ISSN: 0973-7510
E-ISSN: 2581-690X
https://dx.doi.org/10.22207/JPAM.13.1.58 | © The Author(s). 2019
This study investigated the antibacterial resistance profiles of E. coli pathotypes isolated from children under five years and drinking water samples collected from the North West Province of South Africa and ascertained the clonality of the isolates. Two hundred and forty-one E. coli isolates were recovered from stool samples of diarrhoeic and non-diarrhoeic children under five years old, and drinking water, using the Colilert-18® Quanti-tray/2000 and Eosin methylene blue agar. The presence of enteropathogenic (eaeA), enterohaemorrhagic (eaeA,stx1, stx2 and flicH7), enteroaggregative (eagg), enteroinvasive (ipaH) and enterotoxigenic (ST and LT) E. coli pathotypes were also investigated using PCR. Antibiotic susceptibility was carried out through the disk diffusion method. The presence of blaCTX-M, blaSHV, blaCMY, and blaDHA genes that code for b-lactamases was investigated using real-time PCR. Similarities between human and water isolates were tested using ERIC-PCR. Overall, EHEC (35.8%), EPEC/EHEC (22%), ETEC (21.6%) and EIEC (20.2%) were detected. The highest antibiotic resistance was detected to Clarithromycin (100%) and Erythromycin (100%) while the lowest resistance was against Gentamicin (0.4%). Also, 100% sensitivity was recorded to imipenem and meropenem. Multi-antibiotic resistance was observed in all the pathotypes, and the ESBL genes were detected in 71.6% of the pathotypes. ERIC-PCR indicated 100% similarities in some water and human samples. Pathogenic E. coli is amongst the diarrhoea-causing agents in the North West Province with EHEC being the most identified pathotype. The clonal relatedness of the human and water isolates suggests that domestic water might be a route of transmission.
E. coli, Stool samples, Antibiotic resistance, children.
Escherichia coli is a normal flora of warm-blooded animals, including humans. However, some strains have acquired pathogenic potentials allowing them to cause diarrhoea (intestinal pathotypes) and other systemic illnesses involving the urinary tract, brain and blood (extra intestinal pathotypes)1. Intestinal or diarrhoeagenic E. coli (DEC) strains constitute the primary diarrhoea aetiologic agents in children younger than five years old2,3 and in most cases, they are associated with growth defects4. About 760,000 children lose their lives every year to diarrhoeal diseases worldwide5. Diarrhoeagenic E. coli pathotypes are ubiquitous in the environment, especially in aquatic environments5,6 and are most often responsible for waterborne diarrhoeal disease outbreaks both in developed and developing countries6–8. For example, they were responsible for the second largest waterborne diarrhoea outbreak in the world reported in 2011 in Germany, which recorded over 3,000 hospitalisations and 36 fatalities8. DEC strains are separated into different pathotypes based on specific virulence factors, their invasive capability and the clinical symptoms produced. These pathotypes include the EPEC (Enteropathogenic E. coli), STEC (Shiga toxin-producing E. coli or EHEC (Enterohaemorrhagic E. coli)), ETEC (Enterotoxigenic E. coli), EIEC (Enteroinvasive E. coli), EAEC (Enteroaggregative E. coli)9 and recently, DAEC (Diffusely-adherent E. coli) 10. Each pathotype has its unique virulence-associated gene(s) (VGs) contributing to its disease-causing potentials11.
EPEC members are a significant cause of infantile diarrhoea and are frequently associated with public places like schools and hospitals12. Members of this pathotype are responsible for approximately 5-10% of pediatric diarrhoea episodes in developing countries13. An infectious dose of 108 CFU can trigger infection even in a healthy adult14. EPEC strains contain a cluster of VGs on a the Locus Enterocyte Effacement (LEE) which is a pathogenicity island involved in severe diarrhoea15,16. EPEC members could be classified into atypical EPEC ((aEPEC; lacking the adherence effector plasmid (EAF) but containing the LEE) or typical EPEC (tEPEC; possessing the EAF plasmid). Characteristic EPEC pathotypes produce peculiar histopathological lesions on intestinal epithelial cell surfaces17. The mechanism of infection in EPEC involves numerous virulence factors, including intimin, bundle-forming pilus (BFP), paa and Long polar fimbriae (LPF)5.
The eae-encoded intimin proteinis an essential EPEC virulence gene which allows for attaching and effacing (A/E) lesions on the host’s intestinal epithelial cell surface5,18. Other virulence factors found in tEPEC are the bfpA and perABC genes which encode the bundle-forming pili used for localised attachment to epithelial cells15,16. STEC, also referred to as EHEC, consists of pathotypes that can cause hemorrhagic colitis (HC)9. In most severe cases, infection may lead to hemolytic uremic syndrome (HUS)19. The most notorious serotype in this group is the O157:H7 serotype, often implicated in most foodborne diseases outbreaks globally2. The pathogenesis of STEC infections involves two Shiga-like toxins (stx1 and stx2), and other VGs that aid in attachment like the intimin (eae) gene. STEC/EHEC members are the primary cause of foodborne illnesses and can initiate foodborne diarrhoea even at minimal infective doses20.
ETEC pathotypes are the cause of childhood diarrhoea in many developing countries. Also, a few foodborne outbreaks due to ETEC have been recorded in some developed countries5,11,21,22. ETEC pathotypes induce infection by attaching to the intestinal cell surface, and secreting either the heat stable (ST) or labile (LT) enterotoxin (or both) which are plasmid encoded23. The ST toxin is a 2kDa single peptide toxin; its mode of action is the activation of guanylyl cyclase C to increase the cyclic guanosine monophosphate (cGMP) level. An increase in cGMP leads to a surge in bicarbonate and chloride ion secretion which inhibits the intake of sodium and chloride ions resulting in loose diarrhoea24. LT, on the other hand, is an 86kDa protein. It deregulates the host’s adenylate cyclase, thus, enhancing attachment to intestinal epithelial cells which results in diarrhoea25.
