ISSN: 0973-7510
E-ISSN: 2581-690X
Human consumption of antibiotics has increased their concentrations in many parts of the environment, including rivers, sediments, soil, and wastewater. Consequently, resistant bacteria originating from these environments are distributed to humans, resulting in illness. The aim of this study was to identify mobilized colistin-resistant (mcr) genes and quinolone-resistant (qnr) genes in E. coli strains obtained from clinical samples. Additionally, the study explored the impact of different radiation dosages on the expression of antibiotic-resistance genes. In this study, conducted in Beni-Suef, Egypt, samples from 430 community-acquired urinary tract infection (UTI) cases resulted in the isolation of 85 different strains of E. coli. Conventional microbiological procedures were employed to identify these bacterial isolates. Three bacterial isolates with resistance to both quinolones and colistin underwent examination for their corresponding genetic determinants, which subsequently proved the presence of their respective genes. Furthermore, the expression levels of the mcr-1 and qnr-S genes were assessed using real-time PCR after exposure to gamma irradiation. Remarkably, the use of a sublethal dosage of 3 kGy gamma irradiation treatment on bacterial cells increased their susceptibility to colistin and quinolones post-irradiation. Additionally, there was a notable reduction in the expression levels of both mcr-1 and qnr-S genes, which could be helpful for preventing the storage of antibiotic-resistant E. coli in the environment.
mcr-1 Gene, qnr-S Gene, Antibiotic-resistant E. coli, Gamma Irradiation
The utilization of antibiotics by humans has resulted in an increase in antibiotic concentrations in waterways, sediments, soil, and sewage.1 Antibiotics pose a risk of entering the environment through multiple pathways, including the improper disposal of unused or expired antibiotics and the release of animal and human waste. Because of their limited absorption in the digestive systems of humans and animals, a substantial portion of antibiotics is excreted in feces and urine.2,3 The discharge of antibiotics during industrial antibiotic manufacturing often reaches concentrations of up to 1 mg/l, significantly higher than the amounts observed in human or animal waste.4
The persistence of antibiotic-resistant bacteria in the environment is attributed to the genetic variability of antibiotic-resistance genes.5 Horizontal gene transfer (HGT) plays a pivotal role in disseminating these resistant genes among bacteria. These processes involve the movement or transmission of genes responsible for antibiotic resistance from a pathogenic bacterium to other bacteria. Consequently, the proliferation of these resistance genes is driven by environmental selection.6 HGT can be facilitated by three distinct processes: conjugation, transduction, and transformation. Among these mechanisms, conjugation stands out as the primary means by which bacteria exchange genes for antibiotic resistance. During conjugation, genetic material, specifically DNA, is transferred among a broad range of bacterial species.7
The development of antibiotic resistance in environments is a complicated process because although antibiotic compounds might be present in nature, it is crucial to understand their contribution to antibiotic resistance.8
An intriguing aspect is the existence of numerous plasmid-mediated mobile colistin resistance (mcr) genes. According to Sun et al.,9 unknown selective pressures were responsible for the continuous evolution of the mcr gene, leading to the emergence of various mcr variants. Remarkably, within a relatively short timeframe, the mcr genes exhibited numerous variations. For instance, mcr-1 has thirteen variants, each differing from mcr-1 by one amino acid (mcr-1.1 to mcr-1.13).10
Moreover, recent discoveries indicate that mobile genetic components, including plasmids, can serve as mediators for quinolone resistance. Plasmid-encoded genes (qnr) mediate quinolone resistance and belong to the pentapeptide repeat proteins. These proteins act as a protective shield for DNA gyrase and topoisomerase IV against quinolone drugs.11 There are three primary categories of qnr elements: qnr-A, qnr-B, and qnr-S.12,13
Gamma irradiation is a form of electromagnetic radiation characterized by a low wavelength produced by radioactive isotopes as the unsteady nucleus disintegrates and decays into a stable form.14 Gamma rays can damage DNA either directly or by inducing ionization of water molecules. Within solutions, ionizing radiation produces three distinct free radicals: hydroxyl radicals (•OH), solvated electrons (e-eq), and hydrogen atoms (•H). This phenomenon occurs through the mechanism of water radiolysis.15 These free radicals can inflict damage based on reactive oxygen species effects on proteins.16 Additionally, they contribute to the occurrence of DNA double-strand breaks or single-strand breaks.15
To solve the issue of increasing the rise of E. coli antibiotic resistance genes, new strategies and approaches must be devised. Our study seeks to tackle this issue by exploring the impact of radiation at various dosages as a strategy to limit the dissemination of antibiotic resistance genes in the environment. This spread typically occurs through horizontal gene transfer (HGT),6 and our investigation underscores the efficacy of modern applications of gamma rays in contemporary practices, such as the treatment of blood components and the sterilization of medical equipment.
