ISSN: 0973-7510
E-ISSN: 2581-690X
Bifidobacteria are widely used as probiotics for their application in the development of functional food and prophylactic therapy. This has necessitated the development of a molecular approach for the genera to be widely identified up to species and subspecies level. In the current study, PCR-RFLP of the partial RNA polymerase β-subunit (rpoB) gene fragment was evaluated for differential identification of Bifidobacterium species. The rpoB gene partial sequences of 575 bp were amplified from 93 previously identified isolates collected from various sources of human and animal origin along with 12 standard reference strains. The PCR amplified products were digested with three restriction endonucleases HhaI, HinfI and BanI separately. Dendrograms constructed from the patterns of HhaI, were found to be more discriminatory and successfully differentiated all the twelve species and also at sub-species level in between B. longum subsp. longum and B. longum subsp. infantis. However, B. adolescentis and B. pseudocatenulatum group clusters were not separated and represented by one group. The groups were further discriminated by HinfI restriction digestion. A separate combination thereof may be used for inferring the classification of bifidobacterial species targeted on rpoB PCR-RFLP analysis. To our knowledge, this work is the first report based on use of rpoB PCR-RFLP for discrimination of the isolates of genus Bifidobacterium and also provides insights into specific advantages of this method over hsp60 PCR-RFLP in differentiating B. longum subsp. longum and B. longum subsp. infantis.
Bifidobacterium, PCR-RFLP, HhaI, rpoB gene, probiotics
Gastrointestinal tract comprises of a complex ecosystem where intestinal microbiota has been revealed to exert numerous metabolic, nutritional, physiological and immunological effects to human and animal health.1 The microbiota is further modulated by the effect of probiotics (live supplemented microorganisms) for maintaining and improving the health integrity.2-5 Amongst these microorganisms, genus Bifidobacterium which are generically defined as Gram-positive, anaerobic and bifid shaped bacteria are found in the gastrointestinal tract (GIT) of humans and animals.4-7 Many species were recognized and classified under this genus after it was designated as an independent taxon by Orla-Jensen.8,9 A few of the species among this genus are important contributors to the gut microbiome which have focused interest in characterization up to species and subspecies level due to their potential use as probiotic organisms.10-12 The classification is important in the food and pharmaceutical industries as some of developed products require specific strains. Thus, the immense contributions to human health13 imposed for precise taxonomic identification and molecular based discrimination of genus Bifidobacterium.
Traditional techniques for identification of bifidobacterial species have limited application due to low reproducible and indecisive results.14 The current understanding on the complexity and diversity within the genus Bifidobacterium has targeted the use of sophisticated molecular techniques for the differentiation of species and subspecies. Stackebrandt and Ludwig15 reported that DNA hybridization >70% between two species possessed more than 97% 16S rRNA sequence similarity. However, 16S-based classification of genus Bifidobacterium is sometimes not discriminative enough to allow differentiation between certain species.16 The rate of evolutionary substitution in protein-encoding genes has higher importance and provides improved resolution than those in 16S rRNA genes.17,18 So, modern taxonomic approaches classified bifidobacterial phylogeny based on these highly conserved protein coding genes like tuf, atpD, recA, dnaK.19 In microbial ecological studies, the use of a single copy gene with increased resolution has highlighted enhanced phylogenetic relationships and diversity measurements at subspecies level in comparison to 16S rRNA gene.20 Among these conserved macromolecules, rpoB gene encoding the RNA polymerase β-subunit has come up into view as one of the major genes which is correlated with distinct phylogenetic analysis and identification of firmly related bacterial species.21 Adekambi et al.22 proposed entire rpoB gene sequencing as a suitable approach to DNA-DNA hybridization technique for differentiation of bacterial genus and species. In recent decades, PCR-RFLP technique of rpoB gene has been implemented for species identification in genera viz. Legionella,23 Mycobacterium,24 Cronobacter,25 Leptospira,26 Lactic acid bacteria27 etc. Kim et al.28 reported that rpoB gene sequences are appropriate molecular markers for differentiation and determining the relationships of bifidobacterial species. Mainly this gene is responsible for rifaximin resistance in Bifidobacterium species.29 So far, no information is available with reference to the context of PCR-RFLP of rpoB gene in genus Bifidobacterium. Therefore, in the current study we have tried to build up a PCR-RFLP based identification strategy targeting partial rpoB gene sequence of bifidobacterial isolates, isolated from different sources of human and animal origin.