EIEC pathotypes, like Shigella, cause of watery diarrhoea or bacillary dysentery in severe cases26. The process of infection depends on some essential virulence chromosomal-encoded and plasmid-encoded determinants, including the ipaH gene27. They are highly invasive pathogens capable of attaching to intestinal epithelial cells and replicating to the large intestine28. EAEC pathotypes are the most commonly encountered aetiologies of mild and persistent diarrhoea29. They are frequently isolated from both diarrhoeic and non-diarrhoeic individuals, especially in low-resource settings3.
EAEC members have been reported to cause inflammation of the intestines in children and impairing their growth and negatively impacting their development even without diarrhoea4. EAEC virulence genes are heterogeneous. The pathogenesis and genes encoding virulence factors in these strains are not uniform but show a considerable degree of variability among strains4,14. Nonetheless, the plasmid-borne aggR gene has been characterised and reported as the most common gene associated with EAEC pathotypes, and it regulates the virulence genes encoded in the chromosomes, plasmid and the type VI secretion system4.
Most diarrhoea cases due to DEC are self-limiting. Oral rehydration and electrolyte replacement have been advised and reported to be effective in some paediatric cases30. Antibiotics such as fluoroquinolones, azithromycin and rifampicin have been used in severe diarrhoea cases and have reportedly reduced the duration of diarrhoea symptoms31. However, because of the excessive use of numerous antibiotics against these pathogens, resistant strains have emerged21,31. There is a global increase in antibiotic-resistance among pathogenic E. coli strains against commonly prescribed antibiotics, even to newly discovered and more potent antibiotics2,31. Multi-drug resistant strains are another public health hazard associated with these DEC pathotypes. It has been reported that E. coli infections caused by multi-drug resistance strains are problematic, as they are usually severe and take a longer time to clear especially in infections in children21,32,33.
Phenotypic multi-drug resistance of some organisms including pathogenic E. coli emerged because of a metallo-b-lactamase gene possessed by these strains. This gene plays an active role in hydrolysing all b-lactam antibiotics except aztreonam34 and it is said to confer resistance to other antibiotics in E. coli11,21. In pathogenic E. coli, the most essential resistance mechanism against b-lactams involves plasmid-encoded extended-spectrum b-lactamases (ESBLs)33,35. ESBL-producing E. coli hydrolyses penicillin, extended spectrum Cephalosporins and the monobactams19,21 but cannot efficiently degrade Cephamycin, Carbapenems and b-lactamase inhibitors19,36. However, resistant strains have emerged11. There are different groups of ESBLs including CTX-M, CMY, SHV and DHA37–39. The CTX-M are reported to be a product of plasmid transfer from pre-existing chromosomal ESBL genes38. They are the most frequently reported group in most parts of the world21,40.
Drinking poorly treated or untreated polluted water or consuming raw or partially-cooked meat and vegetables have been implicated in the transmission of pathogenic E. coli strains to humans41,42. Molecular techniques like the enterobacterial repetitive intergenic consensus-PCR (ERIC-PCR) have been used to detect similarity between Gram-negative bacteria by comparing their genetic fingerprints40,43. This genotyping approach ensures strain-specific fingerprinting which allows for the evaluation of the genetic diversity in pathogenic E. coli and other organisms44,45. The outstanding reproducibility and discriminatory power of this technique in detecting similarity between strains45,46 warranted its application in the current study to evaluate the genetic relatedness of E. coli isolates from human stool and water samples. Given that E. coli has not been identified as a contributing aetiology of diarrhoea in the North West Province of South Africa, especially among the paediatric group, the current study, therefore, sought to investigate if there was any clonal relationship between water and clinical E. coli isolates within the community. These results would identify the possible source of the E. coli, thus stimulating the need for control measure to be implemented to safeguard the health of children within the Province.
Sample collection
Three hundred and fifty (350) stool samples were collected from diarrhoeic and 155 samples from non-diarrhoeal cases in children under five years attending the Brits District Hospital, Oukasie Clinic, Lethabeleng and Bopang Clinics. Also, 88 water samples, including eight directly from a municipal tap and 80 from water storage containers, were collected from different homes around the sampled clinics. Of the stored water samples, 38 were fetched from the municipal tap while 42 were from a well/underground water or rainwater harvested from rooftops. Also, four samples were collected from the Crocodile River. Sampling was done between September 2016 and December 2017 and transported to the Council for Scientific and Industrial Research (CSIR), for processing. Approval to conduct the study was obtained from the College of Agriculture and Environmental Sciences (CAES) UNISA (2016/CAES/033) and North West Department of Health. Written informed consent was obtained from the participants’ parents/guardians. Details of the participants’ age, gender, and clinical signs were recorded.
Isolation of E. coli from water and stool samples
Isolation of E. coli from water and stool samples was carried out using the Colilert-18®/ Quanti-tray/2000 (IDEXX Laboratories, Inc., Johannesburg, South Africa) following the manufacturer’s instructions. Briefly, the Colilert-18® reagent was added to 100 mL of a water sample, mixed properly, sealed in a Quanti-Tray 2000 and incubated for 18 – 24 h. For stool samples, 2g of stool sample was thoroughly mixed with 100 ml of distilled water in a sterile vessel. The supernatant was then extracted and analysed as the water sample. After incubation at 37°C, presumptive E. coli isolates were harvested from fluorescent Quanti-trays wells and streaked onto EMB agar to obtained pure colonies as previously described47. Purified colonies were stored at -80oC in a 50% glycerol for further analysis.
Confirmation of pathogenic potentials of E. coli isolated strains
DNA extraction and detection of virulence genes (VGs)
Two hundred and forty-one randomly selected E. coli isolates (136 human and 105 isolates) were inoculated onto nutrient agar and incubated for 24 h at 37°C. DNA was extracted from selected colonies after 24 h, by the heat lysis method as previously described 48. Extracted DNA templates were first examined for the presence of the malate dehydrogenase (mdh) gene to confirm the isolates as being E. coli47. Isolates harbouring the mdh gene were further separated into five DEC pathotypes, EPEC, EAEC,EIEC, ETEC, EHEC targeting specific VGs through multiplex and singleplex PCR assays and using primers and PCR-cycling conditions described by Abia et al.6. Multiplex PCR Group 1 contained the stx1 and flicH7 genes, Group 2 contained eaeA, ipaH, and eagg genes and Group 3 contained the stx2 gene.
