ISSN: 0973-7510
E-ISSN: 2581-690X
This study aimed to: (1) assess Escherichia coli contamination in polony, beef burgers and traditionally fermented cow milk from the formal and informal markets in Harare, Zimbabwe, (2) determine the antibiotic sensitivity of Escherichia coli isolates, and (3) identify Shiga-toxin producing Escherichia coli isolates using the presence of virulence genes, namely, intimin, enterohemolysin A and Shiga toxins 1 and 2. Ninety-six samples comprising 32 beef polony slices, 32 beef burger patties, and 32 fermented milk specimens were obtained from the informal and formal outlets of the central business district. Escherichia coli occurred in 20 (21%) of the samples, being more prevalent in the informal (29%) than in the formal (13%) market. Of the 20 E. coli isolates, 6 (30%) were Shiga-toxin producing E. coli, and the rest (70%) were negative for virulence genes. The predominance of Escherichia coli was greater in meat products (25%) than in fermented milk (13%). Total Escherichia coli counts were not substantially different between formal and informal markets (t-test: p=0.08). All the E. coli isolates were multidrug-resistant with antimicrobial resistance prevalence ranging from 25% for Sulphamethoxazole to 100% for Penicillin and Erythromycin. The presence of E. coli in food indicates faecal contamination and probable existence of other enteric pathogens. The presence of virulent and antimicrobial-resistant E. coli strains in food threatens food safety and public health. We conclude that ready-to-eat animal products from both informal and formal sectors could result in the dissemination of antimicrobial-resistant Escherichia coli species if corrective measures are not taken.
Shiga-toxin producing E.coli, Escherichia coli, ready-to-eat animal products, antimicrobial resistance
Food contaminated with pathogenic microorganisms is unsafe and represents a danger to food safety and community well-being, especially in developing countries. Escherichia coli (E. coli) is among the most prevalent food-borne pathogenic agents1. A large proportion of resistant and pathogenic E. coli causing human infections are mainly derived from meat and milk products2. Escherichia coli O157:H7 and several other E. coli strains including Shiga-toxin-producing E. coli (STEC) strains (serogroups O45, O121, O26, O111, O103, O145 etc.) are important pathogenic infectious agents causing many morbidities and mortalities in people of both the developed and developing world3.
Pathogenicity of several Escherichia coli species is generally founded on their virulence properties such as enterotoxigenic E. coli (ETEC), enteropathogenic E. coli (EPEC), enterohaemorrhagic E. coli (EHEC) and STEC4. Virulence in Escherichia coli results from genomic or plasmid virulence genes5. Genes exist individually or polygenically in clinical isolates of varying frequency6. The presence of virulence genes such as Shiga-toxins (stx1 and stx2), hemolysin (hlyA) and intimin (eaeA) in E.coli strains of ready-to-eat animal products pose a serious food safety and public health concern. The virulence factors are responsible for E.coli adhesion, colonization and invasion into the gastric epithelial cells7.
Even if isolation and antimicrobial susceptibility testing are done on food samples, some bacterial infections are treated without the establishment of the causative agent, antibiogram and/or assessing virulence of the strains8.Environmental contamination leads to the perpetuation of pathogenic E. coli as well as acquiring of antimicrobial resistance (AMR) and virulence genes by non-pathogenic Escherichia coli. Lack of information on antibiotic resistance and prevalent virulence gene combination profiles of pathogenic microbes compromises the effectiveness of antibiotic therapy9. Several types of research found that insufficient evaluation in the use of antibiotics during the treatment of human / animal infections, prophylactic and growth stimulation in animal production can confer resistance in various bacteria10-14. The emergence of AMR and multidrug-resistant strains render the management of pathogenic bacteria increasingly challenging. Throughout the decades, resistance to cephalosporins among Escherichia coli species has multiplied primarily due to the prevalence of Extended-spectrum β-Lactamases (ESBL).
Virulence and resistance factors are transmitted by horizontal gene transfer among organisms15. The exchange of DNA fragments is possibly the primary genetic mechanism for the spread and co-selection of resistance and virulence genes, though some modes, like the compensatory or adaptive mutations, could also be involved15. Mobile genetic components, like plasmids and transposons, have frequently been correlated with genes that encode AMR and may be interchanged across microbes belong to different evolutionary phenotypes16.Antimicrobial resistance and virulence are acquired by organisms through different evolutional routes but both processes are important for bacterial survival under hostile environments15. Virulence factors are needed for overcoming the host’s immune system and acquisition of AMR is vital for resisting antimicrobial treatments, adaptation and survival in adverse conditions15.