Bifidobacterial isolates and standards used in the study
Previously identified 93 bifidobacterial isolates comprised of 59 isolates from human sources (milk samples and faeces) and 34 isolates from animal origin (rumen samples, chicken faeces) as reported in our earlier study of hsp60 PCR-RFLP analysis 30 were included in this work. Standard strains representing 12 different Bifidobacterium species were procured from Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (DSMZ, Germany) were included for validation, as listed in Table 1. Standard strains and our isolated 93 isolates were cultured and revived in M-58 medium whenever required, and stored supplemented with 10% glycerol (v/v) at -80 °C.
Table (1):
Standard strains of the Bifidobacterium species included in this study.
No |
Species |
Origin |
Strain No. |
|---|---|---|---|
1. |
B. longum subsp. longum |
Intestine of adult |
DSM-20219 |
2. |
B. boum |
Bovine rumen |
DSM-20432 |
3. |
B. pseudocatenulatum |
Sewage |
DSM-20439 |
4. |
B. pullorum |
Chicken faeces |
DSM-20433 |
5. |
B. longum subsp. infantis |
Intestine of infant |
DSM-20088 |
6. |
B. breve |
Intestine of infant |
DSM-20213 |
7. |
B. animalis |
Chicken faeces |
DSM-20105 |
8. |
B. adolescentis |
Intestine of adult |
DSM-20083 |
9. |
B. bifidum |
Breast-fed infant faeces |
DSM-20456 |
10. |
B. catenulatum |
Human faeces |
DSM-16992 |
11. |
B. thermophilum |
Pig faeces |
DSM-20210 |
12. |
B. merycicum |
Rumen of cattle |
DSM-6492 |
Genomic DNA extraction
Genomic DNA extraction was carried out from the cultures following the protocols of Brookman and Nicholson,31 with some modifications. Bacterial strains were grown in Bifidobacterium medium (DSMZ- specification) and conditions as described in our previous study.30 After incubation, the bacterial pellets were collected by centrifugation at 10,000 rpm for15 min, and to it 800 µL of CTAB buffer (Cetyl trimethylammonium bromide) was added and mixed thoroughly. The suspension was kept for 1 h at 70°C and then mixed thoroughly. Then around 500 µL of chloroform: isoamyl alcohol (24:1) was added to the suspended cells and blended to form a white emulsion. In the subsequent stage centrifugation was done at 10,000 rpm for 20 min. The aqueous layer was moved and DNA was precipitated with addition of 300 µL of isopropanol and centrifuged at 10,000 rpm for 10 min). At that point 500 µL of 70% ethanol was added to the white pellet, mixed and afterward kept at 60°C for 10 min in an incubator.
Air dried pellet was dissolved in 60 µL TE buffer [10 mmol Tris-HCl and 1mM EDTA (pH 8.0)]. The quality of the isolated DNA was checked by 0.8% agarose gel electrophoresis and quantified by Nanodrop spectrophotometer (ND-1000; V3.5.2, Nanodrop Technology, Cambridge, UK). For working conditions, measurement was trailed by weakening DNA in Milli Q water. For working conditions, quantification was followed by diluting DNA in Milli Q water.
Amplification of rpoB gene
Amplification of rpoB gene fragment with primers Bilon-rpoB-F- 5’-AGACCGACAGCTTCGATTGG-3’; Bilon-rpoB-R 5’-AACACGATGGCGGACTGCTT-3’ (Sigma, Aldrich) were carried out as mentioned by Makino et al.32 The composition of PCR mixture (50 μL) was 10X buffer (10 mM Tris-HCl (pH 8.3), 1.5 mM MgCl2 (Genei, Bangalore), 50 mM KCl, 200 μM each dNTP (Genei, Bangalore), template DNA (50 ng), primer (20 pmol each), Taq DNA polymerase (1 unit Sigma, Aldrich) and the volume was made up with sterile PCR grade water. The reaction was subjected to 35 cycles of amplification: initial denaturation 95°C for 8 min, denaturation 95°C for 45 sec, annealing 57°C for 30 sec, extension 72°C for 1 min and this was completed by final extension of 72°C for 10 min. Amplified PCR products were separated on 1.2% agarose gel (Sigma, Aldrich) containing 10 µg/mL ethidium bromide in 1X TBE electrophoresis buffer (10.8 g/L Trizma base (Sigma, Aldrich), 5.5 g/L Boric acid (SRL Ltd., Mumbai), 2.0 ml/L 0.5 M EDTA at 100 V. 100 bp DNA ladder (Promega, #G2101) was used as a molecular base size marker. Photographs of the amplified products were captured using Syngene G-Box, UK gel documentation system.