ST and LT genes were detected through Singleplex PCR. The ST gene was amplified using primer sequence Forward: TTTCCCCTCTTTTAGTCAGTCAA and reverse: GCAGGATTACAACACAATTCACAGCAG with the temperature conditions used for the detection of mdh gene6. The PCR assay for LT gene was performed using a 218 bp primer (Forward: GCACACGGAGCTCCTCAGTC)and (Reverse: TCCTTCATCCTTTCAATGGCTTT) with an initial activation at 95°C for 3 min, followed by 35 cycles of denaturation at 94°C for 30 s, annealing at 58°C for 1 min and an extension at 72°C for 1 min. A melt curve was prepared as described for the other VGs using the Rotor-Gene™ real-time analysis software, version 6.1 (build 93) (Corbett Life Science (Pty) Ltd., Sydney, Australia). All reactions were performed in a total volume of 20µl made up of 10µl of 2x SensiFASTTM high-resolution melt (HRM) mix (SF) (Bioline GmbH, Germany), 0.5µl of primer (forward and reverse; final concentration 0.5µM each), 5µl of nuclease-free water (NF H20) and 4µl DNA template. All assays included a positive control consisting of DNA from a reference strain obtained from the Microbiology Laboratory of the NRE, CSIR and previously characterised by Abia et al. (2016)6. A reaction mixture void of template DNA was also added in each PCR assay as a No Template Control.
Determination of the antibiotic-susceptibility of E. coli
Antibiotic sensitivity testing was performed using the disk diffusion method following the CLSI guidelines 33,49. Twenty antibiotics, selected based on their common use in treating Enterobacteriaceae infections, were screened in the current study (Table 1). E. coli ATCC 25922 and ATCC 35218 were used as positive and negative controls respectively. Breakpoints recommended by the CLSI were used49.
Table (1):
Antibiotics used for E. coli susceptibility profile
| Class of antibiotic | Antibiotics | Antibiotics code | Concentration (µg) |
|---|---|---|---|
| Macrolides | Clarithromycin | CLR | 15 |
| Erythromycin | ERY | 15 | |
| Carbapenem | Meropenem | MEM | 10 |
| Imipenem | IMI | 10 | |
| β-lactam/β-lactamase inhibitor combination | Amoxicillin/clavulanic acid | AMC | 30 |
| Penicillin | Ampicillin | AMP | 10 |
| Folate pathway inhibitors | Cotrimoxazole | SXT | 25 |
| Sulfonamides | Trimethoprim | TMP | 5 |
| Nitrofurans | Nitrofurantoin | NIT | 300 |
| Fluoroquinolones | Ciprofloxacin | CIP | 5 |
| Norfloxacin | NOR | 10 | |
| Aminoglycosides | Amikacin | AMK | 30 |
| Gentamicin | GEN | 10 | |
| Streptomycin | STR | 10 | |
| Polypeptides | Colistin sulfate | CST | 25 |
| Tetracycline | Tetracycline | TET | 25 |
| Tigecycline | TGC | 15 | |
| Cephalosporine | Cephazolin | CFZ | 30 |
| Cefuroxime | CXM | 30 | |
| Cephalothin | CPD | 10 |
Determining the Minimum Inhibitory Concentration (MIC) of the Antibiotics
MICs were assessed following the CLSI recommendation and results were interpreted as per the guideline described by the CLSI for Entero-bacteriaceae49
Detection of antibiotic resistance genes
The presence of the genes conferring resistance to the b-lactamase antibiotics: blaCTX-M, blaSHV, blaCMY, and blaDHA was investigated using primer sequences detailed in Table 2.
Table (2):
Gene-specific primers for ESBL detection
| Primer | Sequence (5’- 3’) | Reference |
|---|---|---|
| BlaDHA | F: AACTTTCACAGGTGTGCTGGGT | 37 |
| R: CCGTACGCTTACTGGCTTTGC | ||
| CMY-2 | F: GATTCCTTGGACTCTTCAG | 38 |
| R: TAAAACCAGGTTCCCAGATAGC | ||
| SHV | F: TTAACTCCCTGTTAGCCA | 39 |
| R: GATTTGCTGATTTCGCCC | ||
| CTX-M | F: GGTTAAAAAATCACTGCGTC | 38 |
| R: TTGGTGACGATTTTAGCCGC |
Amplifications of the blaSHV, blaCMY, and blaDHA genes were carried out in 15µL reaction volumes containing 10µL of SF, 0.5µL of each primer sets(Forward and reverse) final concentration, 0.5µM of each primer), 3µl of template DNA and 1µL of NF H20, using the same PCR cycling conditions as for the mdhgene. The blaCTX-M gene was amplified in a 15µL reaction volume and optimized cycling conditions consisting of an initial denaturation at 99°C for 50 s, followed by denaturation at 98°C for 10 s, annealing at 58.5°C for 30 s, extension at 72°C for 15 s and a final extension at 72°C for 1min. A melt curve was prepared as described for the other genes using the Rotor-Gene™ real-time analysis software, version 6.1 (build 93) (Corbett Life Science (Pty) Ltd., Sydney, Australia).
Detection of Clonality using ERIC-PCR
Enterobacterial Repetitive Intergenic Consensus Polymerase Chain Reaction (ERIC-PCR) was performed using primers ERIC1 (52 -ATGTAAG CTCCTGGGGATTCAC-32) and ERIC2 (52 -AAGTAAGTG ACTGGGGTGAGCG-32)50. DNA was extracted using the GeneJET Genomic DNA purification kit (ThermoFisher Scientific) according to the manufacturer’s protocol. PCR was carried out on the T100 Thermal Cycler (Bio-Rad, USA) in anoverall volume of 25µL made up of 12.5µL of DreamTaq Green PCR Master mix (2X) (ThermoFisher Scientific), 0.1µL of 100µM primers ERIC 1 and ERIC 2, 9.3µL of NF H2O and 3µL of template DNA. PCR conditions were optimized to an initial denaturation at 94°C for 3 min, followed by 30 denaturation at 94°C (30 s), annealing at 50°C (1 min), extension at 65°C (8 min) and a final extension at 65°C (16 min). The PCR apliconswere separated through electro-phoresis on a 1.0% (w/v) agarose, after staining in 0.1 mg/ml ethidium bromide for 20 min. The gels were visualised and the images captured using a Gel Doc™ XR+ System (Bio-Rad, South Africa).
