ISSN: 0973-7510
E-ISSN: 2581-690X
Marine-derived actinobacteria are considered as potential sources of bioactive metabolites including antifungal substances. Fifteen out of 155 marine-derived actinobacteria exhibited strong antifungal activity against the rice blast fungus Pyricularia oryzae. Their extracts were further determined for minimum inhibitory concentrations (MIC) and minimum fungicidal concentrations (MFC). Ethyl acetate extract from the strain AMA49 and its subfraction AMA49F1 strongly inhibited hyphal growth of various P. oryzae strains with MICs (8 to 16µg/ml) and MFCs (16 to 128µg/ml) comparable to propiconazole. Both extracts destroyed fungal membrane and organelles, completely inhibited conidial germination, appressorium formation, and were non-toxic to Galleria mellonella. High performance liquid chromatography/mass spectrometry identified oligomycin A and its derivatives as the active components of AMA49F1 besides several diketopiperazines. AMA49 was identified as a Streptomyces sp. based on morphological characteristics and 16S rDNA sequence analysis. The results suggest that the Streptomyces sp. strain AMA49 is a potential biocontrol agent against rice blast pathogen P. oryzae. This is the first report on the inhibitory effect of the marine-derived Streptomyces extract containing oligomycin A and its derivatives on mycelial growth, conidial germination and appressorium formation of P. oryzae.
Marine-derived Streptomyces sp., Pyricularia oryzae, rice blast disease, diketopiperazines, oligomycin A.
Rice is a staple food that feeds almost half of the world population1. The 2018/19 world rice total supply is estimated at 657.9 million tons2. The fungus Pyricularia oryzae (syn: Magnaporthe oryzae) causes the highly destructive rice blast disease, which is found in at least 85 countries worldwide3. The effects of the fungus are so severe with approximately 30% of rice production losses globally that it was placed first in a list of the top ten fungal plant pathogens4. Blast disease is typically controlled by chemical fungicides, including tricyclazole, isoprothiolane, probenazole, propiconazole, carbendazim, blasticidin and kasugamycin3,5,6. However, excessive use of chemical fungicides in agriculture has damaged human health and the environment, and has led to the development of fungicide-resistant pathogens7. Therefore, there is a growing need for new, environmentally safer and more potent fungicides. Antifungal activity has been reported in secondary metabolites from microbes, especially actinobacteria. Many reports have shown that actinobacteria protect plants and exhibit antagonistic effects against several phytopathogenic fungi8,9.
Terrestrial actinobacteria have long been a source of bioactive compounds against fungal plant pathogens10,11,12. In addition, they have been exhaustively investigated, leading to less new compounds discovered. Marine environment covers more than 70% of the earth’s surface. It is also evident that there was higher functional ecosystem diversity in the marine environment than the terrestrial ecosystem13. Therefore, marine-derived actinobacteria have become a potential source of bioactive compounds14. Many new compounds such as bonactin15, chandrananimycins15, marinomycins15, salinosporamide A15, trioxacarcins15 and fluvirucin B616 have been isolated from marine-derived actinobacteria. The majority of secondary metabolites from marine actinobacteria were derived from members of the genus Streptomyces17,18. Most of them have terrestrial origins. However, the actinobacteria from marine habitat developed with unique chemical, physicological, and structural features to survive under varying salinity, pressure, and temperature in marine ecosystems19. Furthermore, the activity of marine-derived actinobacteria against fungal plant pathogens has rarely been reported.
In this study, actinobacteria from marine habitats were isolated and their antifungal activities against the rice blast pathogen P. oryzae were investigated. The effects on fungal cells of the most active crude extract and its subfraction were determined. The active isolate was then identified using the morphological characteristics and molecular methods based on 16S rDNA sequence analysis.
Sample collection and actinobacterial isolation
Forty marine samples were collected around Nakhon Si Thammarat, Trang, Satun, Songkhla and Phuket provinces in southern Thailand. There is no specific permission required in these locations. The samples consisted of 17 sediments, 16 marine animals (nudibranch, sea fan, sea anemones, sea squirts and sponges) and seven algae. Samples were kept in plastic bags containing sea water and placed in ice boxes. The samples were transported back to the laboratory and processed as soon as possible after collection. Sediment samples were placed in Petri dishes and air dried at room temperature for a week. The dried sediments underwent chemical and physical treatment as previously described20. Algae and invertebrate samples were pretreated using a lyophilization process. The diluted samples were inoculated onto chitin agar21, modified soil extract agar (A9)22,23 and starch-yeast extract-peptone-seawater agar (SYP-SW)24. All media were prepared with 60% natural seawater and supplemented with cycloheximide (50 mg/l) and nalidixic acid (20 mg/l)25. All plates were incubated at 25 ± 2°C for four weeks. Actinobacteria were isolated and purified on A9 medium. The pure cultures were stored in 20% glycerol at -80°C.
Source of P. oryzae
Sixteen isolates of P. oryzae, comprising six virulent strains from paddy (PTRC1-6) and ten isolates from rice near-isogenic-lines (NILs) (PTRC7-16) were obtained from the Phattalung Rice Research Center (PTRC), Thailand. They were grown on potato dextrose agar (PDA) slants at 25 ± 2°C for seven days and then stored at 4°C. All isolates were newly subcultured on PDA before use26.
Antifungal assay of actinobacterial culture broth
Actinobacteria were selected based on morphotypes and cultured in ISP2 medium with 60% seawater at 25 ± 2°C for four weeks. The four-week-old actinobacterial culture broths were screened for antifungal activities against P. oryzae PTRC1 by an agar well diffusion method27. Briefly, wells (7 mm in diameter) were punched in four places 0.5 cm from the edge of the fungal colony and each well was filled with 80µl of culture broth. Plates were incubated at 25 ± 2°C for three to five days. The radii (mm) of the fungal colony on the test and control plates were then measured. The percentage of hyphal growth inhibition was calculated using this formula: Inhibition (%) = 100 – [(R2 / r2) x 100], where R and r represent the average of triplicate measurements of radii (mm) of fungal colonies in the test and control plates, respectively28.
Fermentation and extraction of crude extracts
Actinobacteria that inhibited P. oryzae PTRC1 more than 80% in the culture broth screening test were selected for cultivation in ISP2 medium with 60% seawater for four weeks under static conditions at 25 ± 2°C. Fermentation broths were separated into culture filtrate and cells by filtration and extracted with organic solvents20. Three crude extracts were obtained from each isolate: broth ethyl acetate extract (BE), cell ethyl acetate extract (CE) and cell hexane extract (CH).
Antifungal screening test of crude extracts
The actinobacterial crude extracts were dissolved in 100% DMSO to obtain stock solutions of 100 mg/ml. The antifungal activities of the crude extracts at a concentration of 200µg/ml were determined using an agar microdilution method in 24 well-plate29. The standard fungicide propiconazole at a concentration of 20µg/ml and 0.2% DMSO were used as positive and negative controls, respectively. Agar plugs (1.5 mm in diameter), cut from the periphery of three-day old P. oryzae PTRC1 grown on PDA, were placed in the middle of the surface of the agar in each well. The microplates were incubated at 25 ± 2°C for three days. Crude extracts that completely inhibited hyphal growth at this concentration were further determined to quantify their minimum inhibitory concentrations (MIC) and minimum fungicidal concentrations (MFC).