PCR-RFLP and dendrogram construction
Appropriate restriction enzymes were identified using different accessible nucleotide sequences from NCBI database through Cleaver software (http://cleaver.sourceforge.net/) so as to evaluate different restriction polymorphisms. HhaI (GCG↓C), HinfI (G↓ANTC) and BanI (G↓GYRCC) were selected as appropriate enzymes which were expected to give up distinct and sufficient fragment sizes to produce species specific restriction profiles. Restriction digestion was performed in a reaction volume of 20 µL having HhaI (1unit, Gene Mark; #E143) 0.05 µL; 10X Y buffer 2 µL; BSA 2 µL (0.1%); rpoB gene amplified PCR product (1 µg approx). For HinfI and BanI the restriction digestion mixture was prepared having 0.1 µL HinfI (Takara, Shiga, Japan; #1238A) and BanI (1unit, Thermo Scientific; #ER1001), 2 µL 10X H buffer and the rpoB gene amplified PCR product (approximately 1 µg.). A total volume of 20 µL was adjusted with Milli Q water (sterile) for the above reactions and incubated for 2 h at 50°C in HhaI digestion and 37°C for HinfI and BanI digestion respectively. After the incubation period completion, the digested PCR products were separated using 3% agarose gel (1X TBE) for 4 h supplemented with ethidium bromide at a final concentration of 10 µg/mL and photographs of the patterns were visualized under gel documentation system. 50 bp DNA ladder (Thermo Fisher Scientific; #SM0371) was used as a molecular marker for estimation of migration distance of individual bands of DNA fragments. RFLP restriction patterns generated from enzyme digestion of all the isolated bifidobacterial isolates and reference standard strains of DSMZ were analysed.
To estimate the group specific positions of the isolates, and their similarity with standard reference strains, fingerprinting profiles generated with each enzyme (HhaI, HinfI and BanI) were evaluated with the restriction profiles of 12 reference cultures. Based on the RFLP fingerprints, binary matrixes were generated using Gel Quest software for each enzyme separately. The restriction fragments which are shorter than 40 bp were not included in this analysis. The data matrices were transferred to MEGA6 software33 including the standards to estimate distances and the Unweighted Pair Group Method with Arithmetic averages (UPGMA) was implemented for cluster analysis.
Amplification and PCR-RFLP of rpoB gene
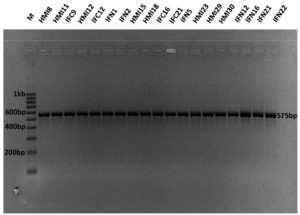
The DNA extracted from the standard cultures amplified with rpoB gene primers (Bilon-rpoB-F and Bilon-rpoB-R) produced amplicons of 575 bp size in all the isolates. The rpoB amplification of bifidobacterial isolates has been depicted in Fig. 1 and PCR-RFLP fingerprinting patterns of standard cultures with HhaI, HinfI and BanI are shown in Fig. 2, 3, 4 respectively.
Fig. 1. Agarose gel photographs showing rpoB gene amplification of bifidobacterial isolates (M-100 bp DNA size ladder; Lane 1-19 respective isolates)
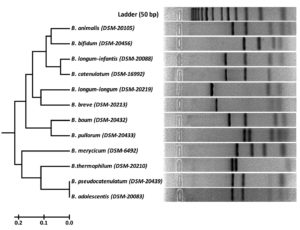
Fig. 2. RFLP fingerprints of rpoB PCR amplified products with HhaI (GCG/C) of standard reference isolates.