The freely available DNA fingerprint gel images analysis tool, GelJ Version 2.0 (https://sourceforge.net/projects/gelj/) was used to analyse the gel images following a previously described protocol51. The normalisation of the DNA fragments was performed using a Quick Load 1 kb DNA molecular weight marker (Inqaba Biotec, South Africa) and the Jacquard and Unweighted Pair Group Method with Arithmetic mean (UPGMA) cluster analysis was used to produce a dendrogram.
Data analysis
Results were analysed using the statistical package for social sciences (SPSS) (IBM SPSS Statistics Version 20) for windows. The Spearman’s correlation was used to assess the relationship between the diarrhoea status and the various DEC pathotypes. All relationships were considered significant at p £ 0.05.
Identification of E. coli pathotypes using the virulence genes
One hundred and thirty-six(136) E. coli isolates were obtained from the stool, of which 125 (91.9%) isolates were positive for at least one of the targeted VGs. The most detected gene was the eaeA gene of EPEC/EHEC (33/125; 26.4%) while the least detected gene was the LT (7/125; 5.6%) gene. None of the human isolates was positive for the stx2 (EHEC) and eagg (EAEC) (Table 3).
Table (3):
Distribution of E. coli pathotypes among human and water samples
Virulence genes (Pathotypes) |
Human samples |
Water samples |
Total |
|---|---|---|---|
eaeA (EPEC/EHEC) |
33 (26.4%) |
14 (16%) |
47 (22.2%) |
Eagg (EAEC) |
0 |
0 |
0 |
ipaH (EIEC) |
25 (20%) |
18 (20.6%) |
43 (20.3%) |
ST (ETEC) |
20 (16%) |
9 (10.3%) |
29 (13.6%) |
LT (ETEC |
7 (5.6%) |
10 (11.4%) |
17 (8%) |
Stx1 (EHEC) |
16 (12.8%) |
14 (16%) |
30 (14.2%) |
Stx2 (EHEC) |
0 |
2 (2.3%) |
2 (2.3%) |
flicH (EHEC) |
24 (19.2%) |
20 (22.95) |
44 (20.7%) |
Total |
125 (91.9%) |
87 (82.8%) |
212 (87.9%) |
For the water samples, 105 isolates were obtained, and 87 of these (82.8%) expressed at least one of the VGs tested. Like the human samples, the eaeA gene was the most detected of all the VGs tested (Table 3). No virulence gene was found in water samples collected directly from the municipal tap.
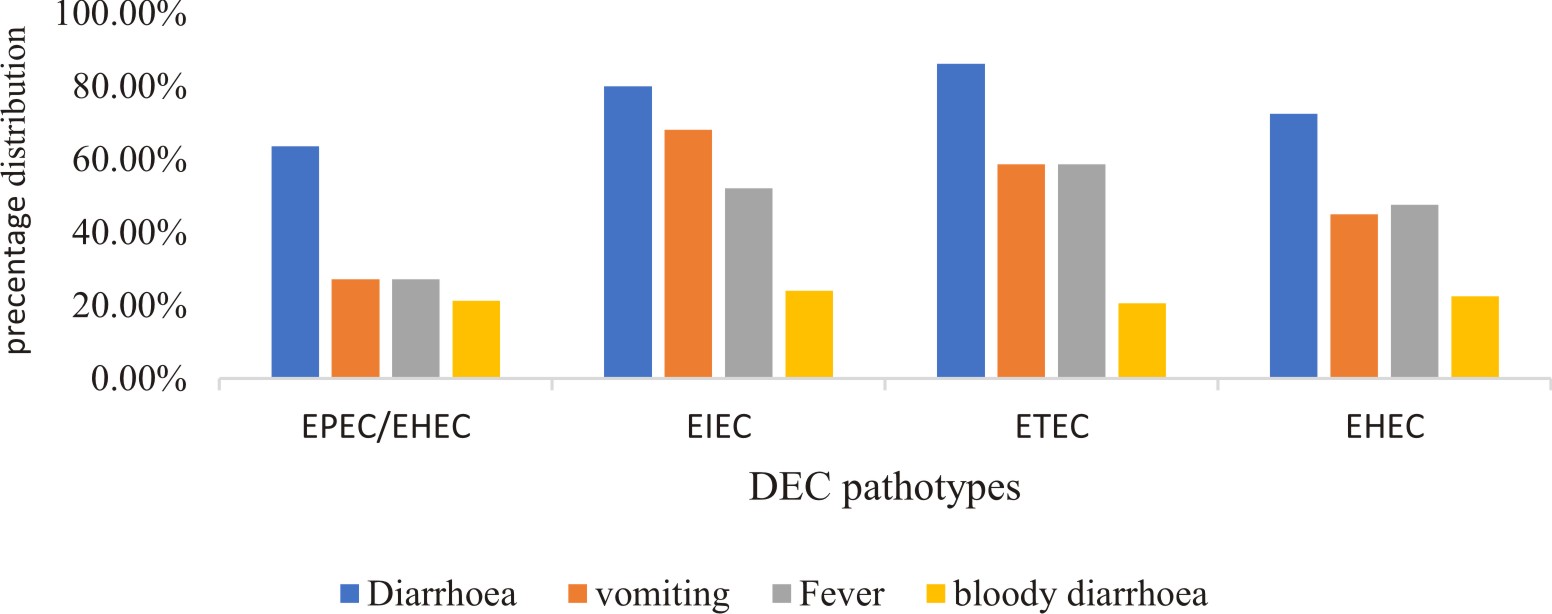
There were differences in the detection of the various DEC pathotypes among the children showing different clinical symptoms (Fig. 1).