Food safety is really a human health concern in Zimbabwe, as demonstrated by repeated outbreaks of food-related diseases17. Like in other developing countries, there is little up-to-date statistics on the incidence, type, and virulence of food borne pathogens causing infections in humans, creating a missing link in developing strategies to control food borne diseases. It is against this background that this work seeks to document the existence of antimicrobial-resistant STEC strains in ready-to-eat animal products from the informal and formal sectors in Harare, Zimbabwe. This information is potentially valuable in developing effective intervention strategies and in educating the food industry and consumers.
The general purpose of this work was to evaluate the microbial safety of ready-to-eat meat products and sour milk from the formal and informal markets in Harare, Zimbabwe,with a focus on Shiga-toxin producing E.coli. To achieve this objective the following specific objectives were addressed:(1) to evaluate the occurrence of Escherichia coli contamination in polony, beef burgers and traditionally fermented cow milk from the formal and informal markets, (2) to assess the susceptibility of Escherichia colis trains against eight commonly used antibiotics (Ampicillin, Gentamycin, Penicillin G, Erythromycin, Tetracycline, Neomycin, Ceftriaxone, and Sulfamethoxazole) and (3) to determine STEC isolates by the existence of four virulence genetic factor, namely, enterohemolysin A (hly A), intimin (eae A) and Shiga toxins 1 and 2 (stx1 and stx2)
Study area
Harare, the commercial and administrative capital of Zimbabwe, does indeed have a populace of around two million citizens18. Apart from the typical formal food outlets (e.g. supermarkets and butcheries), foods like fruits, vegetables, and ready-to-eat meat products, are sold at informal markets dotted around the city centre. Food vendors sell their foods from vending stalls and open space. The socio-economic challenges facing Zimbabwe have compromised adherence to food safety regulations and hygiene practices by both formal and informal food outlets in Harare, potentially exposing consumers to unsafe food.
Sample collection
This study is an observational cross-sectional study in Harare Central Business Centre assessing the presence of Shiga-toxin producing and other E. coli strains from ready-to-eat animal products sold formally and informally. A total of 96 samples of ready-to-eat meat products and sour milk were randomly obtained from informal and formal traders in the central business centre, over four weeks beginning 05 January 2020. A total of 32 beef polony, 32 beef burger patties, and 32 fermented milk samples were collected from both informal (48 samples) and formal (48 samples) markets at bus termini and shopping centres in the city centre. The specimens were placed inside an ice cooler as well as immediately ferried to the Central Veterinary Laboratory for analysis.
Detection of Escherichia coli
A mass of 1 gram of each specimen was applied to 9 milliliters of peptone water broth, homogenized and incubated for 24 hours at 44°C for 24. A culture loop was inoculated separately on Blood agar and MacConkey agar plates and incubated aerobically at for 24 hours. Colonies were examined for their cultural/ morphological characteristics on the media. Large, round, red or pink colonies surrounded by a hazy boundary on MacConkey agar indicated E. coli. Gram staining and various biochemical tests such as catalase, oxidase, indole, motility, citrate, triple iron sugar, lysine, urease and carbohydrates (sugar) fermentation (inositol, lactose, xylose, inulin, glucose, maltose, mannose, sorbitol, mannitol and sucrose) were conducted to confirm whether the bacterial strains were E. coli strains or not.
Escherichia coli enumeration/ Total Escherichia coli count
A mass of 1g sample was suspended in 9ml phosphate-buffered saline, homogenized and serially diluted up to 10-6. A volume of 0.1 ml of each serial dilution was spread over Cefixime Tellurite Sorbitol-MacConkey agar and incubated for 24 h at 44°C. Suspected colonies of Escherichia coli were counted and confirmed using gram staining and biochemical tests as described above.