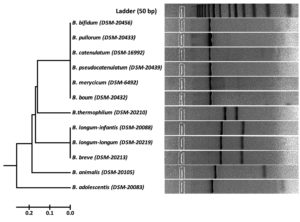
Fig. 3. RFLP fingerprints of rpoB PCR amplified products with HinfI (G/ANTC) of standard reference isolates.
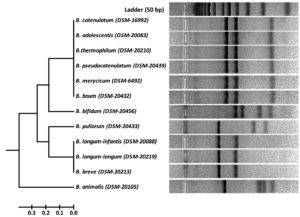
Fig. 4. RFLP fingerprints of rpoB PCR amplified products with BanI (G/GYRCC) of standard reference isolates.
Dendrogram Analysis
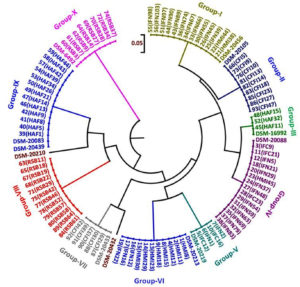
Three dendrograms in respect to each enzyme (HhaI, HinfI and BanI) were constructed on the basis of digested amplified rpoB PCR products. HhaI restriction enzyme based fingerprinting patterns of the isolates with standard cultures produced 10 different clusters (Fig. 5). The study conducted on the constructed dendrogram showed 13 isolates clustering with B. bifidum in group-I. Nine isolates showed similarity pattern with B. animalis in group-II. Three isolates grouped together with B. catenulatum in group-III whereas a total of 15 isolates were clustered under B. longum subsp. infantis in group-IV. Group-V contained 4 isolates which showed similarity outline with B. longum subsp. longum. The digestion performed with standard strains depicted the clear differentiation between B. longum subsp. longum and B. longum subsp. infantis. Eleven isolates with similar RFLP patterns clustered with B. breve in group-VI. Five isolates grouped to B. pullorum in group-VII. In group-IX both B. adolescentis and B. pseudocatenulatum were grouped together. Eight isolates clustered with B. merycicum in Group-X. Group-VIII contained same 12 isolates as obtained in a separate group in hsp60 which were identified as B. ruminantium in our previous study.30 None of our isolates grouped with the restriction patterns of B. boum and B. thermophilum included from the standard isolates list.
Fig. 5. Dendrogram constructed from RFLP fingerprints of amplified rpoB PCR products with HhaI (GCG/C).
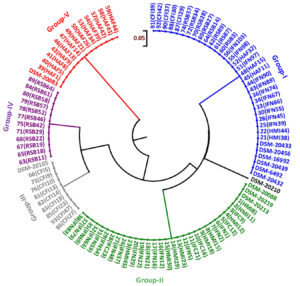
Fig. 6. Dendrogram constructed from RFLP fingerprints of amplified rpoB PCR products with HinfI (G/ANTC).
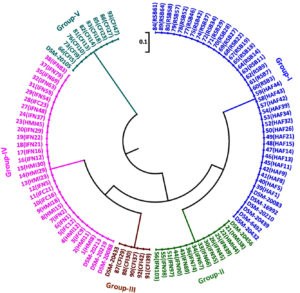
Cluster analysis of HinfI based fingerprinting patterns yielded 5 different clusters (Fig. 6). Dendrogram showed a total of 29 isolates clustering with B. bifidum, B. catenulatum, B. pullorum, B. merycicum, B. boum and B. pseudocatenulatum in Group-I. Thirty isolates grouped mutually with B. breve, B. longum subsp. longum and B. longum subsp. infantis having same restriction patterns in Group-II. Nine isolates showed a same similarity pattern with B. animalis species in Group-III and 13 isolates were clustered under B. adolescentis in Group-V. Group-IV contained 12 isolates which did not cluster with any of the standard strain. It was observed that none of our isolated species grouped with B. thermophilum. Grouping of two to three species in a cluster was found in HinfI restriction digestion. To some extent HinfI restriction endonuclease does not provide sufficient discriminatory power for differentiation of species and subspecies. Most of the species can be identified by using a single enzyme, HhaI except for a few. But genetically it may be possible that close subspecies and species can be distinguished by rpoB PCR-RFLP with the combination of these two enzymes. BanI based fingerprinting patterns of our isolates with standard cultures yielded 5 different clusters (Fig. 7) in the dendrogram. The study conducted on the constructed dendrogram showed 36 isolates clustered with B. catenulatum, B. adolescentis, B. pseudocatenulatum, B. boum, B. merycicum and B. thermophilum in Group-I. Thirteen isolates grouped with B. bifidum in Group-II. Five isolates displayed a same resemblance pattern with B. pullorum in Group-III and 30 isolates were clustered under B. breve, B. longum subsp. infantis and B. longum subsp. longum in Group-IV. Group-V contained 9 isolates which exhibited similarity pattern with B. animalis. Group specific band patterns estimated through Gel Quest software for each enzyme are represented in Table 2.