Determination of minimum inhibitory concentration (MIC) and minimum fungicidal concentration (MFC)
The actinobacterial extracts and propiconazole were diluted with melted PDA to final concentrations from 0.25 to 128µg/ml. The DMSO at 0.2% was used as a negative control. Tests were performed in the same manner as the screening test in four replicates. The MIC was defined as the lowest concentration of extract that completely inhibited fungal growth (100% inhibition). Fungal agar plugs from MIC wells and higher concentrations were transferred to new PDA plates and incubated at 25 ± 2°C for five days. The concentration of extract that completely inhibited hyphal growth on the new plates was recorded as the MFC.
Fractionation of bioactive extract AMA49CE
The most active crude extract AMA49CE against P. oryzae PTRC1 was fractionated by column chromatography over Sephadex LH-20. Elution was performed with 100% methanol. Fractions showing similar chromatograms on TLC were combined and evaporated to dryness under reduced pressure to obtain subfractions. All fractions were checked for antifungal activity against P. oryzae PTRC1. The major component of the most active fraction against P. oryzae PTRC1 was analysed by high performance liquid chromatography/mass spectrometry (HPLC/MS).
Compound identification
The extract AMA49F1 was dissolved in methanol (c = 3 mg/ml) and analysed by high-resolution electrospray ionisation mass spectrometry (HRESIMS) as described in detail previously30. To confirm the identity of oligomycin A, an oligomycin standard provided by the Natural Compound Library (NCL) of the German Centre for Infection Research (DZIF) was analysed in the same manner. Finally, a mixture of the extract AMA49F1 and the oligomycin standard was co-injected.
Effect of active crude extract and its fraction on fungal hyphae
The effect of AMA49CE and AMA49F1 on the hyphal surface of P. oryzae PTRC1 was further studied using scanning electron microscopy (SEM). Their effect on cell components was studied using transmission electron microscopy (TEM)31. Agar plugs from the edge of a three-day-old colony of P. oryzae PTRC1 were treated at 25 ± 2°C for three days with the AMA49CE and AMA49F1 at four times their MICs (4xMIC). All samples were prepared for SEM and TEM at the Scientific Equipment Center, Prince of Songkla University.
Conidial germination inhibition test
Conidia of P. oryzae PTRC1 (3.5 x 105 conidia /ml) were prepared in 0.1% Tween20. Then, 50µl of conidial suspension were mixed with 50µl of the extract to obtain final concentrations of 1xMIC, 2xMIC and 4xMIC in a well-microscopic slide. Propiconazole and DMSO were used as positive and negative controls, respectively. Three replicates per treatment were performed. The test slides were placed in a moist chamber and incubated at 25 ± 2°C for 24 h. After incubation, 2µl of the treated conidia were dropped onto PDA, then further incubated at 25 ± 2°C for three days to check for viability. The percentages of germinated conidia and appressorium formation were determined under a light microscope (100 conidia per replicate)32. The percent reduction of conidial germination and appressorium formation compared to a control was then computed. Germ tube lengths of the germinated conidia were also measured.
Toxicity testing
In vivo toxicity testing was performed using Galleria mellonella larvae33. The use of invertebrates does not need ethical permission according to Thai law and others34,35. G. mellonella larvae (220-250 mg) at the sixth developmental stage were stored in sterile Petri dishes in darkness at 30°C for 24 h before use. Ten healthy G. mellonella larvae with no melanization were used in each replicate. Three replicates were used in each treatment. AMA49CE and AMA49F1 were separately dissolved in DMSO and diluted with PBS to 4xMIC. In the treatment group, 10µl of each extract was injected into the last left proleg and last right proleg, using an Ultra-Fine II syringe. Two control groups were used in this study: members of the first control group were injected in each last proleg with 10µl of 3.2% DMSO in PBS, and the other control group received no injection. Larvae were incubated at 30°C for 72 h without feeding33 and the dead larvae were investigated. Larvae were considered dead if they did not move after being touched with a sterile pipette tip. The percentage mortality of larva was calculated36.
Identification of actinobacteria
The strain AMA49 was characterized by phenotypic characteristics including the color of the colony, aerial mycelium, substrate mycelium, and color of soluble pigment. Spore shape was also considered after 14 days growth at 25 ± 2°C on yeast extract-malt extract agar (ISP2), oatmeal agar (ISP3), inorganic salts-starch agar (ISP4), glycerol-asparagine agar (ISP5), and tyrosine agar (ISP7). Carbon utilization (D-glucose, D-fructose, raffinose, cellobiose, sucrose, rhamnose and mannitol) was also determined37. Each carbon source was added to Pridham and Gottlieb carbon utilization basal medium at a final concentration of 1%. The basal medium plus D-glucose and sugar-free basal medium were used as positive and negative controls, respectively. AMA49 was grown on the carbon utilization medium for 14 days at 25 ± 2°C. The growth was measured and compared with the growth on positive and negative controls media. The 16S rRNA gene was amplified from the genomic DNA using primers 20F (5′-GAGTTTGATCCTGGCTCAG-3′) and 1500R (5′-GTTACCTTGTTACGACTT-3′). Amplified PCR products were sequenced directly by Macrogen Inc., Korea using universal primers. The consensus DNA sequence was compared to sequences in the EzTaxon database with the BLASTN program. Multiple alignments of the sequences were analysed using the program BioEdit Sequence Alignment Editor (version 7.0.4.1)38. The phylogenetic tree was constructed with Neighbour-Joining (NJ) algorithms using Molecular Evolutionary Genetics Analysis version 6.0 (Mega 6.0) software39.
Statistical analysis
The data of conidial germination inhibition, appressorium formation inhibition and germ tube length of P. oryzae PTRC1 were analysed by two-way ANOVA. Tukey HSD post-hoc test (P < 0.05) was used in the analysis. The percent mortality of G. mellonella was analysed using one-way ANOVA with Tukey HSD post-hoc test (P < 0.05). All tests were performed using SPSS statistical software version 11.5.
Actinobacterial isolation and antifungal activity screening test
A total of 525 isolates of actinobacteria were obtained from different sources including mangrove soil (156 isolates), marine organisms (160 isolates), marine sediments (26 isolates) and soil from a lagoon (183 isolates). From these 525 isolates, 155 were selected based on their colonial morphology for antifungal activity screening by an agar well diffusion method. Among them, 64 isolates (41.29%) presented antifungal activity against P. oryzae PTRC1. Of these 64, 15 isolates that exhibited more than 80% hyphal growth inhibition were selected for fermentation and extraction. Among the top 15 active isolates, 9 were isolated from seaweeds, 4 from mangrove soils, and each one from sea fan and marine sediment. Fourteen isolates were identified as Streptomyces sp. and one isolate as Micromonospora sp. using morphological characterization (Table 1).