Antibiotic susceptibility profiles of the human and water E. coli isolates
Two hundred and forty-one (241) E. coli isolates comprising all those screened for VGs, were further assessed for their susceptibility to 20 antibiotics. The highest recorded resistance was against clarithromycin and erythromycin (100%) and total susceptibility (100%) was recorded against meropenem and imipenem (Table 4). Human isolates showed an overall higher resistance compared to the water isolates. Also, a low percentage of isolates showed resistance to Gentamicin (0.4%) and Norfloxacin (2.4%).
Table (4):
Number of E. coli isolates resistant to selected antibiotics
Antibiotics tested |
Human (%) |
Water (%) |
Total resistance (%) |
|---|---|---|---|
CLR |
136 (100%) |
105 (100%) |
241 (100%) |
ERY |
136 (100%) |
105 (100%) |
241 (100%) |
MEM |
0 |
0 |
0 |
IMI |
0 |
0 |
0 |
AMC |
13 (9.5%) |
31 (29%) |
34 (18.2%) |
CPD |
70 (51.4%) |
81 (77%) |
151 (62.5%) |
AMP |
119 (87.5%) |
105 (100%) |
224 (92.9%) |
SXT |
36 (26.5%) |
32 (30.4%) |
68 (28.2%) |
TMP |
35 (25.7%) |
44 (41.9%) |
79 (32.7%) |
NIT |
10 (7.3%) |
3 (2.8%) |
13 (5.3%) |
CIP |
6 (4.4%) |
6 (5.7%) |
12 (4.9%) |
NOR |
4 (2.9%) |
2 (1.9%) |
6 (2.4%) |
AMK |
14 (10.3%) |
8 (7.6%) |
22 (9%) |
GEN |
1 (0.7%) |
0 |
1 (0.4%) |
STR |
31 (22.7%) |
44 (41.9%) |
75 (31%) |
CST |
30 (22%) |
40 (38%) |
70 (29%) |
STR |
10 (7.3%) |
13 (12.3%) |
23 (9.5%) |
TGC |
6 (4.4%) |
12 (11.4%) |
18 (7.4%) |
CFZ |
28 (20.6%) |
35 (33.3%) |
43 (17.8%) |
CXM |
33 (24.2%) |
38 (36%) |
71 (29.4%) |
The antibiotic resistance rate in individual pathotypes is shown in Table 5. From the human samples, resistance to Amoxicillin/clavulanic acid was recorded only in the EHEC and ETEC pathotypes, while, all the water isolates were resistant to Amoxicillin/clavulanic acid. Also, only the EIEC pathotype from the water was resistant to Norfloxacin.
Table (5):
Antibiotic resistance among E. coli pathotypes from human samples
| Antibiotics | E. coli pathotypes | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Human Isolates | Water Isolates | ||||||||
| EHEC | EPEC/ EHEC* | EIEC | ETEC | EHEC | EPEC/EHEC* | EIEC | ETEC | ||
| CLR | 40 | 33 | 25 | 27 | 22 | 7 | 10 | 10 | |
| ERY | 40 | 33 | 25 | 27 | 8 | 1 | 10 | 7 | |
| MEM | 0 | 0 | 0 | 0 | 12 | 8 | 9 | 10 | |
| IMI | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | |
| AMC | 8 | 2 | 1 | 3 | 10 | 6 | 10 | 11 | |
| CPD | 23 | 19 | 18 | 12 | 15 | 7 | 10 | 6 | |
| AMP | 38 | 32 | 25 | 26 | 36 | 14 | 18 | 19 | |
| SXT | 8 | 12 | 7 | 9 | 36 | 14 | 18 | 19 | |
| TMP | 7 | 16 | 3 | 9 | 0 | 0 | 0 | 0 | |
| NIT | 3 | 5 | 0 | 2 | 0 | 0 | 0 | 0 | |
| CIP | 4 | 0 | 2 | 0 | 13 | 8 | 1 | 3 | |
| NOR | 1 | 0 | 1 | 2 | 36 | 14 | 18 | 9 | |
| AMK | 4 | 4 | 3 | 3 | 3 | 0 | 0 | 1 | |
| GEN | 0 | 1 | 0 | 0 | 0 | 0 | 2 | 0 | |
| STR | 12 | 10 | 3 | 7 | 1 | 0 | 4 | 3 | |
| CST | 17 | 3 | 5 | 7 | 0 | 0 | 0 | 0 | |
| TET | 4 | 2 | 1 | 2 | 6 | 2 | 3 | 2 | |
| TGC | 4 | 1 | 0 | 1 | 5 | 3 | 1 | 0 | |
| CFZ | 14 | 4 | 3 | 6 | 11 | 7 | 7 | 5 | |
| CXM | 17 | 7 | 3 | 6 | 14 | 7 | 8 | 5 | |
*Isolates that were only positive for the eaeA gene which is present in both EPEC and EHEC
Determination of Multiple-Antibiotic Resistance (MAR)
Multi-resistance in this study was defined as the resistance of E. coli to three or more antibiotics 47. MAR was observed in 87.5% and 80.8% of isolates from human and water samples respectively. Twenty-nine (21.3%) isolates from human samples demonstrated simultaneous resistance to four antibiotics while 23.2% from water samples were resistant to five antibiotics concurrently. No isolate was resistant to more than 11 antibiotics (Fig. 2).