Samples collection
This study took place between January 2021 to January 2022. A total of 430 urine specimens were collected from specific laboratories, namely Alfa Dar-Elfoad Laboratory and Dar Elkhebra Laboratory, situated in Beni-Suef, Egypt. These specimens represented cases of community-acquired urinary tract infections. The urine samples were acquired through either catheterization or mid-stream clean-catch methods.
Isolation and purification of E. coli
Approximately 5 milliliters of urine samples underwent centrifugation at a speed of 2000 rpm for 5 minutes. Following centrifugation, a loopful of undiluted urine sediment was aseptically inoculated into suitable culture media, specifically Eosin Methylene Blue and MacConkey agar plates manufactured by Himedia, India. The incubation period lasted for 24 hours at 37°C.
In order to verify the purity of the isolated bacteria, well-isolated individual colonies were subjected to subculturing on the same growth medium. Identification of the isolates was carried out using microscopic and morphological approaches, involving the observation of macroscopic morphology and color on the appropriate agar media. The identification was further confirmed using the Vitek2 computerized system (Vitek 2 GN-card) from bioMerieux SA, France, following the manufacturer’s instructions.17
Antimicrobial susceptibility testing
The susceptibility of E. coli bacterial isolates to seventeen commonly used antibiotics was evaluated using the disc diffusion method. The protocols outlined by the Clinical and Laboratory Standards Institute (CLSI)18 were followed for this assessment. Antibiotic discs for this research were sourced from Oxoid, United Kingdom. They included colistin (CT) 10µg, piperacillin/tazobactam (TZP) 75/10µg, nitrofurantoin (F) 300µg, ciprofloxacin (CIP) 5µg, cefepime (FEP) 30µg, imipenem (IMP) 10µg, ceftazidime (CAZ) 30µg, levofloxacin (LEV) 5µg, norfloxacin (NOR) 10µg, ampicillin/sulbactam (SAM) 20µg, aztreonam (ATM) 30µg, chloramphenicol (C) 30µg, doxycycline (DOX) 30µg, amikacin (AK) 30µg, cefoperazone/sulbactam (SCF) 105µg, sulfamethoxazole/trimethoprim (SXT) 25µg and amoxicillin-clavulanic acid (AMC) 20/10µg. Fresh bacterial cultures were grown on Mueller-Hinton agar (MHA) plates obtained from Himedia, India. Subsequently, antibiotic discs were applied to the agar plates. After 24 hours of incubation at 37°C, the plates were examined, and the diameter of the inhibitory zone was measured and recorded in millimeters.
DNA extraction and detection of antibiotic resistance genes
Through polymerase chain reaction (PCR), we were able to identify the presence of genes encoding resistance to the antibiotics colistin (mcr-1 and mcr-2) and quinolones (qnr-A, qnr-B, and qnr-S), following the protocol outlined by Liu et al.19 The DNA extraction process employed the QIAamp DNA micro kit (Qiagen, USA), according to the manufacturer’s instructions. The PCR Master Mix was prepared in accordance with the recommendations specified by the manufacturer of the Emerald Amp GT PCR master mix (Takara) (Code No. RR310A kit). The primers used in this investigation were sourced from Metabion, Germany, and detailed information on primers and conditions is provided in Tables 1 and 2.