Table (1):
The top 15 active actinobacteria strains against hyphal growth of P. oryzae PTRC1 by an agar well diffusion method
Strains |
Morphological characterization |
Source |
Inhibition (%) |
|---|---|---|---|
AMA15 |
Streptomyces sp. |
Mangrove soila |
88.89 |
AMA16 |
Streptomyces sp. |
Mangrove soila |
88.89 |
AMA49 |
Streptomyces sp. |
Mangrove soila |
82.64 |
AMA50 |
Streptomyces sp. |
Mangrove soila |
87.46 |
AMR5 |
Streptomyces sp. |
Padina sp. (seaweed)b |
80.86 |
AMR6 |
Streptomyces sp. |
Padina sp. (seaweed)b |
82.64 |
AMR9 |
Streptomyces sp. |
Padina sp. (seaweed)b |
80.86 |
AMR29 |
Streptomyces sp. |
Padina sp. (seaweed)b |
86.56 |
AMR64 |
Streptomyces sp. |
Melithaea sp. (sea fan)c |
82.64 |
AMR67 |
Streptomyces sp. |
Acanthophora spicifera (seaweed)c |
85.94 |
AMR69 |
Streptomyces sp. |
Gracilaria sp. (seaweed)c |
85.31 |
AMR71 |
Streptomyces sp. |
Padina sp. (seaweed)c |
87.46 |
AMR75 |
Streptomyces sp. |
Gracilaria sp. (seaweed)c |
84.33 |
AMR76 |
Streptomyces sp. |
Gracilaria sp. (seaweed)c |
82.64 |
ALA2 |
Micromonospora sp. |
Marine sedimentd |
84.22 |
a Samples collected from mangrove forest (5-10 cm from soil surface) of Songkhla Lake, Songkhla, Thailand, b sample collected from the Andaman sea, Trang, Thailand, c samples collected from the Andaman sea, Phuket, Thailand, and d sample collected from marine sediment (6-10 cm from sediment surface) from the Songkhla Lake, Songkhla, Thailand.
Antifungal activity of actinobacterial crude extracts
A total of 45 crude extracts from the 15 most active actinobacteria were screened for their antifungal activity against P. oryzae PTRC1. Only five crude extracts from two isolates, AMA49BE, AMA49CE, AMA49CH, AMA50BE and AMA50CE could inhibit P. oryzae PTRC1. Their MIC/MFC values ranged from 8/32µg/ml to 128/>200µg/ml (Table 2). Cell ethyl acetate crude extract from AMA49 (AMA49CE) showed the best activity. Therefore, AMA49CE was selected for further tested with various pathogenic strains of P. oryzae. In these tests, the MIC and MFC values of AMA49CE were in the range of 8 to 16µg/ml and 16 to 128µg/ml, respectively. Against the same pathogenic strains, the MIC and MFC ranges of propiconazole were 4 to 8µg/ml and 128 to >128µg/ml (Table 3), respectively.
Table (2):
Antifungal activity against P. oryzae PTRC1 of five active crude extracts from marine-derived actinobacteria strains AMA49 and AMA50
Crude extracts |
MIC/MFC (µg/ml) |
|---|---|
AMA49BE |
128/>200 |
AMA49CE |
8/32 |
AMA49CH |
32/64 |
AMA50BE |
64/128 |
AMA50CE |
64/64 |
BE: broth ethyl acetate extract, CE: cell ethyl acetate extract, CH: cell hexane extract, MIC: minimum inhibitory concentration, MFC: minimum fungicidal concentration.
Table (3):
Antifungal activity of cell ethyl acetate crude extract from Streptomyces sp. AMA49 (AMA49CE) against pathogenic strains of P. oryzae
| P. oryzae strains | MIC and MFC (µg/ml) | |||
|---|---|---|---|---|
| AMA49CE crude extract | Propiconazole | |||
| MIC | MFC | MIC | MFC | |
| PTRC 1 | 8 | 32 | 8 | 128 |
| PTRC2 | 16 | 128 | 8 | 128 |
| PTRC 3 | 16 | 32 | 8 | 128 |
| PTRC 4 | 16 | 16 | 8 | 128 |
| PTRC 5 | 8 | 32 | 8 | 128 |
| PTRC 6 | 8 | 64 | 8 | 128 |
| PTRC 7 | 8 | 32 | 8 | 128 |
| PTRC 8 | 8 | 32 | 8 | 128 |
| PTRC 9 | 16 | 32 | 8 | 128 |
| PTRC 10 | 16 | 16 | 8 | >128 |
| PTRC 11 | 8 | 64 | 8 | >128 |
| PTRC 12 | 8 | 32 | 4 | 128 |
| PTRC 13 | 8 | 32 | 8 | 128 |
| PTRC 14 | 16 | 64 | 8 | >128 |
| PTRC 15 | 8 | 128 | 8 | >128 |
| PTRC 16 | 8 | 32 | 4 | 128 |
MIC: minimum inhibitory concentration, MFC: minimum fungicidal concentration
Fractionation and effect of active fraction on P. oryzae PTRC1 hyphae
The most active crude extract AMA49CE against P. oryzae PTRC1 was fractionated by CC over Sephadex LH-20 to yield five subfractions (F1-F5). Subraction F1 (AMA49F1) showed the strongest activity against P. oryzae with MIC/MFC values of 8/16µg/ml, while subfractions F2-F5 were less active (MIC and MFC values ≥ 128µg/ml). The effect of AMA49CE and AMA49F1 on fungal morphology was also studied using SEM. Normal hyphae were observed in untreated samples (Fig. 1a). P. oryzae treated with 4xMIC (32µg/ml) of propiconazole showed obvious hyphal tip swelling (Fig. 1c). Hyphal morphologies of P. oryzae were severely affected by AMA49CE (Fig. 1e) and AMA49F1 (Fig. 1g) at 4xMIC (32µg/ml). The treated hyphae were swollen, expanded and flattened (Fig. 1e and g). Moreover, distorted conidia were also observed in both samples treated with AMA49CE and AMA49F1 (Fig. 1e and g).