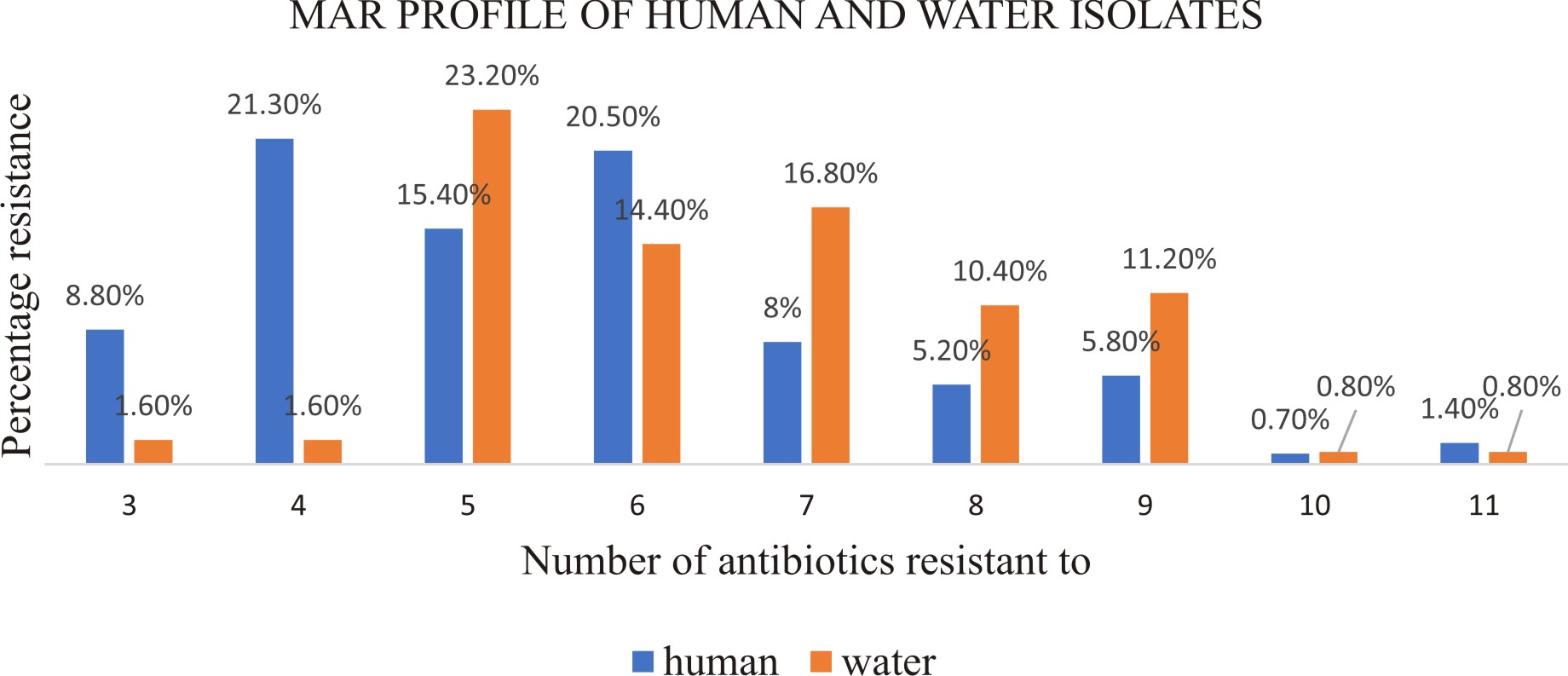
Fig. 2. percentage of MAR E. coli isolates from human and water samples resistant to a different number of antibiotics
Prevalence of ESBLs encoding genes
Identified E. coli pathotypes were tested for the presence of ESBLs encoding genes. Out of the 212 tested, ESBLs was detected in 152 (71.6%). The genes were detected more in water samples (85%) compare to human samples (62.4%). The blaDHA gene was the most detected gene in both human and water samples. ESBL genes were expressed more in EHEC pathotypes compared to other pathotypes (Table 6). Different antibiotic resistance phenotypes were associated with different ESBLs encoding genes. The most frequent resistance pattern observed was to Clarithromycin-Erythromycin-Cephalothin-Ampicillin-Cefuroxime-Cephazolin-Trimethoprim-Colistin.
Table (6):
Distribution of ESBL genes among the E. coli pathotypes
Source |
ESBL gene |
EHEC |
EIEC |
EHEC/EPEC |
ETEC |
Total ESBL genes |
|---|---|---|---|---|---|---|
Human |
CMY |
10 |
2 |
1 |
3 |
16 |
CTX-M |
8 |
1 |
3 |
6 |
18 |
|
DHA |
8 |
10 |
5 |
6 |
29 |
|
SHV |
7 |
3 |
1 |
4 |
15 |
|
Sub-total |
33 |
16 |
10 |
24 |
78 |
|
Water |
CMY |
5 |
6 |
5 |
4 |
18 |
CTX-M |
10 |
3 |
4 |
2 |
20 |
|
DHA |
11 |
1 |
5 |
3 |
21 |
|
SHV |
3 |
3 |
0 |
9 |
15 |
|
Sub-total |
29 |
13 |
14 |
18 |
74 |
Determination of the clonality (ERIC-PCR analysis) of E. coli pathotypes
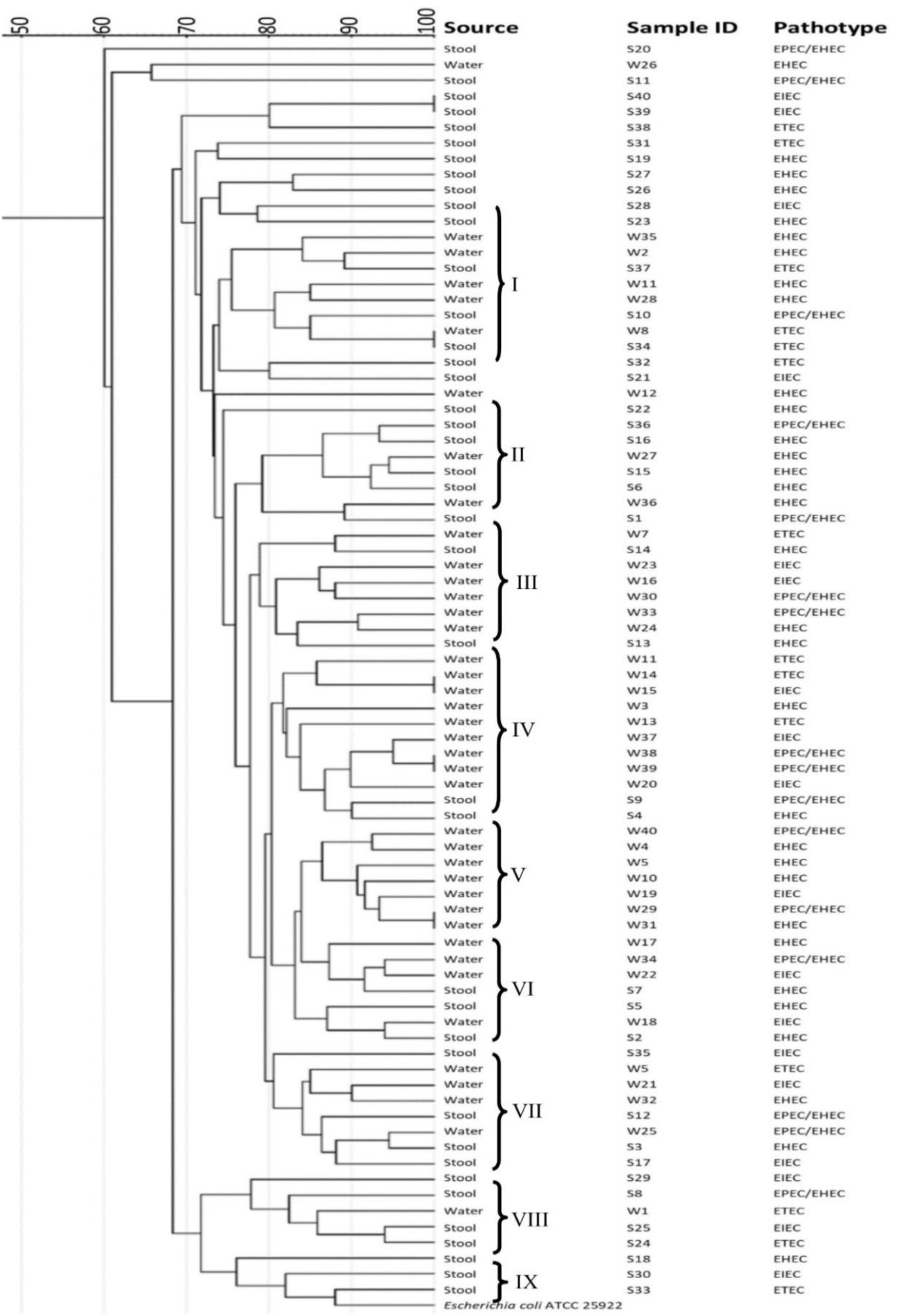
The dendrogram revealed that all the clusters contained mixed isolates from water and human samples, except for cluster V which was made up of water samples alone and cluster IX which contained only stool samples (Fig. 3). There was a 100% similarity between some water samples and human samples (S40, S39, W8, and S34). There was also a 100% similarity between water samples of different clusters (W15, W14, W38, W39 W29 and W31 of cluster IV and V). In all the clusters (apart from the clusters with 100% similarities), the similarity patterns between some of the water and human samples were £ 80%.
Identification and distribution of DEC pathotypes
Diarrhoeagenic E. coli pathotypes are among the most significant aetiologic agents of diarrhoea worldwide, particularly in children from developing countries52. In this study, the frequency, virulence markers and antibiotic resistance patterns of DEC from diarrhoeic and non-diarrhoeic paediatric stools and water samples were evaluated. The frequency of DEC was high in paediatric specimens compared to the water sample. These findings could be because E. coli is a normal inhabitant of the human intestinal tract and the participants included diarrhoeic children. A high frequency of DEC in water samples could have indicated pollution. Several studies have reported a high prevalence of DEC in different aquatic bodies in South Africa6,53. Detection of DEC in water, particularly domestic waters, should be regarded as a public health concern because these waters are used for human and animal consumption and other indoor potable uses such as washing dishes and cooking. This study confirmed that DEC might have contributed to the burden of diarrhoea within the studied communities. The