Table (1):
Oligonucleotide primers sequences
Gene |
Primer sequence (5′-3′) |
Length of amplified product |
Reference |
|---|---|---|---|
pho-A |
CGATTCTGGAAATGGCAAAAG |
720 bp |
20 |
CGTGATCAGCGGTGACTATGAC |
|||
mcr-1 |
CGGT CAGTCCGTTTGTTC |
308 bp |
21 |
CTTGGTCGGTCTGTAGGG |
|||
mcr-2 |
TGGTACAGCCCCTTTATT |
1617 bp |
22 |
GCTTGAGATTGGGTTATGA |
|||
qnr-A |
ATTTCTCACGCCAGGATTTG |
516 bp |
23 |
GATCGGCAAAGGTTAGGTCA |
|||
qnr-B |
GATCGTGAAAGCCAGAAAGG |
469 bp |
23 |
ACGATGCCTGGTAGTTGTCC |
|||
qnr-S |
ACGACATTCGTCAACTGCAA |
417 bp |
23 |
TAAATTGGCACCCTGTAGGC |
Table (2):
PCR conditions and annealing temperatures
Gene |
Primary denaturation |
Secondary denaturation |
Annealing |
Extension |
No. of cycles |
Final extension |
|---|---|---|---|---|---|---|
pho-A |
94°C |
94°C |
55°C |
72°C |
35 |
72°C |
5 min. |
30 sec |
40 sec |
45 sec |
10 min. |
||
mcr-1 |
94°C |
94°C |
60°C |
72°C |
35 |
72°C |
5 min. |
30 sec |
30 sec |
30 sec |
7 min. |
||
mcr-2 |
94°C |
94°C |
55°C |
72°C |
35 |
72°C |
5 min. |
30 sec. |
40 sec. |
1.2 min. |
12 min. |
||
qnr-A |
94°C |
94°C |
55°C |
72°C |
35 |
72°C |
5 min. |
30 sec. |
45 sec |
45 sec |
10 min. |
||
qn-B |
94°C |
94°C |
55°C |
72°C |
35 |
72°C |
5 min. |
30 sec. |
45 sec |
45 sec |
10 min. |
||
qnr-S |
94°C |
94°C |
55°C |
72°C |
35 |
72°C |
5 min. |
30 sec. |
45 sec |
45 sec |
10 min. |
Agarose gel electrophoreses
A sterile flask was prepared, containing 1.5 g of high-purity agarose suitable for electrophoresis. This flask was then combined with 100 ml of Tris-acetate-EDTA (TAE) buffer and subjected to microwave irradiation to ensure the complete dissolution of all the agarose granules. Following this, the mixture was allowed to cool to a temperature of 70°C, after which 0.5 mg/ml ethidium bromide was thoroughly blended.
The warmed agarose was promptly transferred into the gel casting device along with the required comb and allowed to cool to room temperature for polymerization. Subsequently, the comb was removed, and TAE buffer was added to the electrophoresis tank. A volume of 20µl was used for each individual PCR item, and both negative and positive controls were placed into the gel. The power source for the tank was set at a rate of 1-5 volts per centimeter of its length. After approximately 30 minutes, the running process was stopped, and the gel was transferred to the UV cabinet. The gel documentation system was used to capture images, and computer software was employed for the analysis and interpretation of the acquired data.24
Effect of gamma irradiation on bacterial cells
Ionizing radiation, such as gamma rays, has been employed to eradicate bacterial cells.25 Gamma irradiation was carried out at the Egyptian Atomic Energy Research Institute in Cairo, Egypt, using a high-level cobalt (60CO) source. The measured radioactivity of the source was estimated to be around 1.47 x 1017 Bq, equivalent to 397,949 Ci. The bacterial samples were cultured in Mueller Hinton broth medium for 24 hours in sterile test tubes. Subsequently, the samples were exposed to various gamma radiation doses: 0.25, 0.5, 1.0, 1.5, 2.0, 2.5, 3.0, and 3.5 kGy. The dose rate was 0.25 kGy/14.6 minutes during the experiment. Serial dilutions of the irradiated control and non-irradiated control cultures were plated on the surface of Mueller Hinton agar plates (5 µl each) and incubated for 24 hours at 37°C. The viable count was determined and compared.26
Determination of mcr-1 and qnr-S genes expression by SYBR Green real-time PCR after gamma irradiation treatment
The evaluation of antibiotic-resistant gene expression in isolates that were synergistically affected by ciprofloxacin or colistin with ZnO-NPs and exposed to varying doses of gamma radiation was conducted. The SYBR Green quantitative PCR (qPCR) test for the identification of mcr-1 and qnr-S was performed using the Quantitate SYBR green PCR kit (Cat. No. 204141) and RevertAid Reverse Transcriptase (Thermo, Fisher) (200 U/L) for master mix preparation for SYBR Green real-time PCR. (No. Cat. EP0441). Detailed information on primers and conditions is provided in Tables 3 and 4. The strata gene MX3005P software was used to calculate Ct values and amplification curves. Following the computation of gene expression variance in RNA samples using the strata gene MX3005P software, the CT values of each sample were compared with those of the control group using the “Ct” methodology as outlined in the provided reference.27