Table (2):
Group wise estimated band patterns obtained from rpoB PCR-RFLP with respect to each restriction enzyme (approximate DNA fragment size ± 5bp) of the isolated strains.
| Groups | HhaI | HinfI | BanI | |||
|---|---|---|---|---|---|---|
| Isolates | Band patterns | Isolates | Band patterns | Isolates | Band patterns | |
| Group-I | 13 | 157-106-64-52 | 29 | 572 | 36 | 265-203 |
| Group-II | 9 | 221-155-76-52 | 30 | 363-210 | 13 | 203-178-110-76 |
| Group-III | 3 | 272-155-76 | 9 | 464-112 | 5 | 350-131-85 |
| Group-IV | 15 | 272-154-77-51 | 12 | 464-93 | 30 | 352-203 |
| Group-V | 4 | 412-77-52 | 13 | 518-57 | 9 | 282-110-89-76 |
| Group-VI | 11 | 364-76-54 | ||||
| Group-VII | 5 | 159-138-64-56-43 | ||||
| Group-VIII | 12 | 220-153-92-74-41 | ||||
| Group-IX | 13 | 221-166-52 | ||||
| Group-X | 8 | 196-138-85-43 | ||||
In the investigation, attempts have been made to demonstrate the efficacy of rpoB PCR-RFLP method in discrimination of closely related Bifidobacterium up to subspecies level. Baffoni et al.34 reported that B. longum subsp. infantis and B. longum subsp. longum were not identified individually in other genes like hsp60 PCR-RFLP. But in rpoB PCR-RFLP with HhaI enzyme clear differentiation in between B. longum subsp. infantis and B. longum subsp. longum was observed (Fig. 2). In another study it was difficult to differentiate in between both the species of B. longum using 16S rRNA based species specific and group specific primers.35 Sakata et al.36 unified B. infantis, B. longum and B. suis into a single species but Mattarelli et al.37 reclassified the 3 biotypes of B. longum as 3 subspecies. Deletoile et al.38 reported MLSA/MLST method with seven housekeeping genes including rpoB, for strain typing in Bifidobacterium species and showed clearly demarcated clusters and phylogenetic distinctness of B. longum sub-species. Although all the isolates were distinguished into separate groups the two species B. adolescentis and B. pseudocatenulatum in our study displayed identical restriction patterns which needs successive digestion with other enzyme for both species’ differentiation. In our study, B. pseudocatenulatum and B. catenulatum were distinguished from the restriction profiles in HinfI restriction digestion in spite of having more than 90% similarity in 16S rRNA gene sequence in taken into account. Kim et al.28 reported that rpoB and hsp60 gene sequences based phylogenetic trees showed more discriminative patterns than the 16S rRNA gene tree. Their finding was especially discernible among related species such as B. pseudocatenulatum and B. catenulatum, B. saeculare, B. gallinarum and B. pullorum and B. pseudolongum subsp. pseudolongum and B. pseudolongum subsp. globosum. The authors also presented hsp60 based results which provided clear differentiation between these closely related groups. This substantiated the results of Jian et al.43 and the present study with respect to discrimination of B. pseudocatenulatum and B. catenulatum species in different groups.