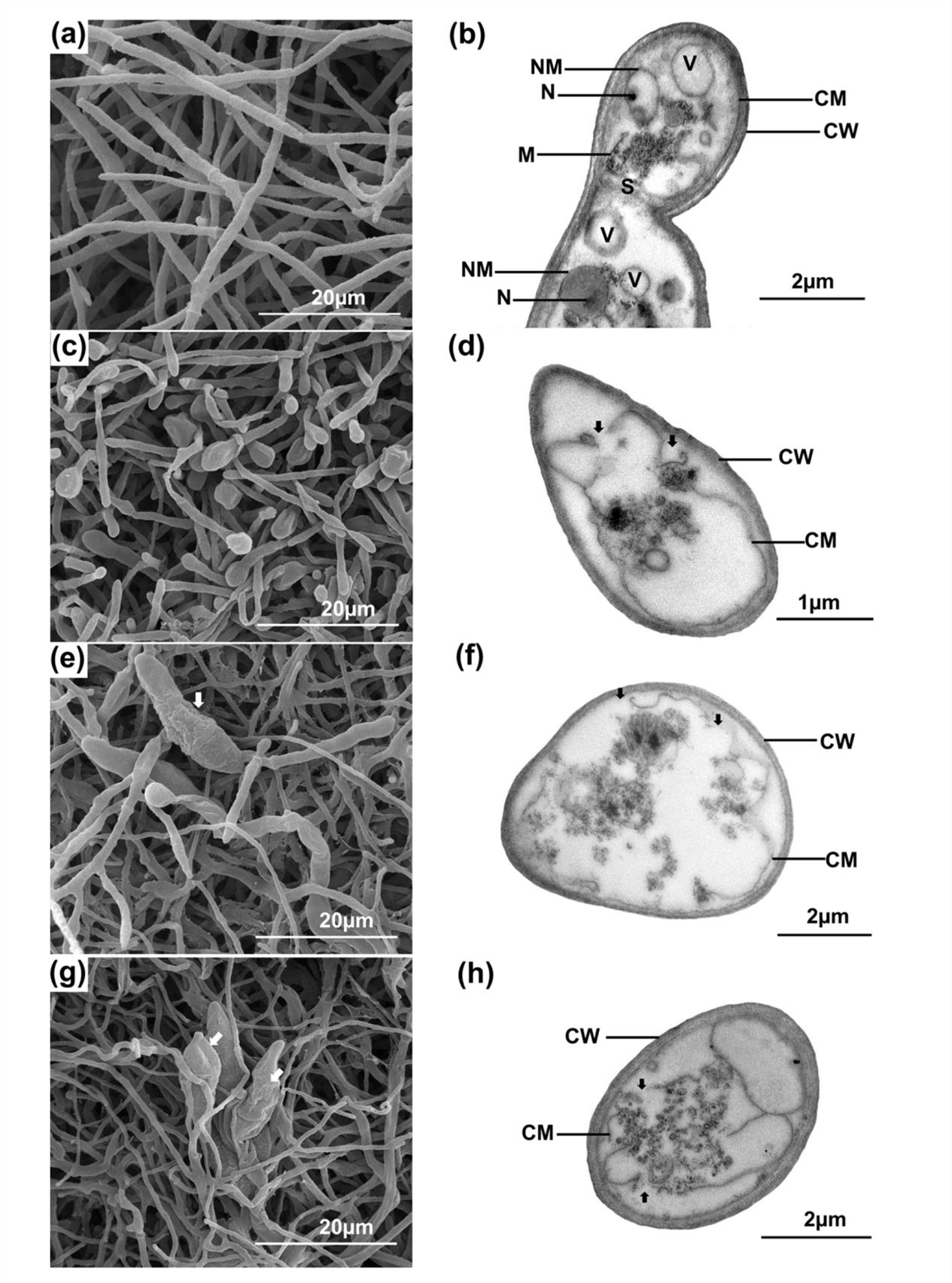
Fig. 1. Scanning electron microscopic (SEM, left) and transmission electron microscopic (TEM, right), images of untreated P. oryzae PTRC1 hyphae (negative control) (a-b), hyphae treated with 32µg/ml (4xMIC) of propiconazole (positive control) (c-d), with 32µg/ml (4xMIC) of AMA49CE (e-f) and hyphae treated with 32µg/ml (4xMIC) of MA49F1 (g-h) after incubation at 25 ± 2°C for three days. CW, cell wall; CM cell membrane; V, vacuole; M, mitochondria; N, nucleous; NM, nuclear membrane; S, septum. White arrows in (e) and (g) indicate the abnormal condia. Black arrows in (d), (f) and (h) indicate the cell membrane damage.
The effects of AMA49CE and AMA49F1 on fungal cell components were further studied with TEM. Untreated hyphae revealed normal intact fungal cell components (Fig. 1b), including cell wall (CW), cell membrane (CM), nucleus (N), nuclear membrane (NM), mitochondria (M), vacuole (V) and septum (S). On the other hand, the hyphae treated with 4xMIC of AMA49CE (Fig. 1f) and AMA49F1 (Fig. 1h) showed cell membrane alterations including membrane ruptures, membrane shrinking and separation from the cell wall. Moreover, the extracts penetrated into cytoplasm and destroyed the organelles. These alterations were similar to those of P. oryzae PTRC1 hyphae treated with propiconazole at 4xMIC (32µg/ml) (Fig. 1d).
Toxicity of active crude extract and its fraction
G. mellonella were injected with AMA49CE and AMA49F1 at 4xMIC (32µg/ml). The mortality rate 72 h after injection with AMA49CE and AMA49F1 were only 6.7%, which was classified as harmless. Propiconazole, a standard antifungal agent, was classified as harmless. No mortality was observed in the control DMSO-injected larvae (Table 4).
Table (4):
Percent mortality of Galleria mellonella treated with extracts from Streptomyces sp. AMA49
| Concentrations | % Mortality± SE | ||||
|---|---|---|---|---|---|
| AMA49CE | AMA49F1 | Propiconazole | 3.2%DMSO | Uninjected | |
| 4xMIC | 6.7 ± 3.3a | 6.7 ± 3.3a | 0.0 ± 0.0a | 0.0 ± 0.0a | 0.0 ± 0.0a |
Conidial germination inhibition
Conidia of P. oryzae PTRC1 germinated well in 0.032% DMSO (negative control). After 24 h of incubation, 98.66% of conidia germinated with long germ tube lengths up to 100 mm and 75.67% produced appressoria. The effects of AMA49CE and AMA49F1 on conidial germination, germ tube elongation and appressorium formation were determined at 1xMIC (8µg/ml), 2xMIC (16µg/ml) and 4xMIC (32µg/ml) after 24 h incubation. AMA49CE and AMA49F1 at 4xMIC completely inhibited conidial germination as well as propiconazole (Fig. 2a). Both extracts at 1xMIC had less effect than propiconazole on conidial germination; however, they could inhibit germ tube elongation (Fig. 2a and c). In addition, AMA49CE and AMA49F1 at 2xMIC and 4xMIC could also completely inhibit appressorium formation (Fig. 2b). Both extracts (AMA49CE, AMA49F1) inhibited conidial germination, appressorium formation and germ tube length in a concentration-dependent manner. Furthermore, AMA49CE (4xMIC) showed sporicidal activity after 24 h treatment, while the conidia were completely killed by AMA49F1 at all concentrations tested (1xMIC, 2xMIC, and 4xMIC) after 24 h treatment (data not shown).

Fig. 2. The effect of extracts AMA49CE and AMA49F1 from Streptomyces sp. AMA49 on P. oryzae PTRC1 condia at 1xMIC (8µg/ml), 2xMIC (16µg/ml) and 4xMIC (32µg/ml) after 24 h incubation anaylsed by two-way ANOVA and Turkey HSD post-hoc test. Propiconazole and DMSO were used ad positive and negative controls, respectively. The data represent mean values based on three replicates in each treatment. Error bars indicate the ± SE. (* P £ 0.001; main effect of concentration. (a) condial germination inhibition, (b) appressorium formation inhibition, (c) germ tube lenght.
Characterization of the major component in the active fraction
AMA49F1 was analysed by HPLC/MS. The UV spectrum showed two major peaks at the retention times of 3.77 to 3.80 min and 4.00 to 4.08 min. The molecular formula of C11H18N2O2, deduced from a peak at m/z at 211.1442 in the HRESIMS spectrum, implied that the peak indicated diketopiperazines (DKPs). Besides, a peak presented at 15.3 min with UV absorption maxima at 224 nm. HRESIMS data displayed a m/z value at [M+H]+ 813.5121, indicating a molecular formula of C45H74O11. A database search within the Dictionary of Natural Products suggested oligomycin A, which was confirmed by HPLC analysis using an oligomycin standard. Several other peaks with m/z at 807.5262, 821.5050, 805.5096 and 791.5311 at retention times of 13.6, 13.8, 14.4 and 15.5 min indicated the presence of further members of the oligomycin family.
