces strains generally do not sporulate [31]. Produces: phospholipase [32], peroxidase [33], chitinolytic enzymes [34], α-amylase [24], strong amylolythic activity, keratinolytic serine proteinase very specific for keratinous substrates such as elastin, keratin and type I collagen; strong keratinolytic activitiy [29] |
|
6 |
KT274753 |
AF: white to light grey C: circular, powdery R: colourless DP: nd ES: strong |
Spore chains of moderate length, straight, flexous to primitive spirals, oval spores |
As no. 2 |
|
7 |
KT274754 |
AF: white to slightly grey C: circular, rough, leathery R: brownish DP: nd ES: strong |
Closed loops and closed spiral chains of spores, spores of oblong shape |
||
8 |
KT274755 |
AF: off-white to slightly willow-green C: circular, leathery R: colourless DP: nd ES: slight |
Moderate length straight to flexous chains of spores, growing in bunches, oval spores |
Isolated from: limestone of Tell Basta tomb, Zagazig City, Egypt, halotolerant [28], deteriorated zones within the ground of Marcherus icon dated back to the Christian period at Church of Samanoud, El-Mahala [35], mouldy hay, garden soil [20]. Produces: laccase [36], protease, glutaminase [24]; amylolythic activity |
|
9 |
KT274756 |
AF: greyish C: irregular, powdery R: brown DP: nd ES: strong |
Straight to flexous chains of spores, growing on short filaments, spores slightly oblong |
Isolated from: air, tomb, Veio [2], Tunisian agricultural soil containing municipal solid waste compost [37], compost of bamboo waste [1]. Produces: cellulase (endoglucanase) [1]; amylolythic activity |
1nd = not detected
2Own studies, data not shown. Production of extracellular amylases was assessed with an in situ plate method on Waksman medium. After seven days of culture, a reaction was developed with Lugol iodine solution.
3Earthy aroma originates from geosmin, an organic compound produced by Streptomyces. Under acidic conditions it decomposes into an odourless substance. The human nose is extremely sensitive to geosmin [38].
4Exopigment, red or blue depending on pH with antibiotic activity against Gram-positive bacteria
5Tolerates saline conditions up to 12 % w/v NaCl
The classical method required careful examination of the organisms growing on agar. The observation of vegetative filaments, arrangement of spores on aerial filaments and their shape was essential. Therefore, the major challenge during the preparation of actinomycetes cultures for microscopic analyses was to keep the filaments and spore structures intact. To achieve this, the slide culture method proved especially useful. The actinomycetes growth on slides was thin and undisturbed, making detailed studies of substrate filaments within the agar (fragmentation, presence of spores etc.), spore shape and spore arrangement on aerial filaments possible.
All of the isolated strains were found to be filamentous, spore-forming bacteria with the prevalence of long chains of spores. Filament fragmentation was not recorded. Repeatedly branched filaments formed small, tough colonies surrounded by a hydrophobic sheath. The colonies assumed a range of colours when the spore formation began. The species produced diffusible pigments of a melanoid and carotenoid nature (Table 2).
Based on the gathered morphological characteristics, the isolates were classified under the genus Streptomyces. Unfortunately, the differentiation of species within this genus remains difficult, due to the fact that the Streptomyces species are still poorly defined. For example, in the key by Holt et al., 2000,9 only two of the studied species are mentioned, namely S. albidoflavus and S. griseoflavus. Authors of the key are furthermore of the opinion that identification based on morphology alone is rarely secure, even with considerable experience. Systematic classification of the isolated species is as follows: Domain: Bacteria; Phylum: Actinobacteria; Order: Streptomycetales; Family: Streptomycetaceae; Genus: Streptomyces.
Molecular techniques
Identification by 16S rRNA gene sequencing
Analysis of 16S rRNA sequences was carried out to verify the systematic position of the examined isolates specified in the above section. The 735–1446 bp long 16S rRNA sequences were determined for 10 samples. They were then compared with the corresponding sequences of the type strains of the same and most closely related species, as resulted from the BLAST analysis (Table 3). The study confirmed that all of the tested strains belonged to the genus Streptomyces and 7 out of 10 strains could be identified to the species level.
