ISSN: 0973-7510
E-ISSN: 2581-690X
The increasing prevalence of multidrug-resistant (MDR) Candida infections emphasises the urgent need for novel antifungal agents. This study evaluates the antifungal properties of Sophora flavescens (S. flavescens) root extract against five MDR Candida species. A total of 120 bioactive compounds, including flavonoids and alkaloids with known antimicrobial properties, were identified using high-resolution liquid chromatography-mass spectrometry. The antifungal efficacy of the extract was assessed through agar well diffusion, minimum inhibitory concentration (MIC), and minimum fungicidal concentration (MFC) assays. The extract exhibited strong antifungal activity, with inhibition zone diameters ranging from 13.33 to 19.66 mm against all tested strains. The MIC values ranged from 6.25 to 25 mg/mL across all tested Candida species, with corresponding MFCs falling within the same range for C. krusei, C. parapsilosis, C. albicans, and C. glabrata. In contrast, C. auris exhibited slightly higher MFC values, ranging from 12.5 to 25 mg/mL. Biofilm inhibition was assessed using crystal violet staining, while morphological alterations in treated Candida cells were visualised using scanning electron microscopy. Molecular docking studies targeting fungal lanosterol 14α-demethylase (CYP51) of C. albicans revealed that ginkgetin (-11.0 kcal/mol) and roxburghine B (-10.7 kcal/mol) displayed stronger binding affinities than the standard antifungal drug Itraconazole (-9.3 kcal/mol). These findings underscore the potential of S. flavescens root extract as a natural and effective antifungal candidate against hospital-acquired Candida infections.
Sophora flavescens, Candida Species, Antifungal Activity, Bioactive Compounds, Molecular Docking, Nosocomial Infections, Drug-resistant Fungi
Nosocomial fungal infections caused by Candida species represent a significant threat to immunocompromised patients, with mortality rates from invasive candidiasis reaching up to 70% in severe cases.1 The rise of resistant strains and the declining effectiveness of conventional antifungal agents, including azoles and echinocandins, underscore the urgent need for novel therapeutic options.2 C. albicans remains the primary cause of biofilm-related infections on medical devices and human tissues; however, multidrug-resistant (MDR) species, such as C. auris and C. glabrata, pose increasing challenges within clinical settings.3 The excessive use of antifungal drugs, extended hospitalisation, and the frequent application of invasive healthcare instruments, including catheters and ventilators, have collectively accelerated the development of antifungal resistance.4
Medicinal plants and other natural products have recently regained prominence as valuable sources of bioactive compounds, offering structural diversity and naturally evolved defence mechanisms against microbes.5 Sophora flavescens (S. flavescens, Fabaceae), commonly known as ‘Kushen’ in China, is native to East Asia, as well as regions of Russia and India. Its roots have traditionally been utilised in Chinese healing practices for managing various conditions, such as cancer, inflammation, skin disorders, dysentery, and parasitic infections.6 S. flavescens displays diverse pharmacological activities, including antibacterial, antipyretic, antiarrhythmic, antiasthmatic, anti-ulcer, anti-HBV, and antineoplastic effects.7 Phytochemical investigations have revealed flavonoids and alkaloids as the major bioactive compounds of S. flavescens. Notably, quinolizidine alkaloids such as matrine and oxymatrine, along with prenylated flavonoids like kurarinone and S. flavanone G, have shown potent antimicrobial activity against bacteria and viruses.8,9
In silico molecular docking predicts the binding affinity of bioactive compounds, providing insights into structure-activity relationships (SAR), protein-ligand interactions, and potential antifungal mechanisms.10 This computational approach is crucial in structure-based drug discovery by simulating compound interactions with biological targets to identify potential drug candidates.11 Additionally, the evaluation of physicochemical properties provides essential information during the early stages of drug development, supporting the optimisation of compounds for therapeutic use.12 However, its antifungal properties against nosocomial Candida infections remain underexplored.
This study bridges this gap by integrating high-resolution liquid chromatography-mass spectrometry (HR-LCMS) phytochemical profiling with molecular docking to evaluate the potential of S. flavescens as an alternative antifungal agent. By combining in vitro antifungal assays with computational docking studies, the bioactive compounds responsible for antifungal activity and their interactions with fungal C. albicans (lanosterol 14α-demethylase (CYP51)), a key enzyme in ergosterol biosynthesis, are elucidated. This approach not only deepens understanding of S. flavescens as a prospective antifungal candidate but also contributes to the advancement of plant-derived antifungal treatments to combat drug-resistant Candida species.
Plant collection and authentication
The roots of S. flavescens were harvested from Baghdad, Iraq. Plant identification and verification were conducted by Dr. Jalal Hameed Ali, Assistant Professor at the College of Agriculture, University of Sumer, Thi-Qar, Iraq. Comprehensive morphological documentation and photographic records were maintained for reference (Supplementary data Figure S1).
Preparation of ethanol extract
A total of 150 g of dried S. flavescens root powder was extracted with 1000 mL of ethanol at 50-60 °C using a Soxhlet apparatus until the extract appeared clear. The resulting filtrate was concentrated under reduced pressure at 40 °C using a rotary evaporator and subsequently stored at 4 °C for further use.13,14 The percentage yield of the extract was calculated using the following formula:
Percentage Yield = [Weight of Dried Extract / Weight of Plant Material] × 100
HR-LCMS analysis for bioactive compound profiling
Secondary metabolites present in the crude extract of S. flavescens root were identified using HR-LCMS performed on an Agilent 1200 LC system. This analysis was conducted at the Sophisticated Analytical Instrument Facility (SAIF) at the Indian Institute of Technology (IIT), Bombay, India. The HR-LCMS system operated over a mass detection range of 50-3200 amu, featuring a high resolution of 40,000 full width at half maximum, which ensured exceptional mass accuracy within one part per million (ppm). The system demonstrated high sensitivity with a signal-to-noise ratio (S/N) of 100:1 for one picogram of reserpine, thereby enabling the detection of trace-level compounds. Atmospheric pressure ionisation was applied using both electrospray ionisation and atmospheric pressure chemical ionisation modes, operating in positive and negative ionisation polarities. Mass analysis was performed via direct infusion in MS and MS/MS modes, allowing detailed fragmentation patterns for accurate compound identification. A UHPLC-PDA mass spectrometer was utilised to obtain a comprehensive profile of the secondary metabolites present in the S. flavescens root extract. Compounds were identified by comparing their m/z values with standard databases, such as METLIN and MassBank, and further confirmed through MS/MS fragmentation patterns.