Morphological and molecular identification of actinobacterium AMA49
AMA49, isolated from mangrove soil from Songkhla Lake, was the most active actinobacterium against P. oryzae in this study. Its aerial mycelia were white in a colony grown on yeast extract-malt extract agar (ISP2) and glycerol-asparagine agar (ISP5), white to brown on oatmeal agar (ISP3) and tyrosine (ISP7), and pale yellow on inorganic salts starch agar (ISP4). No soluble pigment was observed after 14 days of incubation. This strain was able to utilize D-glucose as a carbon source (positive control), but not D-fructose, raffinose, cellobiose, sucrose, rhamnose, or mannitol. The spore chains presented as the spiral type. On the different culture media, the morphological characteristics and spore chains indicated that AMA49 belonged to the genus Streptomyces. The 16S rDNA sequence of strain AMA49 was compared with other sequences using the EzTaxon server. The sequence of AMA49 was most similar to Streptomyces phaeoluteichromatogenes AJ391814 with 99.49% similarity and the phylogenetic tree, constructed with MEGA 6.0 using the Neighbor-Joining method, showed that strain AMA49 was closely related to the genus Streptomyces (Fig. 3). The morphological characteristics and phylogenetic tree of AMA49 were considered confirmation that the strain was Streptomyces sp. The culture of Streptomyces sp. AMA49 was, therefore, deposited at the BIOTEC Culture Collection, Thailand as BCC77514. The 16S rDNA sequence of Streptomyces sp. AMA49 was submitted to the NCBI database by Accession No. KX129898.
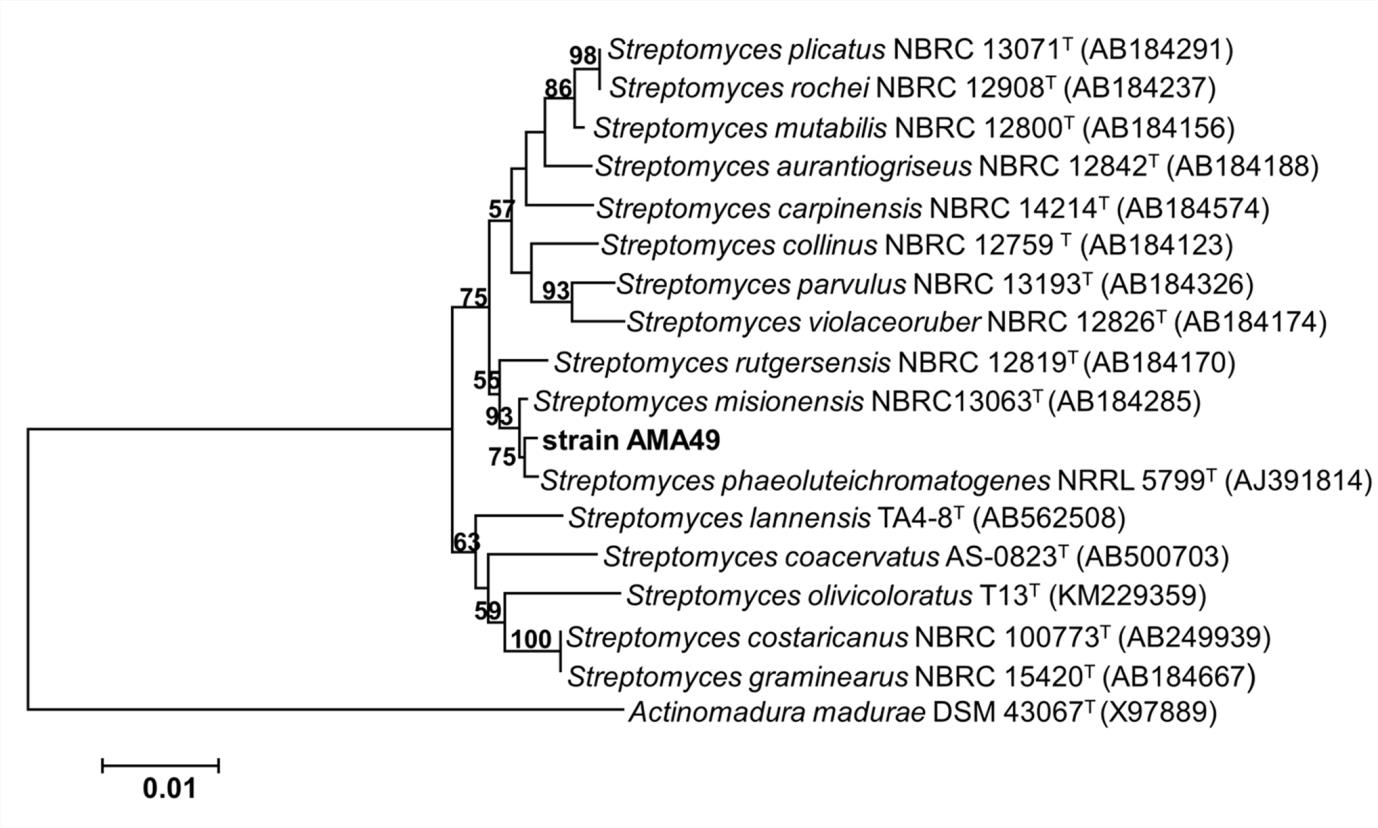
Fig. 3. Phylogentic tree of Streptomyces sp. AMA49 based on 16S rDNA sequecnes. The number on each branch represents bootstrap values (≥ 50%) from Neighbor-joining support with 1000 replications. The scale bar indicates 1% estimated sequence divergence. Actinomadura madurae X97889 was used as root of tree.