Antimicrobial resistance test (AMR test)
A standardized disc diffusion technique was used for antibiotic susceptibility testing of the E. coli isolates on Mueller Hinton agar (Thermo Fisher Scientific, UK) without any replicates following Clinical and Laboratory Standards Institute (CLSI)19. The test was done using eight different antibiotic discs, namely, Ampicillin (AMP 10µg), Gentamycin 30µg (CN30), Penicillin G10µg (PEN G10µg), Erythromycin 15µg (ERY15), Tetracycline 30µg (T30µg), Neomycin 10µg (NEO10), Ceftriaxone 30µg (CRO30), and Trimethoprim/Sulfamethoxazole 1.25/23.75µg (SXT 1.25/23.75) (Oxoid, Germany). The choice for the antibiotics was based on the availability of the antibiotics at the laboratory at the moment the study was done. A microbial suspension of each E. coli isolates(0.5 McFarland organism in normal saline solution) was swabbed with a sterile cotton swab onto 4mm thick Mueller Hinton agar. Disks of the above-named antibiotics were dispensed onto the plates using a disc dispenser. To the nearest millimetre, the inhibition zones were measured after a 24h incubation period at 37°C. CLSI breakpoints were used to interpret the results.
Screening for the virulence genes to detect Shiga-toxin E.coli
Total genomic DNA was extracted from 20 different forms of E. coli isolates using Zymo Research Quick-gDNA TM MiniPrep (Zymo Research Company, South Africa) package. Multiplex PCR was used to screen for the four virulence factors stx1 and 2, eaeA and hlyA. The primer sequences of virulent genes used in Multiplex PCR amplifications were obtained from the literature (Table 1). A master mix with a reaction volume of 25 µl per sample DNA template was prepared to consist of all the requisite components for DNA amplification as per Table 1 and then run on a PCR thermo-cycler (Gene PCR System 24, Perkin Elmer, USA).
Table (1):
Primer sequences and predicted lengths of the amplicons.
Primer |
Direction |
Primer Sequence (5′-3′) |
Band Size (bases) |
Reference |
|---|---|---|---|---|
hly A |
Forward |
ACGATGTGGTTTATTCTGGA |
165 |
7 |
Reverse |
CTTCACGTGACCATACATAT |
7 |
||
stx1 |
Forward |
ACACTGGATGATCTCAGTGG |
614 |
7 |
Reverse |
CTGAATCCCCCTCCATTATG |
7 |
||
Stx2 |
Forward |
CCATGACAACGGACAGCAGTT |
779 |
7 |
Reverse |
CCTGTCAACTGAGCAGCACTTTG |
7 |
||
eaeA |
Forward |
GTGGCGAATACTGGCGAGACT |
890 |
7 |
Reverse |
CCCCATTCTTTTTCACCGTCG |
7 |
The components of the reaction included a commercial master mix kit (2x concentration) (Fermentas) (containing-a PCR buffer, dNTPs, MgCl2, Taq DNA Polymerase) and primers dissolved in nuclease-free water. A total of 35 thermo-cycler reaction cycles were set up with initial denaturation of 3 min at 94°C, denaturation at 94°C for 1 min, annealing at 56°C for 50 s, elongation at 72°C for 1 min and final elongation at 72°C for 1 min. Two percent agarose gel with 5µl of ethidium bromide (10mg/ml) submerged to 4mm depth of 0.5xTBE buffer in a gel electrophoresis tank was used. A 5µl volume of PCR products (Amplicons) and negative control was mixed with 2 µl of the loading/tracking dye and loaded into the wells on the gel. Hyper ladder IV 100bp DNA (0.1 µg/µl, 50 µg) (Bioline) was employed as a molecular weight marker or ladder.
Electrophoresis was run for 1 hour at 120V on a BRL Horizontal Gel Electrophoresis Apparatus (Horizon 11.14 Life Technologies Gibco, USA) and all the gel results were viewed and photographed using the Gel Logic 100 Imaging System (Kodak, EEC) under UV trans-illumination. Analysis of the gel picture was done on the computer to generate the DNA profiles and draw conclusions about the sample by estimating the band size from the DNA ladder/maker.
Table (2):
PCR Master-mix.
Reagents |
1 Reaction (µl) |
|---|---|
RNA free dH2O |
4 |
Master Mix |
12 |
EHEC hly Primer-F |
0.5 |
EHEC hly Primer-R |
0.5 |
stx 1 Primer-F |
0.5 |
stx 1Primer-R |
0.5 |
stx 2 Primer-F |
0.5 |
stx 2 Primer-R |
0.5 |
eae A Primer-F |
0.5 |
eae A Primer-R |
0.5 |
DNA Template |
5 |
Total |
25 |
Statistical analysis
The antimicrobial-resistant test results were analysed using the CLSI breakpoints and WHONET breakpoints20. The independent samplest-test was done in R ver. 3.6.2 to test if total E. coli counts differed significantly between the two trading sectors.