Fungal strains and identification
The antifungal activity of S. flavescens root extract was evaluated against five clinically relevant opportunistic fungal pathogens commonly associated with nosocomial infections: C. auris, C. krusei, C. parapsilosis, C. albicans, and C. glabrata. These clinical isolates were collected from the Clinical Microbiology Laboratory at the Pushpagiri Institute of Medical Sciences and Research Centre, Tiruvalla, Kerala, India. The fungal cultures were maintained on Sabouraud Dextrose Agar (SDA) and Potato Dextrose Agar plates and incubated at 37 °C for 48 h to yield well-developed colonies suitable for experimental analysis.
The Candida species, initially identified by the Clinical Microbiology Laboratory at Pushpagiri Institute, was subsequently confirmed using HiCrome™ Candida Differential Agar (HiMedia Laboratories, India). This chromogenic medium enabled species-level differentiation of Candida by producing distinct colony pigmentation resulting from species-specific enzymatic activities. Each isolate was streaked onto designated segments of the agar plate and incubated at 37 °C for 48 h. Colony morphology and chromogenic profiles were interpreted in accordance with the manufacturer’s guidelines and corroborated with established literature.15-17
Antifungal activity of plant extract
Well diffusion Assay
The antifungal activity of S. flavescens root extract was evaluated using the well diffusion method. The extract residue (50 mg) was re-dissolved in 5 mL of dimethyl sulfoxide (DMSO) and used for the assay. Sterile SDA plates were seeded with freshly prepared fungal cultures adjusted to an optical density (1 OD). Wells with a diameter of 6 mm were punched into the agar using a sterile cork borer, and each well was filled with 200 µL of the plant extract solution, corresponding to a final concentration of 10 mg/mL. Itraconazole (1% w/v) was used as the positive control, while DMSO served as the negative control. The plates were incubated at 37 °C for 48 hours. Antifungal activity was determined by measuring the diameter of the zone of inhibition around each well using a Vernier caliper.14,18
MIC determination
The minimum inhibitory concentrations (MICs) of S. flavescens root extract against fungal pathogens were determined using a 96-well microtiter plate-based antifungal assay employing the resazurin dye method. Each well in columns 1 to 12 of a sterile 96-well plate was initially filled with 100 µL of Sabouraud Dextrose Broth. Subsequently, 100 µL of the plant extract (25 mg/mL) was added to the first well of column 1 and subjected to twofold serial dilutions across columns 1 to 10, with column 1 containing the highest extract concentration and column 10 the lowest. Column 11 served as the positive control (medium with fungal inoculum, without extract), whereas column 12 was designated as the negative control (medium only, without inoculum). A 20 µL aliquot of fungal suspension (1 OD) was added to all wells except those in column 12.
The plates were incubated at 37 °C for 24-48 h. After incubation, 50 µL of 0.01% resazurin solution was introduced to each well, followed by a further 4 h incubation. Fungal viability was assessed by monitoring the colour change of the resazurin dye: a shift to pink indicated viable fungal growth, while no colour change (blue or purple) indicated growth inhibition. The MIC was defined as the lowest extract concentration that completely inhibited visible fungal growth, as evidenced by the absence of colour change.19
MFC determination
To evaluate the minimum fungicidal concentration (MFC), streaks were taken from the four lowest concentrations of S. flavescens root extract that showed no visible growth on the MIC plates. These were then subcultured onto sterile SDA plates. The plates were maintained at 37 °C for 48 h and examined for fungal growth at each concentration. The MFC was defined as the lowest concentration of S. flavescens root extract that prevented fungal growth on the freshly inoculated agar plates, thereby confirming its fungicidal activity.20
Antibiofilm assay by test tube method
The antibiofilm activity of S. flavescens root extract was evaluated using the ring formation method at concentrations of 25, 12.5, 6.25, 3.125, and 1.56 mg/mL. For each assay, 2 mL of yeast extract-peptone-dextrose broth (pH 6.5) was dispensed into sterile test tubes, followed by the addition of 500 µL of the plant extract dissolved in DMSO at 25 mg/mL. Subsequently, 2 mL of fungal cell suspension, adjusted to a 1 OD at 600 nm, was added. The tubes were incubated under static conditions at 37 °C for 72 h to facilitate biofilm formation. After incubation, the liquid contents were gently decanted, and the tubes were rinsed with phosphate-buffered saline (PBS, pH 7.2) to remove non-adherent fungal cells. The tubes were then air-dried at room temperature. Adherent biofilms were stained with 0.1% (w/v) crystal violet for 1 min, followed by rinsing with PBS to remove excess dye.
To quantify the biofilm, the retained stain was solubilised using glacial acetic acid, and the absorbance was measured at 595 nm using a UV-visible spectrophotometer (Orion AquaMate 8100, Thermo Scientific, USA). A reduction in optical density compared to the untreated control indicated inhibition of biofilm formation by the extract.21 The following formula was used to estimate biofilm inhibition:
Inhibition (%) = [ODControl – ODtreated / ODControl] × 100
where ODControl denotes the absorbance of the positive control (untreated biofilm), while ODTreated represents the absorbance of the sample treated with S. flavescens root extract.