absence of E. coli in the municipal tap water samples further strengthened the findings of the study. The detection of E. coli in the stored water samples suggests that poor hygienic practices within the households could have led to the recontamination of the treated water from the municipal tap, as the residual chlorine concentration decreased during storage. In agreement with the present study, Chigo et al. reported a higher prevalence of DEC in human compared to water samples54. In Benin, a high prevalence of DEC was equally observed in humans55. Studies have confirmed the involvement of DEC in diarrhoea in paediatric groups even in the developed world. For example, DEC pathotypes were among the predominant pathogens isolated in a paediatric emergency facility in Washington56. Although DEC pathotypes are a common cause of diarrhoea in children, the high prevalence reported in this study is worrisome as infections with these strains are known to present with some severe health complications such as anaemia, throm-bocytopenia and renal failure57.
The frequency of DEC pathotypes found in our study was similar to the reports from other developing countries6,7,53,58. EHEC was the most detected pathotype found in both human and water samples followed by EPEC/EHEC. These pathotypes were the most detected pathotypes in children with diarrhoea in two studies performed in Iran7,59 and Spain16. EHEC is the primary cause of severe gastroenteritis and HUS outbreak in Germany 60. Usein et al. reported EHEC virulence genes in all the diarrhoea samples screened in their study19. Although the detection rate in this study is slightly high compared to the 30% reported in Indian children 61, however, it should be noted that the present study characterised EHEC pathotype based on the frequency of detection of three different virulence genes. The present study correlates with other studies where the detection rate of EHEC pathotype among children under the age of 5 years was high61. It also agrees with studies that used an equal number of virulence genes to characterise the EHEC pathotype16,41. The overall prevalence of EPEC/EHEC (eaeA gene) found in our study is consistent with other studies18,21,41. A higher frequency of 42.3% has been reported in India15 while over 50% has been reported in South African domestic water6, 67% in Tennessee62 and 47.5% found in Iranian children7.
ETEC is the important diarrhoeic agents in some African countries as reported by the Global Enteric Multicentre Study (GEMS)3. It has been documented as an important agent of diarrhoea in many studies63–65. In this study, DEC pathotypes isolated from the human samples were significantly associated with diarrhoea particularly ETEC (p<0.002) and EIEC (p<0.022). These results highlight the contribution of ETEC and EIEC to the burden of diarrhoea within the studied communities. ETEC and EIEC have been implicated in many diarrhoea and gastro-intestinal outbreaks worldwide3,14,66. EIEC pathotype was the cause of a diarrhoea outbreak in Italy 67, and an important diarrhoeal agent in children in Mexico 52, although it has also been found in stool samples of individuals without diarrhoea26.