Escherichia coli identification and enumeration
A total of 20(21%) morphologically and biochemically confirmed isolates of E. coli were isolated from ready-to-eat animal products. Escherichia coli was higher in the informal sector with a prevalence rate of 29% (14 out of 48) than the formal sector with a prevalence rate of 13% (6 out of 48). There was no statistically significant difference in total E.coli count between formal and informal sectors (Student t-test: p= 0.08).Polony had a higher prevalence rate of about 56% (9 out of 16) in the informal market and about 19% (3 out of 16) prevalence rate from the formal sector. Total E.coli counts ranged from 4.0 x 101 to 1.0 x 103 in the informal market and from 2.0 x 101 to 1.2 x 102 in the formal market. Escherichia coli occurred in 3(19%) of burgers from the informal sector and 1(6%) of those from the formal sector. Total E.coli counts for burgers ranged from 1.6 x 102 to 3.0 x 102 in the informal market and from less than 1.0 x 101 to 9.0 x 101 in the formal market. Escherichia coli was equally prevalent in fermented milk from the informal and formal markets occurring in 2(13%) of the samples. Total E. coli count for fermented milk ranged from 3.0 x 103 to 6.0 x 103 in the informal market and from 3.0 x 101 to 2.0 x 103 in the formal market.
Antibiotic sensetivity testing of Escherichia coli isolates
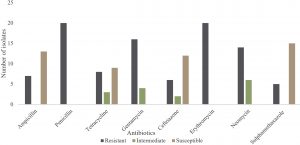
The Zone of inhibition was interpreted as resistant (R), Intermediate (I) and susceptible (S) using CLSI and WHONET breakpoints. All of the 20 Escherichia coli isolates were resistant to PEN G and ERY. About 35%, 40%, 80%, 30%, 70% and 25% of Escherichia coli strains demonstrated resistance to Ampicillin, T, CN, CRO, NEO and SXT, respectively (Fig 1). Escherichia coli isolates showed low resistance rate of 25% to CRO and SXT. Isolates susceptibility to SXT, CRO, AMP and T was 75%, 60%, 65%and 45%, respectively. SXT, CRO, AMP and T showed overlapping zones of inhibition diameter or synergy. Extended-spectrum β-lactamase producing strains constituted 43% prevalence. From the antibiotic sensitivity patterns of the Escherichia coli strains, it was observed that all the isolates were multidrug-resistant and were resistant to 3 or more antibiotics tested.
Detection of Shiga-toxin producing E. coli using virulent genes
The bands corresponding to virulence factors detected in the 14 Escherichia coli isolates are shown in Fig. 2. PCR image indicates two different band sizes of the stx2 gene. Both band 406 and 779 indicate different band sizes of the stx2 gene, which is accountable for the expression of the sxt. Band 406 of the stx2 gene was noticed in 30%(6 out of 20) Escherichia coli starins and band 779 of the stx2 gene was contained only in a single Escherichia coli strain (5%; 1 out of 20) simultaneously with stx2 gene variant band size 406. The 890 bps eae A gene (intimin expression) was present only in one out of 20 E. coli isolates simultaneously with stx2. The 614 (stx1) and the 165-bp (hly A) bands were absent, indicating a lack of the hly A gene in all the 20 E. coli isolates. Nonspecific DNA bands A and B were found below the 100bps and around the 200 bps regions, respectively. Stx2 and eaeA positive isolates have been made reference to as STEC.
Arrows A and B refer to non-specific PCR bands. Lanes A1 – A14 are the 14 E. coli isolates. Each pair of lanes A6.1 and A6.2, A10.1 and A10.2, A12.1 and A12.2 refers to two E. coli isolates differentiated by the colour of the colony but in the same culture plate. The other 6 E.coli isolates were all negative (data not shown). Lane L = 100bp DNA ladder, Lane N = negative control.
Fig. 2. Multiplex PCR of stx 1, stx 2, eae A and hlyA gene in E. coli isolates.