SEM analysis
The antifungal mechanism of S. flavescens root extract was examined through scanning electron microscopy (SEM). Fresh cultures of clinical isolates, including C. albicans, were grown on SDA and adjusted to a 1 McFarland optical density standard. A 100 µL aliquot of C. albicans culture was mixed with 500 µL of the crude extract (10 mg/mL) in 1 mL of SDA. The mixture was kept at 37 °C for 12 h with constant shaking. A control sample, prepared under identical conditions but without the extract, was used for comparison. After incubation, the samples were centrifuged at 10,000 rpm for 10 min to pellet the fungal cells. The collected pellets were washed with sterile Milli-Q water, air-dried, and mounted onto glass slides, sputter-coated with gold using a vacuum evaporator, and examined by SEM (JEOL JSM-6390) to observe surface morphological alterations induced by the extract.22
Molecular docking
Molecular docking studies were conducted using PyRx software, which utilises AutoDock Vina, to investigate the binding interactions between bioactive compounds from S. flavescens root extract and CYP51,23 a key enzyme involved in fungal ergosterol biosynthesis. The docking protocol was validated by calculating the root mean square deviation (RMSD) to ensure accuracy. Ligand flexibility was considered during docking to enhance the reliability of the results. Protein preparation and post-docking interaction analyses were performed using Discovery Studio Visualizer.
Protein preparation
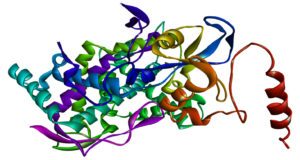
The three-dimensional (3D) crystal structure of CYP51 from C. albicans was retrieved from the Protein Data Bank (PDB ID: 5V5Z, https://www.rcsb.org). Protein preparation was carried out utilising Discovery Studio 2021, wherein all water molecules, heteroatoms, and non-essential ligands were removed to eliminate steric interference (Figure 1). Furthermore, hydrogen atoms were added to optimise the structure, followed by energy minimisation using the CHARMM force field to improve docking accuracy. The active site region was identified based on the co-crystallised ligand and supported by previous literature reports,24 ensuring precise docking simulations.
Figure 1. 3D structure of C. albicans CYP51, PDB ID: 5V5Z, prepared using Discovery Studio. The active site region is highlighted, showing key residues involved in ligand binding
Ligand preparation
Bioactive compounds identified through HR-LCMS analysis were chosen for docking studies. Their two-dimensional molecular structures were retrieved from the PubChem database, converted into 3D structures, and then optimised using Open Babel within the PyRx platform. To obtain the most stable conformations, energy minimisation was performed utilising the universal force field in PyRx, ensuring that the ligands adopted biologically relevant poses.25
Molecular docking analysis
Docking simulations were performed utilising AutoDock Vina 4.2 integrated within the PyRx platform to evaluate the binding affinity and interaction mechanisms of bioactive compounds with CYP51. The docking grid was positioned at the enzyme’s active site, ensuring the inclusion of all essential binding residues within the defined docking area. The grid box dimensions were set at 40 × 40 × 40 Å, providing ample space for ligand flexibility and interactions within the binding pocket. To maintain the balance between computational speed and docking accuracy, the exhaustiveness parameter was adjusted to eight.26
Docking validation and RMSD calculation
To validate the docking protocol, itraconazole was utilised as a reference standard.27 The co-crystallised ligand from the crystal structure of CYP51 was re-docked into its native binding site using PyRx. The RMSD between the docked pose and the crystallographic conformation was calculated with Discovery Studio. An RMSD value below 2 Å was considered indicative of a reliable docking methodology, thereby confirming the accuracy of the simulation.28
Post-docking analysis
The interactions within the docked protein-ligand complexes were evaluated using Discovery Studio Visualizer, focusing on essential molecular interactions, including hydrogen bonding, hydrophobic interactions, and π-π stacking. Compounds exhibiting the lowest binding energy values were shortlisted as top candidates for further assessment. Their docking scores were compared against itraconazole to determine relative binding efficiency. Additionally, structural visualisations of the docked complexes were generated to highlight key ligand interactions within the active site, offering insights into their potential antifungal properties.28
Statistical analysis
To maintain accuracy and consistency, each test was performed in triplicate. Data were analysed using Microsoft Excel, and results are presented as the mean ± standard deviation to reflect the reliability and variability of the measurements.
Identification of bioactive compounds using HR-LCMS
Phytochemical composition
The ethanol extract of S. flavescens root was analysed using HR-LCMS, leading to the detection of 120 bioactive compounds. The predominant chemical groups detected were flavonoids and alkaloids, both known for their antibacterial and antifungal properties. Among the flavonoids, ginkgetin, kurarinone, and Sophora flavanone G were identified, each previously reported to exhibit strong antifungal effects. These compounds are recognised to disrupt fungal cell membrane stability and inhibit ergosterol biosynthesis, a crucial pathway for fungal viability.
Additionally, the analysis confirmed the presence of alkaloids, including matrine and oxymatrine, which are associated with antifungal and immunomodulatory effects. The antifungal efficacy of S. flavescens is likely linked to the ability of these phytochemicals to destabilise fungal membranes, inhibit ergosterol biosynthesis, and impair vital enzymatic functions necessary for fungal survival. These findings offer valuable insights into the metabolite profile of S. flavescens, further reinforcing its potential as a promising source of antifungal agents. Detailed information is available in the Supplementary data (Table S1, Figure S2-S3).
Identification of fungal pathogens
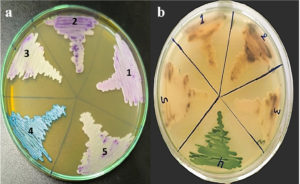
Each isolate exhibited distinct colony colours and morphological characteristics consistent with species-specific profiles (Table 1, Figure 2a and 2b).