Table (3):
Identification by 16S rRNA gene sequencing of the isolated actinomycetes strains
No. |
Sample |
Primers |
Sequenced length |
BLAST closest match |
% identity |
Identification (BLAST + reference strains BI tree) |
|---|---|---|---|---|---|---|
1 |
Pr1 |
9F/1541R |
1436 |
S. flavofungini |
100 |
S. flavofungini |
2 |
Pr2 |
fD1/rP2 |
1396 |
S. coelicoflavus |
100 |
S. coelicoflavus |
3 |
Pr2_2 |
9F/1541R |
1446 |
S. coelicoflavus/S. graminearus |
100 |
S. coelicoflavus |
4 |
Pr3 |
9F/1541R |
1438 |
Streptomyces sp. |
100 |
Streptomyces sp. |
5 |
Pr4 |
fD1/rP2 |
1395 |
S. thermoviolaceus |
100 |
S. thermoviolaceus |
6 |
Pr5 |
fD1/rP2 |
1393 |
S. albidoflavus |
100 |
S. albidoflavus |
7 |
Pr6 |
9F/1541R |
1445 |
S. coelicoflavus/S. graminearus |
100 |
S. coelicoflavus |
8 |
Pr7 |
9F/1541R |
735 |
Streptomyces sp./ S. lienomycyni/ S. rubrogriseus/ S. sclerogranulatus/ S. coelicolor |
100 |
Streptomyces sp. |
9 |
Pr8 |
9F/1541R |
1445 |
Streptomyces sp. |
100 |
S. griseoflavus |
10 |
Pr9 |
9F/1541R |
1440 |
S. griseoaurantiacus |
100 |
S. griseoaurantiacus |
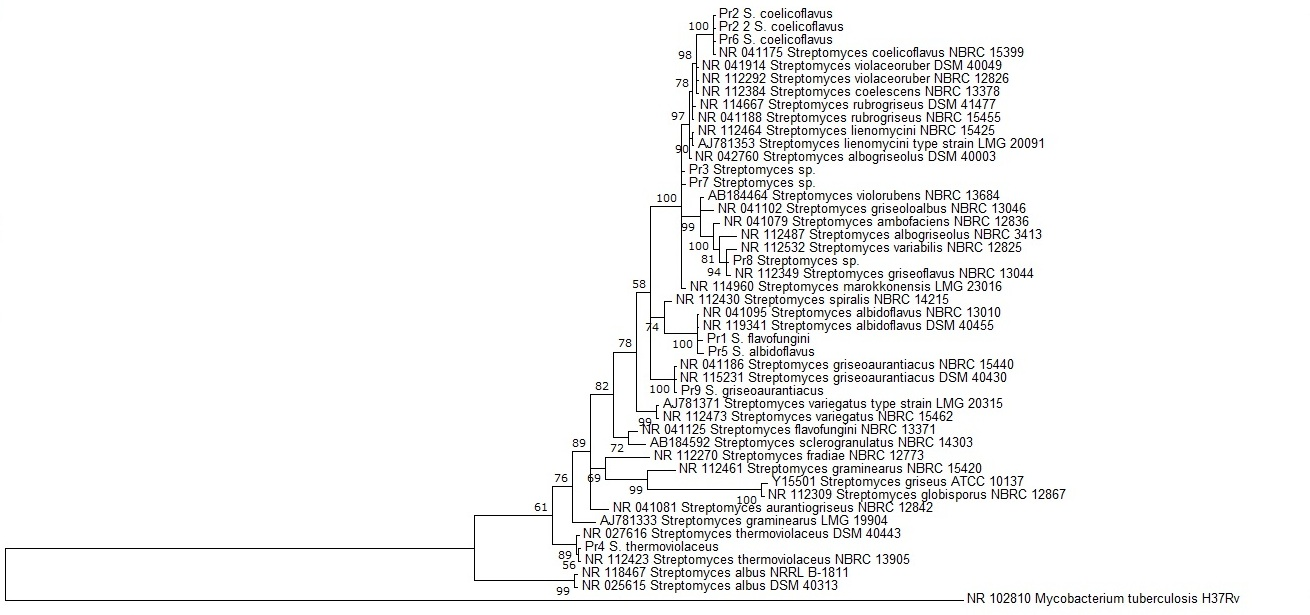
The 16S rRNA gene sequences of a total of 46 taxa, including 10 strains obtained in this study and an outgroup, were analyzed using BI method to determine the taxonomic position of the studied actinomycetes strains. The topology of the resulting BI consensus tree is generally well supported, i.e., by moderate to high posterior probability values (from 56 to 100%). It can be seen from Fig. 1 that 8 strains fall into well supported clades with most closely related taxa. For instance, Pr2, Pr2_2 and Pr6, identified as Streptomyces coelicoflavus by the BLAST search, grouped these three strains into one very well supported clade (100%) with the type strain of this species. Similar situation occurred with Pr9 and Pr4, which prior identification as S. griseoaurantiacus and S. thermoviolaceus, respectively, was followed by their well-supported clustering with these species (posterior probability values of 100 and 89%, respectively). Two out of the 10 isolates, i.e., Pr3 and Pr7 could not be identified to the species level and also could not be clustered with the strains selected for the analysis. The strain Pr8, which could not be identified based on the 16S rRNA gene sequencing, was grouped in a well-supported subclade with S. griseoflavus (94%).