Prevalence of E. coli
One of the purposes of this research was to evaluate the prevalence of Escherichia coli in ready-to -eat meats and sour milk purchased throughout the informal and formal markets of Harare. Our findings indicate a higher frequency of Escherichia coli in ready-to-eat meat products sold in the informal market, especially polony, than those sold in the formal market. The high prevalence of E. coli in the informal sector can be attributed to poor food handling practices in the informal sector and a general lack of compliance to standards enshrined in the National Food Laws & Regulations of Zimbabwe21, 22. Some of the cutlery used such as knives for cutting polony may not be cleaned at all or cleaned after very long intervals resulting in the accumulation and persistence of E. coli. Such unhygienic practices may account for high food prevalence and total counts of E. coli in the informal sector. It has been confirmed that Escherichia coli could even remain alive outside the usual hosts in other environments such as soil and water for more than 90 days23. The high prevalence of E. coli indicates high chances of occurrence of E. coli associated outbreaks.
In general, the total Escherichia coli counts of samples from the formal sector were lower, probably owing to better food handling practices in the sector. Hygienic practices in the formal sector include the provision of sanitary wipes to customers at the entrance and several points within the shops, short intervals for cleaning equipment and utensils, which prevent E. coli from multiplying. Total E. coli count was lower in burgers than polony probably due to the heating of burgers during preparation, which might reduce bacterial load. Though at lower prevalence, the existence of Escherichia coli in some of the samples from the formal sector indicates possible faecal and urine contamination from food handlers.
The fermented milk samples generally showed low contamination by E. coli and this is attributed to the gram-negative lactose fermenters, which produce a bacteriocin-like inhibitory substance with a broad spectrum of antimicrobial activity against enteric pathogens24. The occurrence of Escherichia coli in animal products is a well-thought-out indicator of faecal contamination as well as the presence of other enteric pathogenic bacteria such as Salmonella species, Shigella, Yersinia and many others in the ready-to-eat animal products25. Some of the E. coli isolates were positive for ESBL, therefore, authorities need to act to combat the risk of these serotypes.
Antimicrobial resistance pattern of E. coli isolates
Overall, all Escherichia coli strains were resistant to penicillin and erythromycin. It is notable that no single antibiotic tested was 100% effective against the isolates and all the isolates were multidrug-resistant. These findings corroborate those of the previous studies8; 26. The resistance might have arisen due to the prevalent use of antibiotics, which is proportional to the development of antimicrobial resistance27. The resistance patterns of T, AMP, NEO, ERY and PEN G observed are not astounding. Since its approval in 1948, tetracycline has been commonly used for the management of disease and growth stimulation in various animal production processes28. In recent studies, Escherichia coli resistant to ESBL has been isolated from food and UTI clients in several parts of Zimbabwe26. Due to the variability of the affinity of ESBLs for different substrates, the resistance of isolates to CRO with specific zones of inhibition diameter indicates ESBL production29. Penicillin G displays a fairly low antibiotic efficacy towards Gram-negative bacteria, as the development of ESBL denotes resistance to all beta-lactam antibiotics except certain carbapenems and cephamycins30. ESBL / ampC encoding plasmids may also bear genes that encode resistance to other classes of antibiotics, such as fluoroquinolones, aminoglycosides and sulphonamides31, hence the high resistance to PEN G, ERY, NEO, and AMP, as well as resistance to T, SXT and CRO in some of the isolates32. The synergistic effect observed for SXT, CRO, AMP and T suggests the effectiveness of their combination. Incorporation of plant essential oils in the treatment administration/ regiment together with the conventional antibiotics is required if antimicrobial resistance is to be reduced33.
Detection of STEC using virulence genes
STEC is the most toxic of all DEC (Diarrheagenic Escherichia coli) species and the most dangerous STEC is O157:H7 serotype. Even so, in recent years , the incidence of many non-O157 serotypes in humans associated with the intake of tainted food has increased36. The presence of the Aeromonas species variant of the stx2 (Shiga toxin expressing gene: 406bp) in E. coli indicates gene exchange between E. coli and Aeromonas species9. In several studies,the species of Escherichia coli and Aeromonas were simultaneously isolated from food, faeces and urine samples of patients with uncomplicated UTIs. Despite the presence of stx gene, most of the isolates from this study, are unlikely to cause any serious infection due to lack of hly A and eaeA virulence factors except one Shiga-toxin E.coli with eaeA virulence gene positive. Some virulent genes in addition to stxare needed to cause severe human diseases34,35. The six (30%) strains of STEC identified indicated low prevalence.