Rice blast, caused by P. oryzae fungus, is the most destructive disease of rice and many strains of Streptomyces have had important applications in agriculture as biocontrol agents against various plant pathogens, including P. oryzae40. Since marine environments are a good source of actinobacteria producing bioactive metabolites41, we have used various chemical and physical sample treatments in order to minimize contamination with fungi and other bacteria and to obtain marine-derived actinobacteria as many as possible, such as treatment with liquid nitrogen, 0.05% sodium dodecyl sulfate and 5% yeast extract42, 1.5% phenol, dry heat43 and rehydration and centrifugation44 as well as four selective isolation media including actinomycete isolation agar, modified soil extract agar45,46, humic acid vitamin agar46 and starch nitrate agar47 supplemented with cycloheximide and nalidixic acid. One hundred and fifty five out of 525 isolates were selected based on different morphologies. Among them, 64 of 155 isolates inhibited hyphal growth of P. oryzae PTRC1, exhibiting 34 to 89% inhibition by an agar well diffusion method. Isolate AMA49 isolated from mangrove soil in Songkhla Lake, Thailand, was the most active strain. This strain was identified as Streptomyces sp. using morphological characteristics and molecular identification based on the 16S rDNA sequencing. In addition, culture broth as well as extracts from cells and broth of AMA49 exhibited strong antifungal activities against various strains of P. oryzae. Most reported studies of the anti-P. oryzae activity of Streptomyces strains were based on dual culture methods26,48,49,50. This technique is only a quantitative method and it is difficult to make comparisons with the activities of other strains or antifungal agents because the results from this method depend on the solubility of active metabolites released into the agar medium. In our study, we first screened for potential isolates from their culture broths. After that, extracts from broths and cells were further tested. Although microorganisms produce secondary metabolites intracellularly and extracellularly, the agar well diffusion method used in this study detects mainly the extracellular metabolites that excreted into the fermentation medium. This medium may contain antifungal substances as well as hydrolytic enzymes. However, our preliminary study indicated that this isolate did not produce chitinase enzyme (data not shown). Therefore, the main antifungal activity may only be due to antifungal substances. Cells extracted with ethyl acetate of AMA49 (AMA49CE) and its subfraction (AMA49F1) showed strong activity against P. oryzae PTRC1 with low MIC (8 mg/ml) close to propiconazole, a standard antifungal agent (MIC 8 mg/ml). Ethyl acetate fermentation broth extracts from S. roseobiolascens XAS585 and S. roseofulvus XAS588 isolated from marine mud inhibited hyphal growth of fungal plant pathogens by more than 90% at 150 mg/ml51. In this study, AMA49CE at 8 to 16 mg/ml completely inhibited the growth of P. oryzae (100% inhibition). This suggests that AMA49CE has greater inhibitory activity than the strains S. roseobiolascens XAS585 and S. roseofulvus XAS58851. Conidial germination and appressorium formation are important processes in the penetration of P. oryzae into plant tissues52. This activity has been confirmed in greenhouse conditions by various investigators48,50. AMA49CE and AMA49F1 showed strong inhibition on conidial germination and appressorium formation. They also exhibited sporicidal activity. Streptomyces is the major genus of marine–derived actinobacteria that produced novel bioactive compounds15. There are several parameters that affect growth and metabolite production of marine-derived actinobacteria. Most marine-derived actinobacteria are believed to be terrestrial and have physiologically adapted to a marine environment53. Some isolates require seawater for growth54. In our study, we prepared the isolation media in 60% seawater. However, our obtained isolates grew well in ISP2 agar without seawater. Therefore, AMA49 was further grown in all media without seawater. The sea mud-derived S. roseobiolascens XAS585 and S. roseofulvus XAS588 were also fermented in the production media prepared with distilled water51.
Various types of antifungal compounds effective against plant pathogenic fungi have been isolated from Streptomyces spp. such as 6-prenylindole55, 3-acetonylidene-7-prenylindolin-2-one, 7-isoprenylindole-3-carboxylic acid56, fungichromin57, cyclo(leu-pro)58 and oligomycin A59. Our AMA49CE and its fraction AMA49F1 contained oligomycin A and its derivatives based on HPLC/MS analysis. Oligomycins belong to the polyketide macrolide antibiotics. Oligomycin A has been identified as a potent inhibitor of the mitochondrial ATP synthase. It can bind to a common drug-binding site in the ATP synthase shared with other drugs60. Oligomycin A was obtained from culture broth of Streptomyces libani As1 isolated from sea-mud soils in Korea59. It showed strong antifungal activity across a range of 1xMIC (3-5µg/ml) against the hyphal growth of many phytopathogenic fungi including Magnaporthe grisea. The efficiencies of oligomycin A were similar to those of metalaxyl, chlorothalonil, and tricyclazole. In addition, oligomycin A did not show any phytotoxicity on pepper, cucumber and rice plants. These findings reveal the potential of oligomycins in agriculture. Although oligomycin A was only a minor component in AMA49F1, its potent antifungal activity can explain the observed bioactivity of the extracts. The likewise detected diketopiperazines, cyclic dipeptides, might work co-operatively. One of the diketopiperazines, cyclo(leu-pro) extracted from Streptomyces sp. KH-614 from terrestrial soil, showed antifungal activity against P. oryzae at 1xMIC value of 2.5µg/ml58.
The possible mechanisms of action of AMA49CE and AMA49F1 were revealed by the SEM and TEM studies. Clear alterations to the fungal membrane included swollen, expanded and flattened hyphae, observed by SEM, and membrane shrinking and separation from the cell wall, observed by TEM. In addition, damaged organelles were also observed by TEM. To our knowledge, the effects of the above mentioned antifungal compounds on hyphal morphology have not been reported.
G. mellonella immune system is functionally and structurally similar to the mammalian immune system61. Therefore, the toxicity testing of drugs using G. mellonella larvae as a model has been widely used because of its correlation to toxicity testing using mammalian models such as mice and rats62. Although some cytotoxicity against HeLa cells has been detected for oligomycins A–E63, both AMA49CE and AMA49F1 produced a harmless effect on the G. mellonella model.
In conclusion, Streptomyces sp. AMA49 produced the active antifungal metabolite oligomycin A and diketopiperazines both in the cells and released into the culture media. Extracts from cells of this strain inhibited hyphal growth, conidial germination, and appressorium formation of P. oryzae and were nontoxic. Therefore, the extract AMA49CE and the partially purified fraction AMA49F1 from Streptomyces sp. AMA49 can be a good candidate as a biocontrol agent against the rice blast fungus.
Acknowledgements
We would like to thank the Phatthalung Rice Research Center, Thailand for providing various strains of Pyricularia oryzae and Dr. Patimapon Plodpai from the Department of Pest Management, Faculty of Natural Resources, Prince of Songkla University for providing the Phytophthora spp. Thanks to Drs. Jennifer Herrmann from Helmholtz Institute for Pharmaceutical Research Saarland and Timo Niedermeyer from AG Biogene Arzneistoffe Institut für Pharmazie, Martin-Luther-Universität Halle-Wittenberg, Germany for providing the oligomycin standard. We would like to thank Dr. Korakot Wichitsa-nguan Jetwanna from the Department of Mathematics, Faculty of Science, Prince of Songkla University, Thailand for statistical assistance. The authors are grateful to Mr. Thomas Coyne for English editing of the manuscript.
Conflict Of Interest
The authors declare that there is no conflict of interest.
Author’s Contribution
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Funding
This work was supported by grants PSU2556-003 PSU-Ph.D. Scholarship from Prince of Songkla University; BRN 002G-56 from the National Center for Genetic Engineering and Biotechnology, under the BRT’s Bioresources Utilization Program; FDA-CO-2558-1283-TH, the NSTDA Chair Professor grant of the Crown Property Bureau Foundation and the National Science and Technology Development Agency; grants from the Natural Product Research Center of Excellence, Prince of Songkla University and the Center of Excellence for Innovation in Chemistry (PERCH-CIC).
Data Availability
All datasets generated or analyzed during this study are included in the manuscript.
Ethics Statement
This article does not contain any studies with human participants performed by any of the authors. The study with invertebrates does not need ethical permission.
- FAO. FAO’s director-general on how to feed the world in 2050. Insights from an Expert Meeting at FAO, 2009; 1: 1–35.
- USDA. World agricultural supply and demand estimates. WASDE, 2019; 8: 585.
- Kato H. Rice blast control. Pestic Outlook, 2001; 12: 23-25.
- Dean R., Van Kan J.A.L., Pretorius Z.A., Hammond-Kosack K.E., Di Pietro A., Spanu P.D., Rudd J.J., Dickman M., Kahmann R., Ellis J., Foster G.D. The top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol., 2012; 13: 414-430.