Matsuki et al.35 observed that it is very difficult to differentiate B. adolescentis, B. catenulatum and B. pseudocatenulatum based on the conventional sugar fermentation pattern which puzzled the identification between B. catenulatum group and B. adolescentis. Kim et al.28 reported 351 bp partial rpoB gene sequences to differentiate species of the genus Bifidobacterium which proved to be a suitable marker and based on this B. pseudocatenulatum and B. catenulatum were evidently discriminated. But the authors didn’t state their discrimination with regard to B. longum subsp. longum and B. longum subsp. infantis. Targeted pyrosequencing method of rpoB amplicons (351 bp) revealed the low number of Bifidobacterium species with major incidence of B. longum and B. breve species in the antibiotic-treated infants. 39 In multigenic approach,40 rpoB gene can be a gene of choice for providing detailed molecular analysis due to its easy accessibility and high resolving nucleotide-based molecular typing system.28 Ventura et al.41 reported that single gene trees may not provide adequate phylogenetic relationships because of horizontal gene transfer events, inappropriate mutation rates and variable recombination rates. On the other hand, Vos et al.42 reported that single-copy protein-encoding genes are of benefits in higher sampling efficiency, error-correction and easy survey of homologous recombination rate and mutations effects. But Ventura et al.17 reported that multigene concatenation approach comprising of conserved orthologous proteins to be more discriminatory for differentiation of bifidobacterial species. However, rpoB gene sequences present improved phylogenetic resolution over the 16S rRNA gene.21 So, rpoB gene can be used as a sequence of choice in case of ambiguity when other gene are taken into account for identification.
PCR-RFLP based on rpoB amplified partial gene product can be successfully applied to differentiate in between different species of genus Bifidobacterium as well as at sub-species level. The study also indicates that among the restriction enzymes HhaI, was found to be more discriminatory in differentiating different species of the genus as well as at sub species level. In addition to that whenever a single restriction enzyme fails to differentiate in between different species of the genus additional enzymes thereof may be used for inferring the classification of bifidobacterial species targeted on rpoB PCR-RFLP analysis.
ACKNOWLEDGMENTS
The authors would like to thank ICAR-National Dairy Research Institute, Karnal, Haryana, India for the financial support.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
ETHICS STATEMENT
This article does not contain any studies with human participants or animals performed by any of the authors.
AVAILABILITY OF DATA
All datasets generated or analysed during this study are included in the manuscript.
- Gerritsen J, Smidt H, Rijkers GT, de Vos WM. Intestinal microbiota in human health and disease: the impact of probiotics. Genes and Nutr. 2011;6(3):209-240.
Crossref - Gareau MG, Sherman PM, Walker WA. Probiotics and the gut microbiota in intestinal health and disease. Nat Rev Gastroenterol Hepatol. 2010;7(9):503-514.
Crossref - Walsh CJ, Guinane CM, O’Toole PW, Cotter PD. Beneficial modulation of the gut microbiota. FEBS Lett. 2014;588(22):4120-4130.
Crossref - Jena R, Choudhury PK, Puniya AK, Tomar SK. Efficacy of BOX-PCR fingerprinting for taxonomic discrimination of bifidobacterial species isolated from diverse sources. 3 Biotech. 2021;11(6):270.
Crossref - Singh A, Kumar S, Vinay VV, et al. Autochthonous Lactobacillus spp. isolated from Murrah buffalo calves show potential application as probiotic. Curr Res Biotechnol. 2021;3:109-119.
Crossref - Leahy SC, Higgins DG, Fitzgerald GF, van Sinderen D. Getting better with bifidobacteria. J Appl Microbiol. 2005;98(6):1303-1315.
Crossref - Jaglan N, Kumar S, Choudhury PK, Tyagi B, Tyagi AK. Isolation, characterization and CLA production potential of bifidobacterial isolates from ruminal fluid samples of Murrah buffaloes. Anaerobe. 2019;56:40-45.
Crossref - Orla-Jensen S. La Classification des bacte’ries lactiques. Lait. 1924;4:468-474.
Crossref - Ventura M, Turroni F, Ribbera A, Foroni E, van Sinderen D. Basic biology: Biology of effector organisms for probiotic and replacement therapy. 4. Bifidobacteria: the model human gut commensal, In: Versalovic J, Wilson M, Editors. Therapeutic Microbiology: probiotics and related strategies. American Society for Microbiology, Washington, DC; 2008:35-50.