Presence of E. coli in ready-to-eat meat products and fermented milk from the informal sector suggests poor observance of standard food handling practice, lack of personal hygiene and microbial contamination of the environment. Buying ready-to-eat meaty products and fermented milk from the informal sector poses a higher risk of E. coli infections to consumers. The proportion of antimicrobial-resistant strains of Shiga-toxin producing E. coli and other E. coli isolates from ready-to-eat animal products suggests a major threat to food safety and public health in Zimbabwe. Our findings highlight that there is a higher risk of contracting food-borne E. coli infections from consuming ready-to-eat meat products and fermented milk from the informal market than from the formal market.
Recommendations
Rigorous food safety checks by the food regulatory authorities are critical in both sectors to reduce public health risks.
There is a need to discontinue the use of antibiotics to which E. coli has developed resistance and replace them with their derivatives or natural plants with antimicrobial properties.
ACKNOWLEDGMENTS
We would like to extend our gratitude to Dr Pious Vengesayi Makaya, Managing Director of Central Veterinary Laboratories for providing reagents, and allowing us access to equipment used in this study.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
WMG designed the experiments, performed the experiments, analyzed data,drafted the manuscript, compiled information from the literature, and designed the figures and tables. TM and CG designed the experiments, performed the experiments, analyzed data, drafted the manuscript, compiled information from the literature, supervised and reviewed the manuscript. JM drafted the manuscript, compiled the information from the literature and reviewed the manuscript. TCH reviewed the manuscript. All authors read and approved the manuscript.
FUNDING
None.
ETHICS STATEMENT
This article does not contain any studies with human participants or animals performed by any of the authors.
AVAILABILITY OF DATA
All datasets generated or analyzed during this study are included in the manuscript.
- Osservasalute M. Health status and quality of the Italian regions. Report. 2009;175-177.
- Collignon P, Powers JH, Chiller TM, Aidara-Kane A, Aarestrup FM. World Health Organization Ranking of Antimicrobials According to their Importance in Human Medicine: A Critical Step for Developing Risk Management Strategies for the use of Antimicrobials in Food Production Animals. Clinical Infectious Diseases. 2009;49: 132- 141.
Crossref - FAO/WHO. Food safety risk analysis: A guide for national food safety authorities. FAO, Food and Nutrition Paper. Report. 2006.
- Barnes JH, Vaillancourt JP, Gross WB. Colibacillosis: Diseases of Poultry; Iowa State University Press; Ames: 11th ed. 2003;631-652.
- Dozois CM, Daigle F, Curtiss R. Identification of pathogen-specific and conserved genes expressed in vivo by an avian pathogenic Escherichia coli strain. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:247-252.
Crossref - Delicato ER, de Brito BG, Gaziri LC, Vidotto MC. Virulence-associated genes in Escherichia coli isolates from poultry with colibacillosis. Vet Microbiol. 2003;94:97-103.
Crossref - Farrokh C, Jordan K, Auvray F, et al. Review of Shiga-toxin-producing Escherichia coli (STEC) and their significance in dairy production. International Journal of Food Microbiology. 2013;162:190 – 212.
Crossref - Saidi B, Mafirakureva P, Mbanga J. Antimicrobial resistance of Escherichia coli isolated from chickens with colibacillosis in and around Harare, Zimbabwe. Avian Diseases. 2013;57:152-154.
Crossref - Alperi A, Figueras MJ. Human isolates of Aeromonas spp. possess Shiga toxin genes (stx1 and stx2) highly similar to the most virulent gene variants of Escherichia coli. Clinical Microbiology and Infection. 2010;16:1563-1567.
Crossref - Thomas CM, Nielsen KM. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat Rev Microbiol. 2005;3:711-721.
Crossref - Kelly BG, Vespermann A, Bolton DJ. The role of horizontal gene transfer in the evolution of selected foodborne bacterial pathogens. Food Chem Toxicol. 2009;47:951-968.
Crossref - Spellberg B. The antibiotic crisis: Can we reverse 65 years of failed stewardship? Arch Intern Med. 2011;171:1080-1081.
Crossref - Cho I, S Yamanishi, L Cox, et al. Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature. 2012;488:621–626.
Crossref - CDC (U.S. 2013.Centers for Disease Control and Prevention). Antibiotic resistance from the farm to the table.http://www.cdc.gov/foodsafety/challenges/from-farm-to-table.html(accessed February 07, 2020).
- Beceiro A, Tomas M, Bou G. Antimicrobial resistance and virulence: a successful or deleterious association in the bacterial world? Clin Microbiol Rev. 2013;26:185-230.