Table (1):
Chromogenic agar characteristics of Candida species
No. |
Candida species |
Colony Color |
Morphology |
|---|---|---|---|
1 |
C. auris |
Cream-pale pink, bluish halo |
Smooth, round |
2 |
C. krusei |
Purple |
Fuzzy, spreading |
3 |
C. parapsilosis |
Cream |
Smooth, convex |
4 |
C. albicans |
Light green |
Smooth, raised |
5 |
C. glabrata |
Cream-white |
Smooth, small, round |
Figure 2. Identification of Candida species using HiChrome™ Candida Differential Agar. (a) Distinct colony colours of different species on agar plate; (b) Inverted view of the same plate showing colony characteristics. Species: (1) C. auris, (2) C. krusei, (3) C. parapsilosis, (4) C. albicans, and (5) C. glabrata
Evaluation of antifungal efficacy against nosocomial Candida strains
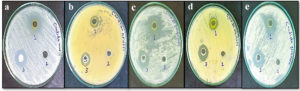
The antifungal efficacy of the ethanol extract of S. flavescens root (10 mg/mL) was evaluated against different Candida species. Comprehensive results are presented in Table 2 and Figure 3. The extract displayed considerable antifungal effects, as reflected by the varying sizes of the inhibition zones.
Table (2):
Antifungal activity of ethanolic root extract of S. flavescens (10 mg/ml) against fungal strains associated with nosocomial infections
| No. | Zone of inhibition (mm) | |||
|---|---|---|---|---|
| Candida species | S. flavescens root extract | Positive Control Itraconazole | Negative Control DMSO | |
| 1 | C. auris | 19.66 ± 0.57 | 20 | 0 |
| 2 | C. krusei | 13.33 ± 0.57 | 14 | 0 |
| 3 | C. parapsilosis | 17.33 ± 1.154 | 16 | 0 |
| 4 | C. albicans | 15.33 ± 0.57 | 18 | 0 |
| 5 | C. glabrata | 17.33 ± 1.154 | 18 | 0 |
Figure 3. Growth inhibition of nosocomial fungal strains following treatment with (1) S. flavescens root extract, (2) negative control (DMSO), and (3) positive control (Itraconazole). The tested fungal species include: (a) C. auris, (b) C. krusei, (c) C. parapsilosis, (d) C. albicans, and (e) C. glabrata.
MIC and MFC of plant extracts
The ethanol extract of S. flavescens root demonstrated noticeable antifungal activity against various Candida species. Inhibition of fungal growth was confirmed through the broth microdilution method, while fungicidal potential was assessed by subculturing on SDA plates. The complete absence of colony growth after incubation signified potent fungicidal activity. The MIC and MFC values observed across the tested isolates are summarised in Table 3 (Figure S4).
Table (3):
MIC and MFC values of S. flavescens ethanol extract against Candida species
Candida species |
MIC (mg/mL) |
MFC (mg/mL) |
|---|---|---|
C. auris |
25-6.25 |
25-12.5 |
C. krusei |
25-6.25 |
25-6.25 |
C. parapsilosis |
25-6.25 |
25-6.25 |
C. albicans |
25-6.25 |
25-6.25 |
C. glabrata |
25-6.25 |
25-6.25 |
Antibiofilm assay
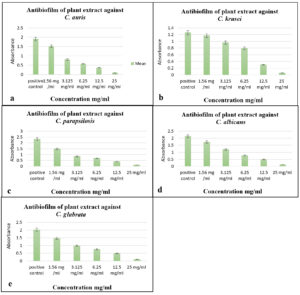
The ethanolic extract of S. flavescens root expressed significant antibiofilm activity against Candida species, as illustrated in Figure 4 and S5. Compared to the untreated control, the extract effectively reduced biofilm formation, suggesting its potential to disrupt fungal adhesion and maturation. The noted decrease in biofilm biomass indicated interference with fungal cell growth and biofilm development. The use of ethanol as a solvent likely facilitated the extraction of bioactive compounds, particularly polar phytochemicals, which may contribute to the observed antibiofilm effect.
Figure 4. Antibiofilm activity of S. flavescens root extract against Candida species. Biofilm inhibition at various concentrations (1.56-25 mg/mL) against (a) C. auris, (b) C. krusei, (c) C. parapsilosis, (d) C. albicans, and (e) C. glabrata. A positive control was included for comparison
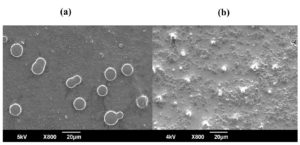
Figure 5. SEM images of C. albicans fungal cells: (a) untreated control and (b) cells after treatment
SEM analysis
SEM analysis revealed pronounced morphological changes in C. albicans following treatment with the ethanol extract of S. flavescens root. In the untreated control, C. albicans appeared as oval-shaped cells with smooth, intact surfaces and well-defined cell walls, characteristic of healthy yeast cells (Figure 5a). In contrast, cells exposed to the ethanol extract showed notable structural disruptions, including damage to the cell wall, membrane roughness, and irregular cell shapes. Treated cells showed shrivelling, surface collapse, and increased roughness, suggesting membrane disruption and potential cytoplasmic leakage (Figure 5b). Fragmentation and lysis were also observed in some cells, indicating strong antifungal activity of the extract. These morphological changes highlight the potential of S. flavescens root extract as an effective antifungal agent, likely targeting the cell wall integrity and membrane stability of C. albicans.
Molecular docking studies of isolated compounds
Molecular docking simulations were performed to investigate the binding interactions and inhibitory potential of bioactive compounds derived from S. flavescens against CYP51 of C. albicans. A total of 120 compounds were identified through HR-LCMS analysis. Of these, 101 compounds, including 100 phytochemicals and the reference antifungal agent Itraconazole, were selected for virtual screening to evaluate their binding affinities, molecular interactions, and SAR. Compounds exhibiting docking scores weaker than -5.0 kcal/mol were considered to have negligible binding affinity and were excluded from further analysis. Consequently, 20 compounds were not included in the final interpretation. Notably, several of the remaining phytochemicals demonstrated higher binding affinities than itraconazole, suggesting their potential as promising antifungal candidates targeting CYP51.