Crossref - Qiao H, Duffy LC, Griffiths E, et al. Immune responses in rhesus rotavirus challenged BALB/c mice treated with bifidobacteria and prebiotic supplements. Paediatric Research. 2002;51(6):750-755.
Crossref - Stanton C, Ross RP, Fitzgerald GF, Van Sinderen D. Fermented functional foods based on probiotics and their biogenic metabolites. Curr Opin Biotechnol. 2005;16(2):198-203.
Crossref - Awasti N, Tomar SK, Pophaly SD, et al. Probiotic and functional characterization of bifidobacteria of Indian human origin. J Appl Microbiol. 2016;120(4):1021-1032.
Crossref - O’Callaghan A, van Sinderen D. Bifidobacteria and their role as members of the human gut microbiota. Front Microbiol. 2016;7:925.
Crossref - Ventura M, van Sinderen D, Fitzgerald GF, Zink R. Insights into the taxonomy, genetics and physiology of bifidobacteria. Antonie van Leeuwenhoek. 2004;86(3):205-223.
Crossref - Stackebrandt E, Ludwig W. The importance of choosing outgroups references organisms in phylogenetic studies: the Atopobium case. Syst Appl Microbiol. 1994;17(1):39-43
Crossref - Miyake T, Watanabe K, Watanabe T, Oyaizu H. Phylogenetic analysis of the genus Bifidobacterium and related genera based on 16S rDNA sequences. Microbiol Immunol. 1998;42(10):661-667.
Crossref - Ventura M, Canchaya C, Del Casale A, et al. Analysis of bifidobacterial evolution using a multilocus approach. Int J Syst Evol Microbiol. 2006;56(12):2783-2792.
Crossref - Ogier JC, Pages S, Galan M, Barret M, Gaudriault S. rpoB, a promising marker for analyzing the diversity of bacterial communities by amplicon sequencing. BMC Microbiol. 2019;19(1):171.
Crossref - Solano-Aguilar G, Dawson H, Restrepo M, Andrews K, Vinyard B, Urban Jr JF. Detection of Bifidobacterium animalis subsp. lactis (Bb12) in the intestine after feeding of sows and their piglets. Appl Environ Microbiol. 2008;74(20):6338-6347.
Crossref - Case RJ, Boucher Y, Dahllof I, Holmstrom C, Doolittle WF, Kjelleberg S. Use of 16S rRNA and rpoB genes as molecular markers for microbial ecology studies. Appl Environ Microbiol. 2007;73(1):278-288.
Crossref - Adekambi T, Drancourt M, Raoult D. The rpoB gene as a tool for clinical microbiologists. Trends in Microbiol. 2009;17(1):37-45.
Crossref - Adekambi T, Shinnick TM, Raoult D, Drancourt M. Complete rpoB gene sequencing as a suitable supplement to DNA-DNA hybridization for bacterial species and genus delineation. Int J Syst Evol Microbiol. 2008;58(8):1807-1814.
Crossref - Ko KS, Hong S-K, Lee K-H, et al. Detection and identification of Legionella pneumophila by PCR-restriction fragment length polymorphism analysis of the RNA polymerase gene (rpoB). J Microbiol Methods. 2003;54(3):325-337.
Crossref - Ong C-S, Ngeow YF, Yap SF, Tay ST. Evaluation of PCR-RFLP analysis targeting hsp65 and rpoB genes for the typing of mycobacterial isolates in Malaysia. J Med Microbiol. 2010;59(11):1311-1316.
Crossref - Vlach J, Javurkova B, Karamonova L, Blazkova M, Fukal L. Novel PCR-RFLP system based on rpoB gene for differentiation of Cronobacter species. Food Microbiol. 2017;62:1-8.
Crossref - Jung LRC, Bomfim MRQ, Kroon EG, Nunes AC. Identification of Leptospira serovars by RFLP of the RNA polymerase beta subunit gene (rpoB). Braz J Microbiol. 2015;46(2):465-476.