Crossref - Wellington EMH, Boxall ABA, Cross P, et al. The role of the natural environment in the emergence of antibiotic resistance in Gram-negative bacteria. Lancet Infect Dis. 2013;13:155 – 165.
Crossref - World Health Organisation. Escherichia coli. WHO response, Report, 2018.
- Zimbabwe National Statistics Agency.2012.Population Census 2012 Reports by Provinces. Accessed: 06 February 2020.
- Hombach M, Bottger EC, Roos M. The critical influence of the intermediate category on interpretation errors in revised EUCAST and CLSI antimicrobial susceptibility testing guidelines. Clin Microbiol Infect. 2013;9:59-71.
Crossref - World Health Organisation. WHONET software. Availablefrom:http//www.who.int/drugresistance/whonetsoftware/en/ Accessed: 20 February 2020.
- Makwanda PN, Woyo E. Food safety violations by food handlers in the food industry in Zimbabwe. American Journal of Nutrition and Food Science. 2014; 1: 25 – 31.
- Government of Zimbabwe. National Food Laws & Regulations. Zimbabwe. Codex Alimentarius. Report, 2016.
- Jang J, Hur HG, Sadowsky MJ, Byappanahalli MN, Yan T, Ishii S. Environmental Escherichia coli: ecology and public health implications-a review. J Appl Microbiol. 2017;123(3):570-581.
Crossref - Katz SE, Pollan M. The art of fermentation: an in-depth exploration of essential concepts and processes from around the world. Dairy Science. 2012;120:12.
- Shar AH, Kazi YF, Kanhar NA, Soomro IH, Zia SM, Ghumro PB. Drinking water quality in Rohri City, Sindh, Pakistan. African Journal of Biotechnology. 2010;9:7102-7107.
- Mbanga J, Ncube V, Magumura A. Detection of extended-spectrum β-lactamase producing Escherichia coli in retail chicken meat and humans in Bulawayo, Zimbabwe. Am J Res Commun. 2016;4:190 – 207.
- Pitout JDD, Gregson DB, Church DL, Elsayed S, Laupland KB. Community-wide outbreaks of clonally related CTX-M-14 beta-lactamase-producing Escherichia coli strains in the Calgary Health Region. J Clin Microbiol. 2005;43: 844-2849.
Crossref - Tadesse DA, Zhao S, Tong E, et al. Antimicrobial Drug Resistance in Escherichia coli from Humans and Food Animals, United States. Emergency Infectious Diseases, 2012; 5: 1950 – 2002.
Crossref - Wayne PA.Clinical and Laboratory Standards Institute: Performance standards for antimicrobial susceptibility testing. Nineteenth Informational Supplement. CLSI document. 2009;M100-S19.
- Rawat D, Nair D. Extended-spectrum β-lactamases in Gram-Negative Bacteria. J Infect Dis. 2010;2:263-274.
Crossref - Abhilash KP, Veeraraghavan B, Abraham OC. Epidemiology and outcome of bacteraemia caused by extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli and Klebsiellaspp in a tertiary care teaching hospital in south India. J Assoc Physicians India. 2010;58:13-17.
- Momtaz H. Serogroups virulence genes and antibiotic resistance in Shiga toxin-producing Escherichia coli isolated from diarrheic and non-diarrheic paediatric patients in Iran. Gut Pathology. 2013;5:39.
Crossref - van Vuuren S, Viljoen A. Plant-based antimicrobial studies-methods and approaches to study the interaction between natural products. Planta Medicine. 2011;77:1168.
Crossref - Brian MJ, Frosolono M, Murray BE, et al. Polymerase chain reaction for diagnosis of enterohemorrhagic Escherichia coli infection and hemolytic uremic syndrome. J Clin Microbiol. 1992;30:1801-1806.
Crossref - Paton AW, Paton JC, Goldwater PN, Manning PA. Direct detection of Escherichia coli Shiga-like toxin genes in primary faecal cultures by a polymerase chain reaction. J Clin Microbiol. 1993;31:3063-3067.
Crossref - Gonzalez-Escalona N, Kase JA. Virulence gene profiles and phylogeny of Shiga toxin-positive Escherichia coli strains isolated from FDA regulated foods during 2010-2017. PLoS ONE. 2019;14(4):e0214620.
Crossref
© The Author(s) 2020. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.