Whereas ΔΔCt = ΔCt reference – ΔCt target
ΔCt target = Ct control – Ct treatment and
ΔCt reference = Ct control- Ct treatment
Table (3):
Oligonucleotide primers in the SYBR Green real-time PCR assay
Gene |
Primer sequence (5′-3′) |
Reference |
|---|---|---|
mcr-1 |
CGGT CAGTCCGTTTGTTC |
21 |
CTTGGTCGGTCTGTAGGG |
||
qnr-S |
ACGACATTCGTCAACTGCAA |
23 |
TAAATTGGCACCCTGTAGGC |
||
16S rRNA |
GCTGACGAGTGGCGGACGGG |
28 |
TAGGAGTCTGGACCGTGTCT |
Table (4):
SYBR green real-time PCR cycling conditions
| Target gene | Reverse transcription | Primary denaturation | Amplification (40 cycles) | ||
|---|---|---|---|---|---|
| Secondary denaturation | Annealing | Extension (Optics on) | |||
| mcr-1 | 50°C | 94°C | 94°C | 60°C | |
| 30 min. | 15 min. | 15 sec. | 30 sec. | ||
| qnr-S | 55°C | 72°C | |||
| 30 sec. | 40 sec. | ||||
| 16S rRNA | 55°C | ||||
| 30 sec. | |||||
Bacterial isolates and antibiotic susceptibility
In Beni-Suef, Egypt, 85 different strains of E. coli were isolated from 430 urine samples obtained from outpatients experiencing urinary tract symptoms. The identification of bacterial isolates utilized standard microbiological techniques and was further validated using the Vitek2 computerized system (Vitek 2 GN-card) from bioMerieux SA, France, following the manufacturer’s instructions.
The investigation aimed to assess the susceptibility of E. coli isolates to seventeen commonly used antibiotics spanning different groups. The resistance rates of E. coli isolates to the tested drugs are detailed in Table 5.
Table (5):
Resistance rates of [85] E. coli isolate against [17] antimicrobial agents
No. |
Antimicrobial agent |
No. (%) |
|---|---|---|
1. |
Colistin (CT) 10 µg |
3 (3.5%) |
2. |
Nitrofurantoin (F) 300 µg |
3 (3.5%) |
3. |
Ciprofloxacin (CIP) 5 µg |
5 (5.8%) |
4. |
Levofloxacin (LEV) 5 µg |
5 (5.8%) |
5. |
Imipenem (IMP) 10 µg |
5 (5.8%) |
6. |
Norfloxacin (NOR) 10 µg |
10 (11.7%) |
7. |
Amikacin (AK) 30 μg |
14 (16.4%) |
8. |
Ampicillin/Sulbactam
(SAM) 20 µg |
19 (22.9%) |
9. |
Ceftazidime (CAZ) 30 µg |
21 (24.7%) |
10. |
Doxycycline (DOX) 30 μg |
21 (24.7%) |
11. |
Chloramphenicol (C) 30 µg |
22 (25.9%) |
12. |
Cefoperazone/Sulbactam (SCF) 105 µg |
28 (32.9%) |
13. |
Aztreonam (ATM) 30 μg |
31(36.4%) |
14. |
Amoxicillin-clavulanic acid
(AMC) 20/10µg |
34 (40%) |
15. |
Cefepime (FEP) 30 μg |
34 (40%) |
16. |
Piperacillin/Tazobactam (TZP) 75/10 μg |
35 (41.2%) |
17. |
Sulfamethoxazole/Trimethoprim (SXT) 25µg |
35 (41.2%) |
The results overall indicated a high prevalence of multidrug resistance among the tested isolates. Specifically, the study revealed a significant proportion of E. coli isolates with elevated resistance levels to piperacillin/tazobactam (41.2%), sulfamethoxazole/trimethoprim (41.2%), amoxicillin-clavulanic acid (40%) and cefepime (40%). Furthermore, a similar study by Nader et al.29 indicated that E. coli isolated from UTIs demonstrated high resistance to the most common antibiotic used in treating UTIs, with the highest resistance rates observed for trimethoprim/sulfamethoxazole (68.75%).
In contrast, the antibiotics with the lowest levels of resistance in the present study were colistin (3.5%), ciprofloxacin (5%), levofloxacin (5%), imipenem (5%), and nitrofurantoin (5%). In previous research, Mashouf et al.30 observed that imipenem, cefepime, and ciprofloxacin demonstrated the most significant antibacterial effects on all tested Gram-negative isolates. Additionally, Randrianirina et al.31 noted a reduced susceptibility to trimethoprim/sulfamethoxazole and ciprofloxacin, and the strains exhibited minimal susceptibility to ceftriaxone, with a documented resistance rate of 53%.