Docking scores and binding affinities
Among the docked compounds, ginkgetin (-11.0 kcal/mol), roxburghine B (-10.7 kcal/mol), Mulberroside F (-10.5 kcal/mol), and Austinol (-10.5 kcal/mol) exhibited the highest binding affinities, all outperforming itraconazole (-9.3 kcal/mol), which functioned as the standard antifungal control (see Supplementary data, Table S2, Figures S6). These findings suggest that these compounds could serve as promising CYP51 inhibitors, potentially disrupting ergosterol biosynthesis in fungal pathogens and exerting remarkable antifungal effects.
Interaction analysis
Molecular interaction analysis provided a further understanding of the binding mechanisms of the top-ranked compounds. Ginkgetin (-11.0 kcal/mol) formed strong hydrogen bonds with Tyr118 and Phe228, two crucial residues involved in CYP51 inhibition. Similarly, Mulberroside F (-10.5 kcal/mol) established π-π stacking interactions with His377 and Tyr132, enhancing its stability within the active site. Additional hydrophobic interactions with Arg96 and Glu150 were also observed, further strengthening the binding affinities of these compounds (Figure S6). The high binding stability demonstrated by these compounds suggests their potential to effectively inhibit CYP51 activity, thereby interfering with fungal membrane synthesis.
Structure-Activity Relationship (SAR) evaluation
SAR analysis highlighted that flavonoid-based compounds displayed strong interactions with the CYP51 active site, primarily attributed to the presence of hydroxyl (-OH) functional groups. Notably, kaempferol 3 (-9.3 kcal/mol) and isorhamnetin 3-O-β (-8.7 kcal/mol) formed multiple hydrogen bonds with active site residues (Figure S6). The presence of electron-donating groups within the flavonoid structure appears to enhance binding efficiency, further reinforcing the potential of flavonoids as natural antifungal agents.
The use of HiCrome™ Candida Differential Agar proved effective in distinguishing clinically significant Candida species based on their unique colony appearances. In this study, each species displayed characteristic pigmentation: C. albicans formed light green colonies, C. krusei showed rough purple colonies, C. glabrata produced smooth cream-white colonies, and C. parapsilosis appeared as smooth cream colonies, which are consistent with previous findings.15,17,29,30 Notably, C. auris presented cream to pale colonies, a variation attributed to strain-specific morphological differences documented in recent literature.31,32 These findings underscore the value of chromogenic media for the presumptive identification of Candida species, supporting its implementation in routine diagnostics and surveillance, particularly in settings requiring rapid differentiation of pathogenic yeasts.
The ethanolic extract of S. flavescens exhibited potent antifungal effects against drug-resistant Candida strains, with C. auris showing the largest zone of inhibition (19.66 ± 0.57 mm). This aligns with prior studies highlighting the efficacy of flavonoids and alkaloids present in S. flavescens, such as kurarinone and matrine, against fungal pathogens.6,9 Notably, the extract showcased potent activity against fluconazole-resistant strains, with MIC values ranging from 6.25 to 25 mg/mL. While these MICs are higher than those of synthetic drugs like fluconazole (MIC >64 µg/mL for resistant isolates),33 this discrepancy likely reflects differences in purity, as plant extracts contain complex mixtures of bioactive compounds. Nevertheless, the extract’s multi-target mechanism may compensate for the need for higher concentrations by mitigating the risk of resistance.5 Comparatively, extracts from Artemisia annua and Glycyrrhiza glabra have shown antifungal activity, albeit with a narrower spectrum, primarily against fluconazole-susceptible Candida strains, thereby underscoring the distinctive potential of S. flavescens in targeting resistant fungal pathogens.34 These results support its potential as a broad-spectrum antifungal agent, especially in light of the rising resistance to azoles.35
This study offers insights into the antifungal activity of compounds from S. flavescens against C. albicans CYP51. The high binding affinities observed for ginkgetin (-11.0 kcal/mol) and roxburghine B (-10.7 kcal/mol) suggest their strong potential as inhibitors, surpassing those of standard antifungal drugs like itraconazole (-9.3 kcal/mol). These findings align with previous studies demonstrating the effectiveness of natural flavonoids and alkaloids as antifungal agents.36
HR-LCMS identified 120 bioactive compounds, including Kurarinone (binding affinity of -9.8 kcal/mol to CYP51) and S. flavanone G (-8.6 kcal/mol to β-glucan synthase). These results are consistent with earlier reports on the quinolizidine alkaloids and prenylated flavonoids of S. flavescens, which have been shown to disrupt ergosterol biosynthesis.37
Meanwhile, prior studies have established that compounds exhibiting docking scores weaker than -5.0 kcal/mol generally exhibit negligible binding affinity and are typically excluded from further analysis.38 In line with this criterion, 20 compounds were omitted from the final interpretation in this study.