Fig. 1. The Bayesian phylogeny showing the relative position of acquired strains on the background of 45 Streptomyces spp. sequences
Identification by MALDI TOF MS
Six (two unidentified: Pr3, Pr7 and four already identified: Pr1, Pr2, Pr6, Pr8) Streptomyces strains were subject to identification by measuring the unique molecular fingerprints of the organisms. The fastidious growth of the isolates on the solid medium yielded only small amount of material that could be harvested for analysis. As a result, only one strain, i.e., Pr3 was identified as Streptomyces sp., and matching against the Biotyper database yielded low score of 1.79 (identification valid at the genus level). The use of cell suspension from shaken liquid cultures provided a sufficient amount of test material, nevertheless in this case, all the analysed strains were identified as one species, namely Streptomyces violaceoruber. The data from the other two, above described, methods indicated however that all of the strains represented different species and none of them could be classified as S. violaceoruber.
The lack of success in obtaining reliable identification results can be attributed to two causes. The first is the absence of an adequate reference spectrum in the Biotyper database. The original Bruker database contains 4,706 bacterial spectra of which only 14 belong to the Streptomyces species. Loucif et al., 20144 point out that the availability of the Streptomyces spectra in the database is an extremely statistically significant (p < 0.0001) factor in obtaining a correct identification at the species level. The second possible cause is the absence of a sufficient protein signal needed to build a spectrum that can be compared to the Biotyper database. In turn, failure to obtain a spectrum can be explained by the structural properties of the cell wall of certain bacteria or their fastidious growth that yields an insufficient amount of material for analysis. Thus, sometimes even in spite of the presence of some reference spectra, results cannot be obtained.16
Based on this studies it can be stated that the use of MALDI-TOF MS for the identification of actinomycetes requires a standardised sample preparation method and a sound, elaborated reference spectra database. As Biswas and Rolain, 20137 indicate, the same microorganism can give different mass spectra, resulting from different culturing conditions or extraction methods. Loucif et al., 20144 developed a protocol based on ethanol–trifluoroacetic acid (E-TFA) extraction of the whole-cell proteins, allowing for a correct identification with high score of >2. The authors point out that with this promising procedure it will be possible to identify new Streptomyces isolates, the spectra of which are not yet available in the MALDI database. Strains will be identified by 16S rRNA sequencing and their spectra added to the database, gradually enlarging it. Thus, MALDI-TOF MS has the potential to reduce the need for identification techniques such as 16S rRNA gene sequencing. Up till now, however, these methods often yielded inconsistent results and the DNA analysis has remained the basic and reference tool for actinomycetes identification.
Microorganisms may cause the almost complete degradation of a material structure, even though symptoms of drastic microbial damage may not be apparent macroscopically. It is essential to identify microorganisms that take part in this process in order to properly protect objects from biodeterioration. Taxonomic studies gain particular importance in the case of damaged valuable historic materials. Given the frequency of occurrence and a number of features that enable Streptomyces to growth on materials (they are mesophilic aerobes, they have desiccation-resistant spores, outstanding metabolic activity and the majority of them tend to develop best under neutral or slightly alkaline conditions), more studies should be undertaken on methods allowing for their proper and rapid identification. This research describes the strengths and weaknesses of several approaches that may be chosen for actinomycetes taxonomic studies.
Literature data collected in Table 2 prove that the studied Streptomyces species possess outstanding adaptation abilities to various environmental conditions. They can be found in the whole range of environments ranging from atmospheric precipitations through soil and desert sand to deep-sea ecosystems. Moreover, the data indicate that the species produce a multiplicity of enzymes and are capable of utilizing various energy sources. All the above implies that none of the objects can be recognized as safe, regardless of its components and environment they belong to. Streptomyces thermoviolaceus together with S. albidoflavus were found to produce the widest range of enzymes as well as to utilize the largest variety of carbon sources, thus may be considered most dangerous to historical objects.