Detection of antibiotic resistance genes
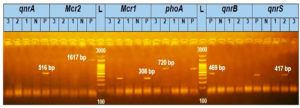
The findings from the polymerase chain reaction (PCR) study revealed that among the investigated E. coli isolates, three isolates (3.5%) were found to possess either a qnr or mcr gene. Notably, a single isolate, E. coli A2, was shown to harbor both the mcr-1 and qnr-S genes, while none of the E. coli isolates exhibited the presence of qnr-A, qnr-B, or mcr-2 genes, as shown in Figure 1.
Figure 1. The Agarose gel electrophoresis image showed that E. coli strain A2 has mcr-1 (308 bp) and qnr-S (417 bp) genes together. (Notes: (1,2,3) are samples numbers, (N) is a negative control and (P) is a positive control.)
In another investigation, four isolates of E. coli that displayed resistance to colistin based on their phenotype revealed that 3 (75%) contained the chromosomal mcr-1 genes.32
Li et al.33 reported that out of 9 (7.3%) E. coli isolates resistant to colistin, PCR screening revealed that 7 (5.7%) harbored the mcr-1 gene. In another study, Rezazadeh et al.34 reported that qnr-S1 was present in 4 (2.9%) of the 136 isolates of E. coli resistant to quinolones. The clinical isolates in this investigation lacked the qnr-A and qnr-B genes. Foda et al.35 isolated E. coli from clinical samples resistant to colistin with MICs ranging from 6.25 to >200µg/ml, and these strains contained the mcr-1 gene.
Effect of gamma irradiation on bacterial cells
Radiation is commonly employed in the processing of blood components, sterilization of medical equipment, preservation of food, and preparation of tissue allografts. This use of radiation eliminates the need for elevated temperatures that might pose potential harm to these items.36 Additionally, it plays a role in sterilizing water and managing waste in clinical environments. Various factors influence bacterial sensitivity to radiation-induced death, including the rate of cell replication, intracellular water content, DNA content, medium composition, temperature, pH, oxygenation level, and the capacity to repair radiation-induced DNA damage.25,37-38
The findings of our investigation indicate that bacterial cells displayed sensitivity to a dose of 3 kGy, which could be classified as either a sensitive or a sublethal dose. Notably, they became susceptible to colistin and quinolones after irradiation.
These findings align with a previous study documenting the impact of gamma radiation at 2.8 kGy and 3.6 kGy on the viability of stationary phase cells of S. epidermidis and E. coli, respectively.39 Another study revealed that gamma radiation affected E. coli resistance to some antibiotics at doses of 2, 3, and 4 Gy, without inhibiting it. However, following exposure to 5 Gy, a small zone of inhibition was observed.40
Furthermore, our results are consistent with those of Aziz et al.,41 where a dose of 3 KGy ultimately reduced the viable count of Gram-positive short-rod bacteria isolates.
Determination of mcr-1 and qnr-S genes expression after gamma irradiation treatment by SYBR Green real-time PCR
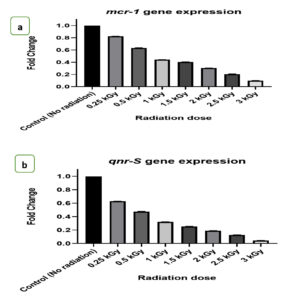
The gene expression levels of mcr-1 and qnr-S genes were quantitatively measured using the sensitive technique of quantitative RT-PCR following gamma irradiation treatment. Transcriptional alterations in these genes were determined from the fold change, as illustrated in Figures 2a and b.
Figures 2. a) The effect of radiation doses on fold changes of mcr-1 genes expression, b) The effect of radiation doses on fold changes of qnr-S genes expression
Our study’s results concerning the clinical isolate E. coli A2 indicate that exposure to gamma irradiation led to a decrease in the expression levels of the mcr-1 and qnr-S genes. These genes are known contributors to the antibiotic-resistant properties of bacteria, particularly against colistin and quinolones antibiotics. Consequently, the susceptibility of the bacteria to these antibiotics would be enhanced.