Kurarinone demonstrated a strong affinity for CYP51 (-9.8 kcal/mol), equivalent to that of fluconazole (-8.6 kcal/mol), suggesting its potential for competitive inhibition.39 Additionally, oleuropein, identified in the present study, has previously been reported to possess antifungal activity in Olea europaea extracts by destabilising fungal membrane integrity,40 thereby supporting its possible role in fungal suppression. The structural diversity of these bioactive compounds enables multi-pathway targeting, an approach increasingly emphasised in combating antifungal resistance.41
Several studies have documented the inhibitory effects of natural products on CYP51. Baicalein (-9.1 kcal/mol), a flavone derivative, has demonstrated antifungal activity against C. albicans biofilms by interfering with ergosterol biosynthesis.42 For instance, a study utilised genome mining to discover biosynthetic gene clusters responsible for the production of restriction and lanomycin, both of which act as inhibitors of CYP51.43
Kean and Ramage’s review emphasises the clinical consequences of antifungal resistance in C. auris, offering an in-depth analysis of the mechanisms responsible for resistance to azoles, polyenes, and echinocandins.44 Furthermore, Berkow et al. report C. auris isolates exhibiting resistance to four major classes of antifungal agents, underscoring the pathogen’s capacity to develop MDR.45 Nevertheless, the extract’s MFC/MIC ratio ≤4 confirms its fungicidal action, comparable to that of amphotericin B. Although natural products often necessitate higher concentrations than synthetic drugs due to their complex matrices, their multi-target mechanisms reduce the likelihood of resistance development.5 For instance, thymol derived from oregano shows similar MIC ranges (15-30 mg/mL) against C. albicans.46
SEM micrographs revealed cell wall collapse and cytoplasmic leakage in treated Candida, consistent with membrane-targeting activity.47 Similar structural damage has been observed for Melaleuca alternifolia oil, known to induce pore formation.48 The extract exhibited dose-dependent biofilm inhibition (IC50: 18.5 mg/mL), paralleling findings for curcumin, which disrupts C. albicans biofilms via agglutinin-like sequence protein 3 (Als3) suppression.49 Biofilm reduction is particularly important, as approximately 60% of fungal infections involve biofilm-mediated resistance.50 Notably, the anti-adhesion properties of oleuropein51 may act synergistically with the membrane-disrupting effects of kurarinone, thereby enhancing biofilm penetration.
The multifaceted antifungal properties of S. flavescens, largely attributed to its prenylated flavonoids, render it a promising candidate for both topical and systemic therapies. These compounds have demonstrated potent antifungal activity, suggesting potential therapeutic benefits.52 Moreover, the combination of S. flavescens with fluconazole has shown enhanced antifungal efficacy. A clinical study observed that patients treated with a combination of Sophora gel and fluconazole capsules experienced higher cure rates and lower recurrence rates of mycotic vaginitis compared to those receiving fluconazole alone. Specifically, the combination therapy achieved a cure rate of 90.7% and a recurrence rate of 2.6%, whereas fluconazole monotherapy resulted in a cure rate of 71.4% and a recurrence rate of 20.0%.53 Advancements in green extraction methods, such as subcritical water extraction, could enhance compound yield and promote sustainability.54 Nanoparticle encapsulation and combinatorial therapies may improve bioavailability while reducing therapeutic doses.55 The structural diversity of S. flavescens compounds enables multi-target antifungal mechanisms, lowering drug resistance risks and making it a promising alternative against azole-resistant Candida strains.35
This study demonstrated the strong antifungal activity of S. flavescens root extract against MDR Candida species, highlighting its potential as a natural therapeutic agent for nosocomial fungal infections. HR-LCMS analysis identified 120 bioactive compounds, among which several flavonoids and alkaloids showed strong antifungal properties. In vitro assays confirmed broad-spectrum activity, with notable inhibition of biofilm formation and fungicidal effects observed across all tested Candida strains. SEM revealed significant structural disruption in fungal cells, corroborating the extract’s membrane-targeting mechanism. Molecular docking further identified key compounds, such as ginkgetin and roxburghine B, possessing high binding affinities toward the fungal CYP51 enzyme, surpassing those of the reference antifungal itraconazole. These results support the therapeutic relevance of S. flavescens, especially in the context of emerging drug-resistance. Future investigations are warranted to explore in vivo efficacy, pharmacokinetics, and formulation strategies, including nanoencapsulation, to enhance its clinical applicability and bioavailability
Additional file: Additional Table S1-S2 and Figure S1-S6.
ACKNOWLEDGMENTS
The authors gratefully acknowledge the School of Biosciences, Mar Athanasios College for Advanced Studies Tiruvalla, Kerala, India, for providing the necessary laboratory facilities to carry out this study, and the Indian Council for Cultural Relations (ICCR) for their generous financial support. The authors deeply appreciate the Department of Microbiology, Pushpagiri Institute of Medical Sciences and Research Centre, Tiruvalla, Kerala, India, for supplying the clinical isolates essential for this research. The authors also extend their gratitude to the College of Agriculture, University of Sumer, Thi-Qar, Iraq, for their valuable collaboration. Furthermore, we thank the Sophisticated Analytical Instrument Facility (SAIF) at the Indian Institute of Technology (IIT), Mumbai, India, for conducting the HR-LCMS analysis, which significantly contributed to our findings. We also acknowledge the School of Chemical Sciences, Mahatma Gandhi University, Kottayam, Kerala, India, for providing access to the SEM facility used in this work.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
MMA and SV conceptualized the study. MMA applied methodology, performed visualization, experiments, Investigation and data curation. TM performed molecular docking data analysis. SV performed supervision, project administration and data validation. MMA wrote the manuscript. SV reviewed and edited the manuscript. All authors read and approved the final manuscript for publication.
FUNDING
This work was supported by a scholarship from the Indian Council for Cultural Relations (ICCR).
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript and/or in the supplementary files.
ETHICS STATEMENT
Not applicable.
- Pappas PG, Lionakis MS, Arendrup MC, Ostrosky-Zeichner L, Kullberg BJ. Invasive candidiasis. Nat Rev Dis Primers. 2018; (1):1-20.
Crossref - Perlin DS, Rautemaa-Richardson R, Alastruey-Izquierdo A. The global problem of antifungal resistance: Prevalence, mechanisms, and management. Lancet Infect Dis. 2017;17(12):e383–e392.
Crossref - Kumamoto CA. Candida biofilms. Curr Opin Microbiol. 2002;5;(6):608-611.
Crossref - Vitiello A, Ferrara F, Boccellino M, et al. Antifungal Drug Resistance: An Emergent Health Threat. Biomedicines. 2023;11(4):1063.