- Feng-Jen, Ch., Chi-Wen, L., Yet-Po, I., Chih-Hung, W., Ding-Hsuan, Ch. Hydrolysis of bamboo cellulose and cellulase characteristics by Streptomyces griseoaurantiacus ZQBC691. Journal of the Taiwan Institute of Chemical Engineers, 2012, 43: 220–225.
- Giacobini, C., De Cicco, M.A., TigHe, I., Accardo, G. Actinomycetes and biodeterioration in the field of fine art., Houghton D. R. (ed.). Biodeterioration, 1988, 7: 418-423.
- Lartigue, M.F. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry for bacterial strain characterization. Infection, Genetics and Evolution, 2013, 13: 230–235.
- Loucif, L., Bendjama, E., Gacemi-Kirane, D., Rolain, J.M. Rapid identification of Streptomyces isolates by MALDI-TOF MS. Microbiological Research, 2014; 169: 940–947.
- Bizzini, A., Durussel, C., Bille, J., Greub, G., Prod’hom, G. Performance of Matrix-Assisted Laser Desorption Ionization Time of Flight mass spectrometry for identification of bacterial strains routinely isolated in a clinical microbiology laboratory. J. Clin. Microbiol., 2010, 48: 1549–1554.
- Ruelle, V., EI Moualij, B., Zorzi, W., Ledent, P., Pauw, E.D. Rapid identification of environmental bacterial strains by Matrix-Assisted Laser Desorption/Ionization Time-of-Flight mass spectrometry. Rapid Commun. Mass Spectrom., 2004, 18: 2013–2019.
- Biswas, S., and Rolain, J.M. Use of MALDI-TOF mass spectrometry for identification of bacteria that are difficult to culture. Journal of Microbiological Methods, 2013, 92(1): 14–24.
- Nonomura, H. Key for classification and identification of 458 species of the Streptomycetes included in ISP. J. Ferment. Technol., 1974, 52: 78-92.
- Holt, J.G., Krieg, N.R., Sneath, P.H., Staley, J.T., Williams, S.T. Bergey’s manual of determinative bacteriology. Ninth edition. Lippincott Williams and Wilkins, Philadelphia, 2000.
- Weisburg, W.G., Barns, S.M., Pelletier, D.A., Lane, D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol., 1991, 173: 697-703.
- Tamura, K., Stecher, G., Peterson, D., Filipski, A., Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Molecular Biology and Evolution, 2013, 30: 2725-2729.
- Huelsenbeck, J. P., Ronquist, F., Nielsen, R., Bollback, J.P. Bayesian inference of phylogeny and its impact on evolutionary biology. Science, 2001, 294: 2310-2314.
- Ronquist, F., and Huelsenbeck, J.P. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics, 2003, 19: 1572-1574.
- Darriba, D., Taboada, G.L., Doallo, R., Posada, D. jModelTest 2: more models, new heuristics and parallel computing. Nature Methods, 2012, 9(8): 772.
- Colonna-Preti, K., and Eeckhout, P. The bacteriological contamination of archaeological ceramics: an example from Pachacamac (Peru), 2013, 205-213.
- Bizzini, A., Jaton, K., Romo, D., Bille, J., Prod’hom, G., Greub, G. Matrix-Assisted Laser Desorption Ionization–Time of Flight Mass Spectrometry as an alternative to 16S rRNA gene sequencing for identification of difficult-to-identify bacterial strains. J. Clin. Microbiol., 2011, 49(2): 693–696.
- BByskal, B. Gymnoascus arxii’s potential in deteriorating woollen textiles dyed with natural and synthetic dyes. International Biodeterioration and Biodegradation, 2014, 86(C): 349–357.
- Mavengere, N.R. Isolation, identification and characterization of novel actinomycetes from Antarctic soil samples, academic dissertation in biotechnology. University of the Western Cape, 2008.
- ATCC http://www.lgcstandards-atcc.org (accessed 17 January 2017).
- CBS http://www.cbs.knaw.nl/Collections (accessed 17 January 2017).
- Raghava Rao, K.V., Raghava Rao, T. Molecular characterization and its antioxidant activity of a newly isolated Streptomyces coelicoflavus BC 01 from mangrove soil. Journal of Young Pharmacists, 2013, 5(4): 121–126.
- Geng, P., and Bai, G. Two novel aminooligosaccharides isolated from the culture of Streptomyces coelicoflavus ZG0656 as potent inhibitors of a-amylase. Carbohydrate Research, 2008; 343: 470–476.