Another study that examined the effects of radiation on E. coli reported a radioprotective system against the initial instance of DNA harm or damage and to promote long-term survivability, the dna-K gene was strongly activated in response to gamma radiation.42 and in contrast to the findings of research, this is in agreement with Yamaguchi et al.43 who showed that mutations in the dna-K genes increased the susceptibility of the bacteria to levofloxacin and that the dna-K genes affect the antibacterial action of the fluoroquinolone in E. coli. Another study found that a mutation in the dna-K gene improved the susceptibility of methicillin-resistant Staphylococcus aureus to oxacillin and methicillin.44
An additional study investigating the impacts of gamma radiation on uropathogenic E. coli expressing colibactin and Cnf-1, aimed at investigating their impact on cytotoxicity, revealed a significant rise in the expression of clbA and clbQ genes in cultures subjected to irradiation at doses resembling these received during pelvic radiotherapy.45
An extra study was conducted to examine the impact of radiation on the treatment of abdominal tumors through radiotherapy. The findings of this study indicated alterations in the microbiota composition, resulting in a reduction in diversity. Specifically, there was a decrease in the presence of Lactobacillus spp. and Bifidobacterium spp., while Staphylococcus spp. and E. coli exhibited an increase. The increased number of Pathogenic E. coli results in the repositioning of claudin-1, occludin, and ZO-1 within tight junctions. This condition exacerbates radiation enteritis and enhances the expression of inflammatory factors.46
According to the results obtained from our investigation, it was observed that gamma irradiation had a beneficial effect on the antibiotic resistance of E. coli. This was evidenced by a reduction in the expression levels of both mcr-1 and qnr-S genes, leading to an increased susceptibility to colistin and quinolones antibiotics. Consequently, gamma irradiation shows promise in limiting the spread of antibiotic resistance genes within the environment, which occurs through horizontal gene transfer (HGT). This proves the effectiveness of modern applications of gamma rays in contemporary practices, such as the treatment of blood components and sterilization of medical equipment.
ACKNOWLEDGMENTS
The authors would like to thank members of the Materials Science and Nanotechnology Department, Faculty of Postgraduate Studies for Advanced Sciences (PSAS), Beni-Suef University, Beni-Suef, Egypt, and members of gamma irradiation unit, the Egyptian Atomic Energy Research Institute, Cairo, Egypt, for their help to complete this project.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
GMES conceptualized and supervised the study. AGM collected resources and analyzed the data. AOEG, MSA and HMEK applied methodology. AFA and AGM performed the experiments. GMES performed formal analysis. AOEG performed investigation. HMEK performed visualization. MSA and HMEK data curation. AGM wrote the manuscript. AGM, AOEG and GMES reviewed the manuscript. AGM and GMES edited the manuscript. All authors read and approved the final manuscript for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
Not applicable.
- Ma J, Zhai G. Antibiotic Contamination: A Global Environment Issue. Journal of Bioremediation & Biodegradation. 2014;5(5):e157.
Crossref - Sarmah AK, Meyer MT, Boxall AB. A global perspective on the use, sales, exposure pathways, occurrence, fate, and effects of veterinary antibiotics (VAs) in the environment. Chemosphere. 2006; 65(5), 725-759.
Crossref - Larsson DG. Antibiotics in the environment. Ups J Med Sci. 2014; 119(2), 108-112.
Crossref - Kristiansson E, Fick J, Janzon A, Grabic R, Rutgersson C, Weijdegard B, Larsson D G. Pyrosequencing of antibiotic-contaminated river sediments reveals high levels of resistance and gene transfer elements. PLoS One. 2011; 6(2), e17038.
Crossref - Pal C, Bengtsson-Palme J, Kristiansson E, Larsson DG. The structure and diversity of human, animal and environmental resistomes. Microbiome. 2016; 4(1), 54.
Crossref - Thomas CM, Nielsen KM. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nature Reviews Microbiology. 2005; 3(9), 711-721.
Crossref - Canton R, Gonzalez-Alba JM, Galan JC. CTX-M Enzymes: Origin and Diffusion. Frontiers in Microbiology, 2012; 3, 110.
Crossref - Kümmerer K. Resistance in the environment. Journal of Antimicrobial Chemotherapy, 2004; 54(2), 311-320.
Crossref - Sun J, Zhang H, Liu YH, Feng Y. Towards understanding MCR-like colistin resistance. Trends in microbiology. 2018; 26(9) 794-808.
Crossref - Wang Y, Tian GB, Zhang R, Shen Y, Tyrrell JM, Huang X, Shen J. Prevalence, risk factors, outcomes, and molecular epidemiology of mcr-1-positive Enterobacteriaceae in patients and healthy adults from China: an epidemiological and clinical study. The Lancet infectious diseases.2017; 17(4), 390-399.