Crossref - Newman DJ, Cragg GM. Natural Products as Sources of New Drugs over the Nearly Four Decades from 01/1981 to 09/2019. J Nat Prod. 2020;83(3):770-803.
Crossref - Al-Hayanni HSA, Alnuaimi MT, AL-Lami RA, Zaboon SM. Antibacterial effect of silver nanoparticles prepared from Sophora flavescens root aqueous extracts against multidrug-resistant Pseudomonas aeruginosa and Staphylococcus aureus. J Pure Appl Microbiol 2022;16(4):2880-2890.
Crossref - Cao X, He Q. Anti-tumor activities of bioactive phytochemicals in Sophora flavescens for breast cancer. Cancer Manag Res. 2020;12:1457-1467.
Crossref - Li J-J, Zhang X, Shen X-C, et al. Phytochemistry and biological properties of isoprenoid flavonoids from Sophora flavescens Ait. Fitoterapia. 2020;143:104556-104562.
Crossref - Li J-C, Zhang Z-J, Liu D, Jiang M-Y, Li R-T, Li H-M. Quinolizidine alkaloids from the roots of Sophora flavescens. Nat Prod Res. 2022;36(7):1781-1788.
Crossref - Ferreira LG, Santos RND, Oliva G, Andricopulo AD. Molecular Docking and Structure-Based Drug Design Strategies. Molecules.2015;20(7):13384-13421.
Crossref - Agu PC, Afiukwa CA, Orji OU, et al.Molecular docking as a tool for the discovery of molecular targets of nutraceuticals in disease management. Sci Rep. 2023;13:13398.
Crossref - National Research Council. Physicochemical properties and environmental fate. In A Framework to Guide Selection of Chemical Alternatives. National Academies Press (US). 2014.
Crossref - Bennour N, Mighri H, Eljani H, Zammouri T, Akrout A. Effect of solvent evaporation method on phenolic compounds and the antioxidant activity of Moringa oleifera cultivated in Southern Tunisia. S Afr J Bot. 2020;129:181-190.
Crossref - Mostafa AA, Al-Askar AA, Almaary KS, Dawoud TM, Sholkamy EN, Bakri MM. Antimicrobial activity of some plant extracts against bacterial strains causing food poisoning diseases. Saudi J Biol Sci. 2018;25(2):361-366.
Crossref - Perry JL, Miller GR. Umbelliferyl-labeled galactosaminide as an aid in identification of Candida albicans.J Clin Microbiol. 1987;25(12):2424-2425.
Crossref - Divya K, Smitha V, Jisha MS. Antifungal, antioxidant and cytotoxic activities of chitosan nanoparticles and its use as an edible coating on vegetables. Int J Biol Macromol. 2018;114:572-577.
Crossref - Rousselle P, Freydiere AM, Couillerot PJ, de Montclos H, Gille Y. Rapid identification of Candida albicans by using Albicans ID and fluoroplate agar plates. J Clin Microbiol. 1994;32(12):3034-3036.
Crossref - Gizaw A, Marami LM, Teshome I, et al. Phytochemical screening and in vitro antifungal activity of selected medicinal plants against Candida albicans and Aspergillus niger in West Shewa Zone, Ethiopia. Adv Pharmacol Pharm Sci. 2022(1):3299146.
Crossref - Barnes L, Heithoff DM, Mahan SP, House JK, Mahan MJ. Antimicrobial susceptibility testing to evaluate minimum inhibitory concentration values of clinically relevant antibiotics. STAR Protocols. 2023;4(3):102512.
Crossref - Witasari LD, Wahyu KW, Anugrahani BJ, et al. Antimicrobial activities of fungus comb extracts isolated from Indomalayan termite (Macrotermes gilvus Hagen) mound. AMB Express. 2022;12(1):14.
- Viju N, Punitha SMJ, Satheesh S. Antibiofilm activity of symbiotic Bacillusspecies associated with marine gastropods. Ann Microbiol. 2020;70:11.
Crossref - Vijayan S, Divya K, Varghese S, Jisha MS. Antifungal efficacy of chitosan-stabilized biogenic silver nanoparticles against pathogenic Candida spp. isolated from human. BioNanoSci. 2020;10:974-982.
Crossref - Masood MM, Irfan M, Khan P, et al. 1, 2, 3-Triazole–quinazolin-4 (3H)-one conjugates: evolution of ergosterol inhibitor as anticandidal agent. RSC Adv. 2018;8(69):39611-39625.
Crossref - Hamdy R, Hamoda AM, Al-Khalifa M, Menon V, El-Awady R, Soliman SSM. Efficient selective targeting of CandidaCYP51 by oxadiazole derivatives designed from plant cuminaldehyde. RSC Med Chem. 2022;13(11):1322-1340.
Crossref - Khanum A, Bibi Y, Khan I, et al. Molecular docking of bioactive compounds extracted and purified from selected medicinal plant species against covid-19 proteins and in vitro evaluation. Sci Rep. 2024;14(1):3736.
Crossref - Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem, 2010;31(2):455-461.
Crossref - Boreak, N, Al Mahde RZ, Otayn WA, et al. Exploring Plant-Based Compounds as Alternatives for Targeting Enterococcus faecalis in Endodontic Therapy: A Molecular Docking Approach. Int J Mol Sci. 2024;25(14):7727.
Crossref - DegfieT, Endale M, Tafese T, Dekebo A, Shenkute K. In vitro antibacterial, antioxidant activities, molecular docking, and ADMET analysis of phytochemicals from roots of Hydnora johannis. Appl Biol Chem. 2022;65(1):76.
Crossref - Abbas HH, Atiyah MM. Anti-fungal activities of aqueous and alcoholic leaf extracts of Moringa oleifera Lam. on Candida albicans isolated from diabetic foot infections. AIP Conf Proc. 2023;2414(1):020008.