- Geng, P., Sun, T., Zhong, Q., Li, X., Shi, L., Bai, F., Bai, G. Two novel potent ±-amylase inhibitors from the family of acarviostatins isolated from the culture of Streptomyces coelicoflavus ZG0656. Chem. Biodivers., 2013; 10(3): 452-459.
- NCBI database: http://www.ncbi.nlm.nih.gov/protein (accessed 17 January 2017).
- Tsujibo, H., Hatano, N., Mikami, T., Hirasawa, A., Miyamoto, K., Inamor, Y. A Novel b-N-Acetylglucosaminidase from Streptomyces thermoviolaceus OPC-520: Gene Cloning, Expression, and Assignment to Family 3 of the Glycosyl Hydrolases. Applied and Environmental Microbiology, 1998; 64(8): 2920–2924.
- Garg, A.P., McCarthy, A.J., Roberts, J.C. Biobleaching effect of Streptomyces thermoviolaceus xylanase preparations on birchwood kraft pulp. Enzyme and Microbial Technology, 1996; 18(4): 261–267.
- Sudipta, R., and Debdulal, B. Broad spectrum antibacterial activity of granaticinic acid, isolated from Streptomyces thermoviolaceus NT1; an endophyte in Catharanthus roseus (L.) G. Don. Journal of Applied Pharmaceutical Science, 2015, 5(7): 6-11.
- Abdulla, H.M., May, E., Baghat, M., Dewedar, A. Characterization of actinomycetes isolated from Ancient stone and their potential for deterioration. Polish J. Microbiology, 2008, 57: 213-220.
- Bressollier, P., Letourneau, F., Urdaci, M., Verneuil, B. Purification and Characterization of a Keratinolytic Serine Proteinase from Streptomyces albidoflavus. Applied and Environmental Microbiology, 1999; 2570–2576.
- Sarmiento-Vizcaíno, A., Braña, A.F., González, V., Nava, H., Molina, A., Llera, E., Fiedler, H.P., Rico, J.M., García-Flórez, L., Acuña, J.L., García, L.A., Blanco, G. Atmospheric dispersal of bioactive Streptomyces albidoflavus strains among terrestrial and marine environments. Microb. Ecol., 2016; 71: 375–386.
- Yagüe, P., López-García, M.T., Rioseras, B., Sánchez, J., Manteca, Á. Pre-sporulation stages of Streptomyces differentiation: state-of-the-art and future perspectives. FEMS Microbiol. Lett., 2013; 342(2): 79–88.
- Sugimori, D., Kano, K., Matsumoto, Y. Purification, characterization, molecular cloning and extracellular production of a phospholipase A1 from Streptomyces albidoflavus NA297. FEBS Open Bio, 2012; 2: 318–327.
- Jaouadi, B., Rekik, H., Badis, A., Zaraî Jaouadi, N., Belhoul, M., Hmidi, M., Kourdali, S., Fodil, D., Bejar, S. Production, purification, and characterization of a highly thermostable and humic acid biodegrading peroxidase from a decolorizing Streptomyces albidoflavus strain TN644 isolated from a Tunisian off-shore oil field. International Biodeterioration and Biodegradation, 2014; 90: 36-44.
- Broadway, R.M., Williams, D.L., Kain, W.C., Harman, G.E., Lorito, M., Labeda, D.P. Partial characterization of chitinolytic enzymes from Streptomyces albidoflavus. Lett. Appl. Microbiol., 1995; 20(5): 271-276.
- Tishchenko, S., Gabdulkhakov, A., Trubitsina, L., Lisov, A., Zakharova, M., Leontievsky, A. Crystallization and X-ray diffraction studies of a two-domain laccase from Streptomyces griseoflavus. Acta Crystallogr. F. Struct. Biol. Commun., 2015, 71(9): 1200-1204.
- Tishchenko, S., Gabdulkhakov, A., Trubitsina, L., Lisov, A., Zakharova, M., Leontievsky, A. Crystallization and X-ray diffraction studies of a two-domain laccase from Streptomyces griseoflavus. Acta Crystallogr. F. Struct. Biol. Commun., 2015, 71(9): 1200-1204.
- Mokni-Tlili, S., Jedidi, N., Hassen, A. Studies on the ecology of actinomycetes in an agricultural soil amended with organic residues. Identification of the dominant groups of Actinomycetales. World J. Microbiol. Biotechnol., 2011, 27: 2239-2249.
- Polak, E.H., and Provasi, J. Odor sensitivity to geosmin enantiomers. Chemical Senses, 1992; 17: 23.
© The Author(s) 2017. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.