Crossref - Strahilevitz J, Jacoby GA, Hooper DC, Robicsek A. Plasmid-mediated quinolone resistance: a multifaceted threat. Clinical microbiology reviews. 2009; 22(4) 664-689.
Crossref - Kim HB, Park CH, Kim CJ, Kim EC, Jacoby GA, Hooper DC. Prevalence of plasmid-mediated quinolone resistance determinants over a 9-year period. Antimicrobial agents and chemotherapy. 2009; 53(2), 639-645.
Crossref - Minarini LA, Poirel L, Cattoir V, Darini ALC, Nordmann P. Plasmid-mediated quinolone resistance determinants among enterobacterial isolates from outpatients in Brazil. Journal of Antimicrobial Chemotherapy.2008; 62(3), 474-478.
Crossref - Mendonca AF, Romero MG, Lihono MA, Nannapaneni R, Johnson MG. Radiation resistance and virulence of Listeria monocytogenes Scott A following starvation in physiological saline. Journal of food protection.2004; 67(3), 470-474.
Crossref - Neill PO. The Chemical Basis of Radiation Biology. International Journal of Radiation Biology and Related Studies in Physics, Chemistry and Medicine, 1987; 52:6, 976.
Crossref - Lee SJ, Choi SA, Lee KH, Chung HY, Kim TH, Cho CK, Lee YS. Role of inducible heat shock protein 70 in radiation-induced cell death. Cell stress & chaperones.2001; 6(3), 273-281.
- Valenza G, Ruoff C, Vogel U, Frosch M, Abele-Horn M. Microbiological evaluation of the new VITEK 2 Neisseria-Haemophilus identification card. Journal of clinical microbiology. 2007; 45(11), 3493-3497.
Crossref - CLSI. M100: Performance Standards for Antimicrobial Susceptibility Testing. 28th informational supplement Wayne PA, Clinical and Laboratory Standards Institute. 2018.
- Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, Shen, J. Emergence of plasmid-mediated colistin resistance mechanism mcr-1 in animals and human beings in China: a microbiological and molecular biological study. The Lancet infectious diseases.2016; 16(2), 161-168.
Crossref - Hu Q, Tu J, Han X, Zhu Y, Ding C, Yu S. Development of multiplex PCR assay for rapid detection of Riemerella anatipestifer, Escherichia coli, and Salmonella enterica simultaneously from ducks. Journal of microbiological methods.2011; 87(1), 64-69.
Crossref - Newton-Foot M, Snyman Y, Maloba MRB., Whitelaw AC. Plasmid-mediated mcr-1 colistin resistance in Escherichia coli and Klebsiella spp. Clinical isolates from the Western Cape region of South Africa. Antimicrobial Resistance & Infection Control.2017; 6(1), 1-7.
Crossref - Xavier BB, Lammens C, Ruhal R, Kumar-Singh S, Butaye P, Goossens H, Malhotra-Kumar S. Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium, June 2016. Euro. Surveillance.2016; 21(27), 30280.
Crossref - Robicsek A, Strahilevitz J, Jacoby GA, et al. Fluoroquinolone-modifying enzyme: a new adaptation of a common aminoglycoside acetyltransferase. Nature medicine.2006; 12(1), 83-88. http://www.nature.com/naturemedicine
- Sambrook J, Fritscgh EF, Mentiates. Molecular cloning. A laboratory manual, 1-82., Cold spring Harbor Laboratory Press, New York. 1989.
- Thayer DW, Boyd G. Elimination of Escherichia coli O157: H7 in meats by gamma irradiation. Applied and Environmental Microbiology.1993; 59(4), 1030-1034.
Crossref - ASTM. Standards on dosimetry for radiation processing: ISO/ASTM 51607:2004. ASTM International. West Conshohocken, Pennsylvania. 2004.
- Yuan JS, Reed A, Chen F, Stewart CN. Statistical analysis of real-time PCR data. BMC bioinformatics2006; 7(1) 1-12. http://www.biomedcentral.com/1471-2105/7/85
- Tivendale KA, Allen JL, Ginns CA, Crabb BS, Browning GF. Association of iss and iucA, but not tsh, with plasmid-mediated virulence of avian pathogenic Escherichia coli. Infection and immunity. 2004; 72(11), 6554-6560.
Crossref - Nader AN, Rania MK, Mohammed A et al. Bacterial Pattern of Community Acquired Urinary Tract Infections: A Challenge for Antimicrobial Resistance, Egyptian Journal of Medical Microbiology. 2021; 30(3), 153-162.