Crossref - Claisse O, Renouf V, Lonvaud-Funel A. Differentiation of wine lactic acid bacteria species based on RFLP analysis of a partial sequence of rpoB gene. J Microbiol Methods. 2007;69(2):387-390.
Crossref - Kim BJ, Kim H-Y, Yun Y-J, Kim B-J, Kook Y-H. Differentiation of Bifidobacterium species using partial RNA polymerase β-subunit (rpoB) gene sequences. Int J Syst Evol Microbiol. 2010;60(12):2697-2704.
Crossref - Vitali B, Turroni S, Serina S, et al. Molecular and phenotypic traits of in-vitro-selected mutants of Bifidobacterium resistant to rifaximin. Int J Antimicrob Agents. 2008;31(6):555-560.
Crossref - Jena R, Choudhury PK, Puniya AK, Tomar SK. Isolation and species delineation of genus Bifidobacterium using PCR-RFLP of partial hsp60 gene fragment. LWT-Food Science and Technology. 2017;80:286-293.
Crossref - Brookman JL, Nicholson MJ. Anaerobic fungal populations. In: Makkar HPS, McSweeney CS, Editors. Methods in Gut Microbial Ecology for Ruminants, IAEA, Netherlands; 2005:139-150.
Crossref - Makino H, Kushiro A, Ishikawa E, et al. Transmission of intestinal Bifidobacterium longum subsp. longum strains from mother to infant, determined by multilocus sequencing typing and amplified fragment length polymorphism. Appl Environ Microbiol. 2011;77(19):6788-6793.
Crossref - Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30(12):2725-2729.
Crossref - Baffoni L, Stenico V, Strahsburger E, et al. Identification of species belonging to the Bifidobacterium genus by PCR-RFLP analysis of a hsp60 gene fragment. BMC Microbiol. 2013;13:149.
Crossref - Matsuki T, Watanabe K, Tanaka R, Oyaizu H. Rapid identification of human intestinal bifidobacteria by 16S rRNA-targeted species and group specific primers. FEMS Microbiol Lett. 1998;167(2):113-121.
Crossref - Sakata S, Kitahara M, Sakamoto M, Hayashi H, Fukuyama M, Benno Y. Unification of Bifidobacterium infantis and Bifidobacterium suis as Bifidobacterium longum. Int J Syst Evol Microbiol. 2002;52(2):1945-1951.
Crossref - Mattarelli P, Bonaparte C, Pot B, Biavati B. Proposal to reclassify the three biotypes of Bifidobacterium longum as three subspecies: Bifidobacterium longum subsp. longum subsp. nov., Bifidobacterium longum subsp. infantis comb. nov. and Bifidobacterium longum subsp. suis comb. nov. Int J Syst Evol Microbiol. 2008;58(4):767-772.
Crossref - Deletoile A, Passet V, Aires J, et al. Species delineation and clonal diversity in four Bifidobacterium species as revealed by multilocus sequencing. Res Microbiol. 2010;161(2):82-90.
Crossref - Fouhy F, Guinane CM, Hussey S, et al. High-throughput sequencing reveals the incomplete, short-term recovery of infant gut microbiota following parenteral antibiotic treatment with ampicillin and gentamicin. Antimicrob Agents Chemother. 2012;56(11):5811-5820.
Crossref - Stackebrandt E, Frederiksen W, Garrity GM, et al. Report of the ad hoc committee for the reevaluation of the species definition in bacteriology. Int J Syst Evol Microbiol. 2002;52(3):1043-1047.
Crossref - Ventura M, Canchaya C, Fitzgerald GF, Gupta RS, van Sinderen D. Genomics as a means to understand bacterial phylogeny and ecological adaptation: the case of bifidobacteria. Ant Van Leeuwenhoek. 2007;91(4):351-372.
Crossref - Vos M, Quince C, Pijl AS, de Hollander M, Kowalchuk GA. A comparison of rpoB and 16S rRNA as markers in pyrosequencing studies of bacterial diversity. PLoS ONE. 2012;7(2):e30600.
Crossref - Jian W, Zhu L, Dong X. New approach to phylogenetic analysis of the genus Bifidobacterium based on partial hsp60 gene sequences. Int J Syst Evol Microbiol. 2001;51(5):1633-1638.
Crossref
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.