Crossref - Willinger B, Manafi M. Evaluation of CHROMagar Candida for rapid screening of clinical specimens for Candida species. Mycoses. 1999;42(1-2):61-65.
Crossref - Singh P, Srivastava S, Malhotra R, Mathur P. Identification of Candida auris by PCR and assessment of biofilm formation by crystal violet assay. Indian J Med Microbiol. 2023;46:100421.
Crossref - Borman AM, Szekely A, Johnson EM. Comparative pathogenicity of United Kingdom isolates of the emerging pathogen Candida auris and other key pathogenic Candida species. MSphere. 2016;1(4):10-1128.
- Kronvall G, Karlsson I. Fluconazole and voriconazole multidisk testing of Candida species for disk test calibration and MIC estimation. J Clin Microbiol. 2001;39(4):1422-8.
Crossref - Fatima A, Gupta VK, Luqman S, et al. Antifungal activity of Glycyrrhiza glabra extracts and its active constituent glabridin. Phytother Res. 2009;23(8):1190-1193.
Crossref - Fisher MC, Alastruey-Izquierdo A, Berman J. et al.Tackling the emerging threat of antifungal resistance to human health. Nat Rev Microbiol. 2022;20;557-571.
Crossref - Roy A, Khan A, Ahmad I, et al. Flavonoids a Bioactive Compound from Medicinal Plants and Its Therapeutic Applications. Biomed Res Int. 2022;2022(1):5445291.
Crossref - An JX, Wang R, Li AP, et al. Prenylated flavonoids isolated from the root of Sophora flavescens as potent antifungal agents against Botrytis cinerea.J Agric Food Chem. 2024;72(36):19618-19628.
Crossref - Kitchen DB, Decornez H, Furr JR, Bajorath J. Docking and scoring in virtual screening for drug discovery: methods and applications. Nat Rev Drug Discover. 2004;3(11):935-949.
Crossref - do Nascimento JET, Rodrigues ALM, de Lisboa DS, et al. Chemical composition and antifungal in vitro and in silico, antioxidant, and anticholinesterase activities of extracts and constituents of Ouratea fieldingiana (DC.) Baill. Evid Based Complement Alternat Med. 2018(1):1748487.
Crossref - Davidova S, Galabov AS, Satchanska G. Antibacterial, antifungal, antiviral activity, and mechanisms of action of plant polyphenols. Microorganisms. 2024;12(12):2502.
Crossref - Jan S, Iram S, Bashir O, et al. Unleashed Treasures of Solanaceae: Mechanistic Insights into Phytochemicals with Therapeutic Potential for Combatting Human Diseases. Plants. 2024;13(5):724.
Crossref - Zhou X, Zeng M, Huang F, Qin G, Song Z, Liu F. The potential role of plant secondary metabolites on antifungal and immunomodulatory effect. Appl Microbiol Biotechnol. 2023;107(14):4471-4492.
Crossref - Liu N. Targeted Genome Mining for the Discovery and Study of Sterol Pathway Fungal Natural Product Drugs. University of California, Los Angeles. 2020.
- Kean R, Ramage G. Combined antifungal resistance and biofilm tolerance: the global threat of Candida auris. Msphere. 2019;4(4):10-1128.
Crossref - Murray CJ, Ikuta KS, Sharara F, et al. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet. 2022;399(10325):629-655.
- Bakkali N, Ott L, Triquet C, Cottencin O, Grynberg D. Learning from others’ experience: Social fear conditioning deficits in patients with severe alcohol use disorder. Alcohol Clin Exp Res. 2023;47(8):1603-1613.
Crossref - Vitiello A, Ferrara F, Boccellino M, et al. Antifungal Drug Resistance: An Emergent Health Threat. Biomedicines. 2023;11(4):1063.
Crossref - Nova BGV, dos Santos Silva L, da Silva Andrade M, et al. The essential oil of Melaleuca alternifolia incorporated into hydrogel induces antimicrobial and anti-inflammatory effects on infected wounds by Staphylococcus aureus. Biomed Pharmacother. 2024;173:116389.
Crossref - Bonincontro G, Scuderi SA, Marino A, Simonetti G. Synergistic Effect of Plant Compounds in Combination with Conventional Antimicrobials against Biofilm of Staphylococcus aureus, Pseudomonas aeruginosa, and Candida spp. Pharmaceuticals. 2023;16(11):1531.
Crossref - Zafer MM, Mohamed GA, Ibrahim SRM, Ghosh S, Bornman C, Elfaky MA.Biofilm-mediated infections by multidrug-resistant microbes: a comprehensive exploration and forward perspectives. Arch Microbiol. 2024;206(3):101.
Crossref - Omar SH. Oleuropein in olive and its pharmacological effects. Sci Pharm. 2010;78(2):133-54.
Crossref - Kong S, Liao Q, Liu Y, et al. Prenylated Flavonoids in Sophora flavescens: A Systematic Review of Their Phytochemistry and Pharmacology. Am J Chin Med. 2024;52(4):1087-1135.
Crossref - Wang NM, Cui L, Ma CF, Wang HX. [Clinical observation on treatment of mycotic vaginitis with Sophora gel combined with Fluconazole capsules. Zhongguo Zhong Yao Za Zhi. 2015;40(5):978-80
- Diaz-Reinoso B, Rivas S, Rivas J, Dominguez H. Subcritical water extraction of essential oils and plant oils. Sustain Chem Pharm. 2023;36:101332.
Crossref - Shaikh J, Ankola DD, Beniwal V, Singh D, Kumar MNVR. Nanoparticle encapsulation improves oral bioavailability of curcumin by at least 9-fold when compared to curcumin administered with piperine as absorption enhancer. Eur J Pharm Sci. 2009;37(3-4):223-230.
Crossref
© The Author(s) 2025. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.