- Yamaguchi I. Overview on the chemical control of rice blast disease, pp. 1-13. In Kawasaki, S (eds.), Rice blast: Interaction with rice and control, Dordrecht: Springer, 2004.
- Skamnioti P., Gurr S.J. Against the grain: safeguarding rice from rice blast disease. Trends Biotechnol., 2009; 27: 141-150.
- Sharma H., Parihar L. Antifungal activity of extracts obtained from actinomycetes. J. Yeast Fungal Res., 2010; 1: 197-200.
- Oskay M. Antifungal and antibacterial compounds from Streptomyces strains. Afr. J. Biotechnol., 2009; 8: 3007-3017.
- Nonoh J.O., Lwande W., Masiga D., Herrmann R., Presnail J.K., Schepers E., Okech M.A., Bagine R., Mungai P., Nyende A.B., Boga H.I. Isolation and characterization of Streptomyces species with antifungal activity from selected national parks in Kenya. Afr. J. Microbiol. Res., 2010; 4: 856-864.
- Alharbi S.A., Arunachalam C., Murugan A.M., Wainwright M. Antibacterial activity of actino-mycetes isolated from terrestrial soil of Saudi Arabia. J. Food Agric Environ., 2012; 10: 1093-1097.
- Awla H.K., Kadir J., Othman R., Rashid T.S., Wong M.Y. Bioactive compounds produced by Streptomyces sp. isolate UPMRS4 and antifungal activity against Pyricularia oryzae. Am. J. Plant Sci., 2016; 7: 1077-1085.
- Ahsan T., Chen J., Zhao X., Irfa M., Wu Y. Extraction and identification of bioactive compounds (eicosane and dibutyl phthalate) produced by Streptomyces strain KX852460 for the biological control of Rhizoctonia solani AG-3 strain KX852461 to control target spot disease in tobacco leaf. AMB Expr., 2017; 7: 54 (doi:10.1186/s13568-017-0351-z).
- Boudouresque C.F., Ruitton S., Bianchi C.N., Chevaldonné P., Fernandez C., HarmelinVivien M., Ourgaud M., Pasqualini V., Perez T., Pergent G., Thibaut T., Verlaque M. Terrestrial versus marine diversity of ecosystems. Proceedings of the 5th Mediterranean Symposium on Marine Vegetation, 2014; 11-25.
- Manivasagan P., Venkatesan J., Sivakumar K., Kim S.K. Marine actinobacterial metabolites: current status and future perspectives. Microbiol. Res., 2013; 168: 311-332.
- Lam K.S. Discovery of novel metabolites from marine actinomycetes. Curr. Opin. Microbiol., 2006; 9: 245-251.
- Leutou A.S., Yang I., Le T.C., Hahn D., Lim K.M., Nam S.J., Fenical W. Fluvirucin B6, a new macrolactam isolated from a marine-derived actinomycete of the genus Nocardiopsis. J. Antibiot., 2018; 71: 609-612.
- Kathiresan K., Balagurunathan R., Selvam M.M. Fungicidal activity of marine actinomycetes against phytopathogenic fungi. Indian J. Biotechnol., 2005; 4: 271-276.
- Fenical W., Jensen P.R. Developing a new resource for drug discovery: marine actinomycete bacteria. Nat. Chem. Biol., 2006; 2: 666-673.
- Jose P.A., Jha B. Intertidal marine sediment harbours actinobacteria with promising bioactive and biosynthetic potential. Sci Rep, 2017; 7: 10041 (doi: 10.1038/s41598-017-09672-6).
- Sangkanu S., Rukachaisirikul V., Suriyachadkun C., Phongpaichit S. Evaluation of antibacterial potential of mangrove sediment-derived actino-mycetes. Microb. Pathog., 2017; 112: 303-312.
- Bredholt H., Fjaervik E., Johnsen G., Zotchev S.B. Actinomycetes from sediments in the Trondheim Fjord, Norway: Diversity and biological activity. Mar. Drugs, 2008; 6: 12-24.
- Hamaki T., Suzuki M., Fudou R., Jojima Y., Kajiura T., Tabuchi A., Sen K., Shibai H. Isolation of novel bacteria and actinomycetes using soil-extract agar medium. J. Biosci. Bioeng., 2005; 99: 485-492.
- Zhang J., Zhang L. Improvement of an isolation medium for actinomycetes. Mod. Appl. Sci., 2011; 5: 124-127.
- Kennedy J., Baker P., Piper C., Cotter P.D., Walsh M., Mooij M.J., Bourke M.B., Rea M.C., O’Connor P.M., Ross R.P., Hill C., O’Gara F., Marcheri J.R., Dobson A.D.W. Isolation and analysis of bacteria with antimicrobial activities from the marine sponge Haliclona simulans collected from Irish waters. Mar. Biotechnol., 2009; 11: 384-396.
- Sabarathnam B., Manilal A., Sujith S., Kiran G.S., Selvin J., Thomas A., Ravji R. Role of sponge associated actinomycetes in the marine phosphorous biogeochemical cycles. Am-Euras J. Agric. & Environ. Sci., 2010; 8: 253-256.
- Boukaew S., Prasertsan P. Factors affecting antifungal activity of Streptomyces philanthi RM-1-138 against Rhizoctonia solani. World J. Microbiol. Biotechnol., 2014; 30: 323–329.
- Aghighi S., Shahidi Bonjar G.H., Rawashdeh R., Batayneh S., Saadoun I. First report of antifungal spectra of activity of Iranian actinomycetes strains against Alternaria solani, Alternaria alternate, Fusarium solani, Phytophthora megasperma, Verticillium dahliae and Saccharomyces cerevisiae. Asian J. Plant Sci., 2004; 3: 463-471.
- Gamliel A., Katan J., Cohen E. Toxicity of chloronitrobenzenes to Fusarium oxysporum and Rhizoctonia solani as related to their structure. Phytoparasitica, 1989; 17: 101-106.
- Golus J., Sawicki R., Widelski J., Ginalska G. The agar microdilution method – a new method for antimicrobial susceptibility testing for essential oils and plant extracts. J. Appl. Microbiol., 2016; 121: 1291-1299.
- Surup F., Chauhan D., Niggemann J., Bartok E., Herrmann J., Keck M., Zander W., Stadler M., Hornung V., Müller R. Activation of the NLRP3 inflammasome by hyaboron, a new asymmetric boron-containing macrodiolide from the myxo-bacterium Hyalangium minutum. ACS Chem. Biol., 2018; 13: 2981–2988.
- Boukaew S., Plubrukam A., Prasertsan P. Effect of volatile substances from Streptomyces philanthi RM-1-138 on growth of Rhizoctonia solani on rice leaf. Bio Control, 2013; 58: 471-482.
- Zhang Y.L., Li S., Jiang D.H., Kong L.C., Zhang P.H., Xu J.D. Antifungal activities of metabolites produced by a termite-associated Streptomyces canus BYB02. J. Agric. Food Chem., 2013; 61: 1521-1524.
- Maguire R., Duggan O., Kavanagh K. Evaluation of Galleria mellonella larvae as an in vivo model for assessing the relative toxicity of food preservative agents. Cell Biol Toxicol, 2016; 32: 209-216.