- Mashouf RY, Babalhavaeji H, Yousef J. Urinary tract infections: bacteriology and antibiotic resistance patterns. Indian pediatrics. 2009; 46(7).
- Randrianirina F, Soares JL, Carod JF, Ratsima E, Thonnier V, Combe P, Talarmin A. Antimicrobial resistance among uropathogens that cause community-acquired urinary tract infections in Antananarivo, Madagascar. Journal of Antimicrobial Chemotherapy.2007; 59(2), 309-312.
Crossref - Karki D, Dhungel B, Bhandari S, Kunwar A, Joshi PR, Shrestha B, Banjara MR. Antibiotic resistance and detection of plasmid mediated colistin resistance mcr-1 gene among Escherichia coli and Klebsiella pneumoniae isolated from clinical samples. Gut pathogens.2021; 13(1), 1-16.
Crossref - Li B, Ke B, Zhao X, et al. Antimicrobial resistance profile of mcr-1 positive clinical isolates of Escherichia coli in China from 2013 to 2016. Frontiers in microbiology. 2018; 9, 2514.
Crossref - Rezazadeh M, Baghchesaraei H, Peymani A. Plasmid-mediated quinolone-resistance (qnr) genes in clinical isolates of Escherichia coli collected from several hospitals of Qazvin and Zanjan Provinces, Iran. Osong Public Health and Research Perspectives.2016; 7(5), 307-312.
Crossref - Foda AM, Kalaba MH, El-Sherbiny GM, Moghannem SA, El-Fakharany EM. Antibacterial activity of essential oils for combating colistin-resistant bacteria, Expert Review of Anti-infective Therapy. 2022; 20:10, 1351-1364,
Crossref - Kainer MA, Linden JV, Whaley DN, et al. Clostridium infections associated with musculoskeletal-tissue allografts. New England Journal of Medicine.2004; 350(25), 2564-2571.
Crossref - Thayer DW, Boyd G. Effect of irradiation temperature on inactivation of Escherichia coli O157: H7 and Staphylococcus aureus. Journal of Food Protection.2001; 64(10), 1624-1626.
Crossref - Thayer DW, Rajkowski KT, Boyd G, Cooke PH, Soroka DS. Inactivation of Escherichia coli O157: H7 and Salmonella by gamma irradiation of alfalfa seed intended for production of food sprouts. Journal of Food Protection.2003; 66(2): 175-181.
- Trampuz A, Piper KE, Steckelberg JM, Patel R. Effect of gamma irradiation on viability and DNA of Staphylococcus epidermidis and Escherichia coli. Journal of medical microbiology 2006; 55(9), 1271-1275.
Crossref - Ogunniran AO, David OM, Isinkaye MO, Olawale AK. Effects of sub-lethal gamma irradiation on antibiotic susceptibility profile and population dynamics of Enterococcus faecalis and Escherichia coli in water. Sri Lanka Journal of Aquatic Sciences2019; 24(1):11-16
Crossref - Aziz NH, Khalil OA, Swelam M, Aly N. Characterization and effect of gamma irradiation on indigenous chloroaromatic degrading bacteria. Isotope and Radiation Research2005; 37(4), 1139-1158.
- Calini V, Urani C, Camatini, M. Overexpression of HSP70 is induced by ionizing radiation in C3H 10T1/2 cells and protects from DNA damage. Toxicology in vitro.2003; 17(5-6), 561-566.
Crossref - Yamaguchi Y, Tomoyasu T, Takaya A, Morioka M, Yamamoto T. Effects of disruption of heat shock genes on susceptibility of Escherichia coli to fluoroquinolones. BMC microbiology.2003; 3(1), 1-8. http://www.biomedcentral.com/1471-2180/3/16
- Singh VK, Utaida S, Jackson LS, Jayaswal RK, Wilkinson BJ, Chamberlain NR. Role for dnaK locus in tolerance of multiple stresses in Staphylococcus aureus. Microbiology.2007; 153(9), 3162-3173.
Crossref - Morgan RN, Farrag HA, Aboulwafa MM, Saleh SE. Effect of subinhibitory concentrations of some antibiotics and low doses of gamma radiation on the cytotoxicity and expression of colibactin by an uropathogenic Escherichia coli isolate. Current Microbiology. 2021; 78, 544-557.
Crossref - Jian Y, Zhang D, Liu M, Wang Y, Xu ZX. The impact of gut microbiota on radiation-induced enteritis. Frontiers in cellular and infection microbiology. 2021; 11, 586392.
Crossref
© The Author(s) 2023. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.