- Kavanagh K., Sheehan, G. The use of Galleria mellonella larvae to identify novel antimicrobial agents against fungal species of medical interest. J. Fungi, 2018; 4: 113 (doi: 10.3390/jof4030113).
- Lange A., Schäfer A., Bender A., Steimle A., Beier S., Parusel R., Frick J.S. Galleria mellonella: a new invertebrate model to distinguish intestinal symbionts from pathobionts. Front Immunol., 2018; 9: 2114 (doi: 10.3389/fimmu.2018.02114).
- Bisht K., Mishra V.K., Yadav S.K., Kumar R. Efficacy of some essential oils against the greater wax moth (Galleria mellonella L.) under storage condition. Environ. Ecol., 2017; 35: 2760-2763.
- Shirling E.B., Gottlieb D. Methods for characterization of Streptomyces species. Int. J. Syst. Bacteriol., 1966; 16: 313–340.
- Hall T.A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids Symp. Ser., 1999; 41: 95-98.
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA 6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol., 2013; 30: 2725-2729.
- Law J.W.F., Ser H.L., Khan T.M., Chuah L.H., Pusparajah P., Chan K.G., Goh B.H., Lee L.H. The potential of Streptomyces as biocontrol agents against the rice blast fungus, Magnaporthe oryzae (Pyricularia oryzae). Front Microbiol., 2017; 8: 3 (doi:10.3389/fmicb.2017.00003).
- Solanki R., Khanna M., Lal R. Bioactive compounds from marine actinomycetes. Indian J. Microbiol., 2008; 48: 410-431.
- Hayakawa M., Nonomura H. A new method for the intensive isolation of actinomycetes from soil. Actinomycetologica, 1989; 3: 95–104.
- Hayakawa M., Sadakata T., Kajiura T., Nonomura H. New methods for the highly selective isolation of Micromonospora and Microbispora from soil. J. Ferment Bioeng., 1991; 72: 320–326.
- Hayakawa M., Otoguro M., Takeuchi T., Yamazaki T., Iimura Y. Application of a method incorporating differential centrifugation for selective isolation of motile actinomycetes in soil and plant litter. An.t Van. Leeuwenhoek, 2000; 70: 171–185.
- Hamaki T., Suzuki M., Fudou R., Jojima Y., Kajiura T., Tabuchi A., Sen K., Shibai H., Isolation of novel bacteria and actinomycetes using soil-extract agar medium. J. Biosci. Bioeng., 2005; 99: 485–492.
- Zhang J., Zhang L. Improvement of isolation medium for actinomycetes. Mod. Appl. Sci., 2011; 5: 124–127.
- Ghanem N.B., Sabry S.A., El-Sherit Z.M., Abu El-Ela G.A. Isolation and enumeration of marine actinomycetes from seawater and sediments in Alexandria. J. Gen. Appl Microbiol., 2000; 46: 105–111.
- Ningthoujam D.S., Sanasam S., Tamreihao K., Nimaichand S. Antagonistic activities of local actinomycete isolates against rice fungal pathogens. Afr. J. Microbiol. Res., 2009; 3: 737–742.
- Zarandi M.E., Bonjar G.H.S., Dehkaei F.P., Moosavi S.A.A., Farokhi P.R., Aghighi S. Biological control of rice blast (Magnaporthe oryzae) by use of Streptomyces sindeneusis isolate 263 in greenhouse. Am. J. Appl. Sci., 2009; 6: 194–199.
- Li Q., Jiang Y., Ning P., Zheng L., Huang J., Li G., Jiang D., Hsiang T. Suppression of Magnaporthe oryzae by culture filtrates of Streptomyces globisporus JK-1. Biol. Control, 2011; 58: 139-148.
- Jianyou L., Jianrong X., Yongheng C. Isolation and identification of two marine-derived Streptomyces from marine mud of coast and offshore Zhuhai, and bioactive potential for plant pathogenic fungi. Afr. J. Biotechnol., 2011; 10: 11855-11860.
- Martin-Urdiroz M., Oses-Ruiz M., Ryder L.S., Talbot N.J. Investigating the biology of plant infection by the rice blast fungus Magnaporthe oryzae. Fungal Genet. Biol., 2016; 90: 61-68.
- Imada C., Koseki N., Kamata M., Kobayashi T., Hamada-Sato N. Isolation and characterization of antibacterial substances produced by marine actinomycetes in the presence of seawater. Actinomycetologica, 2007; 21: 27-31.
- Mincer T.J., Jensen P.R., Kauffman C.A., Fenical W. Widespread and persistent populations of a major new marine Actinomycete taxon in ocean sediments. Appl. Environ. Microbiol., 2002; 68: 5005–5011.
- Sasaki T., Igarashi Y., Ogawa M., Furumai T. Identification of 6-prenylindole as an antifungal metabolite of Streptomyces sp. TP-A0595 and synthesis and bioactivity of 6-substituted indoles. J. Antibiot., 2002; 55: 1009-1012.
- Zhang J., Wang J.D., Liu C.X., Yuan J.H., Wang X.J., Xiang W.S. A new prenylated indole derivative from endophytic actinobacteria Streptomyces sp. neau-D50. Nat. Prod. Res., 2014; 28: 431–437.
- Human Z.R., Moon K., Bae M., de Beer Z.W., Cha S., Wingfield M.J., Slippers B., Oh D.C., Venter S.N. Antifungal Streptomyces spp. Associated with the Infructescences of Protea spp. in South Africa. Front Microbiol., 2016; 7: 1657 (doi: 10.3389/fmicb.2016.01657).
- Rhee K.H. Purification and identification of an antifungal agent from Streptomyces sp. KH-614 antagonistic to rice blast fungus, Pyricularia oryzae. J. Microbiol. Biotechnol., 2003; 13: 984–988.
- Kim B.S., Moon S.S., Hwang B.K. Isolation, identification, and antifungal activity of a macrolide antibiotic, oligomycin A, produced by Streptomyces libani. Can. J. Bot., 1999; 77: 850–858.
- Symersky J., Osowski D., Walters D.E., Mueller D.M. Oligomycin frames a common drug-binding site in the ATP synthase. Proc. Natl. Acad. Sci., 2012; 109: 13961–13965.
- Wojida I. Immunity of the greater wax moth Galleria mellonella. Insect. Sci., 2017; 24:342-357.
- Ignasiak K., Maxwell A. Galleria mellonella (greater wax moth) larvae as a model for antibiotic susceptibility testing and acute toxicity trials. BMC Res. Notes, 2017; 10: 428 (doi:10.1186/s13104-017-2757-8).
- Kobayashi K., Nishino C. Oligomycin E., a new antitumor antibiotic produced by Streptomyces sp. MCI-2225. J. Antibiot., 1987; 40: 1053–1057.
- Hassan S.A., Bigler F., Bogensch tz H., Boller E., Brun J., Chiverton P., Edwards P., Mansour F., Naton E., Oomen P.A., Overmeer W.P.J., Polgar L., Rieckmann W., Samsרe Petersen L., Stהubli A., Sterk G., Tavares K., Tuset J.J., Viggiani G., Vivas A.G. Results of the fourth joint pesticide testing programme carried out by the IOBC/WPRS Working Group “Pesticides and Beneficial Organisms”. J. Appl. Ent., 1988; 105: 321-329.
© The Author(s) 2019. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.