Antibiotic susceptibility profiles of the human and water E. coli isolates
Antibiotic resistance has been recorded in human as well as aquatic environments, and the findings of the present study correspond with the findings reported in other studies2,21,53,68. The result of the antibiotic-resistance profile showed varying degrees of resistance in human and water samples. The results further revealed a higher resistance rate to the most frequently used antibiotics in South Africa including Clarithromycin, Erythromycin and Ampicillin69. Thus, high resistance rate of DEC to these frequently used antibiotics might pose a severe threat to the public health, because the increase in resistant strains will negatively impact on the efficacy of these antibiotics which might in the long term contribute to higher morbidity and mortality31. Other studies have also reported on E. coli resistance to these drugs. For example, a high resistance of DEC isolates was observed to fluoroquinolones, Ampicillin, Cotrimoxazole, and Nalidixic Acid in India2. In Iran, over 50% of the paediatric patients were resistant to Ampicillin, ceftriaxone, tetracycline, Clotrimazole, and Cefixime7. In the UK, E. coli isolates were resistant to Ampicillin, Cefotaxime, streptomycin, sulphonamide and Oxytetracycline70 and in South Africa, high resistance was observed in E. coli pathotypes isolated from harvested rainwater that was intended for domestic purposes53. Antibiotic resistance is increasing worldwide as most microorganisms now exhibit resistance to a large number of antibiotics. The presence of antibiotic-resistant bacteria in water (environment and household) is a health hazard to combat because even if antibiotic consumption were reduced, the existing concentration of antibiotics and the resulting selective pressure on the bacterial communities could persist for an extended period68. This study, therefore, recommends the continuous monitoring of antibiotic-resistant microorganisms in humans and water.
Detection ESBLs gene in the study
ESBL-positive E. coli strains are highly resistant to an array of antibiotics and infections caused by these strains are difficult to treat71. In the present study, ESBL genes were found in a considerable proportion of the pathotypes screened. Isolates from water samples frequently expressed the ESBL genes (85%) compared to human samples (62.4%). The rate of expression of ESBL genes reported in clinical isolates in this study is low compared to the 85.24% reported in Egyptian children40 and 80% found in China 33. The reason might be the difference in geographical location. However, these results indicate the presence of these genes in the studied communities. The blaDHA and blaCTX-M genes were the most expressed genes, followed by blaSHV and blaCMY genes. The presence of the blaDHA gene was higher in human compared to water samples. Similar to the present study, Osinska et al. (2017) reported a high prevalence of blaCTX-M and lower prevalence of blaSHV in b-lactam resistant strains isolated from various sources68. Also, 25% and 23% of blaCTX-M and blaSHV have been reported in Egyptian clinical isolates40. ESBL genes are often plasmid encoded and can easily be transferred between bacteria36. This possibility of transfer, therefore, highlights the need for better control mechanisms and antibiotic stewardship to prevent the spread of these resistant bacteria within communities, especially resource-poor settings like those included in the current study.
Clonal relatedness between stool and water isolates
ERIC-PCR technique was used to amplify the diverse regions of DNA extracted from the pathotypes identified from water and human stool samples in the present study. Studies have shown that a high degree of sequence similarities between isolates usually reflects descent from a common ancestor72,73. The dendrogram (Fig. 3) showed 100% similarities between some isolates from water and human samples (Cluster I), and 100% similarities in some water isolates that belonged to different clusters. The overall remarkable similarities of 80-90% between the ERIC profile for the isolates strongly suggests that the domestic water played a significant role in the transmission of E. coli within the studied population. Thus, there is a need to develop strategies that will help to reduce pathogenic E. coli infections in humans, particularly from domestic waters within communities.
The DEC isolates screened in this study revealed the presence of E. coli pathotypes in children and household drinking water. The tested water samples could serve as a reservoir to not only DEC but also antibiotic resistant E. coli strains including MAR and ESBL-producing strains. These findings suggest that the containers used in storing the tested water samples might be reservoirs of these pathogens as water samples collected directly from the municipal tap did not harbour any pathogenic strain, and water from these storage containers should not be used without appropriate treatment. In case of DEC infection due to the identified pathotypes within the studied communities, antibiotics such as imipenem and meropenem might be an effective treatment as DEC isolates in this study were 100% susceptible to these antibiotics. Gentamicin could also be used as a second option.
None
The authors declare that there is no conflict of interest.
- Youmans BP, Ajami NJ, Jiang Z. Characterization of the human gut microbiome during travelers diarrhea Characterization of the human gut microbiome during travelers diarrhea. 2015; 6(2): 110-119.
- Tilak GP, Mudaliar JLG. Role of enteropathogenic Escherichia coli in paediatric diarrhoeas in South India. Mater Sociomed., 2012; 24(3):178-181.
- Kotloff KL, Nataro JP, Blackwelder WC. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): A prospective, case-control study. Lancet, 2013; 382(9888): 209-222.
- Rogawski ET, Guerrant RL, Havt A. Epidemiology of enteroaggregative Escherichia coli infections and associated outcomes in the MAL-ED birth cohort. PLoS Negl Trop Dis., 2017; 11(7): e0005798.
- Yang SC, Lin CH, Aljuffali IA, Fang JY. Current pathogenic Escherichia coli foodborne outbreak cases and therapy development. Arch Microbiol. 2017; 199(6): 811-825.
- Abia ALK, Schaefer L, Ubomba-Jaswa E, Le Roux W. Abundance of pathogenic Escherichia coli virulence-associated genes in well and borehole water used for domestic purposes in a peri-urban community of South Africa. Int J Environ Res Public Health, 2017; 14 (3): 320.
- Alikhani MY, Hashemi SH, Aslani MM, Farajnia S. Prevalence and antibiotic resistance patterns of diarrheagenic Escherichia coli isolated from adolescents and adults in Hamedan, Western Iran. Iran J Microbiol., 2013; 5(1): 42-47.
- Navarro-Garcia F. Escherichia coli O104:H4 Pathogenesis: an Enteroaggregative E. coli/Shiga Toxin-Producing E. coli Explosive Cocktail of High Virulence. Microbiol. Spectr., 2014; 2(6): 1-19.
- Kaper JB, Nataro JP, Mobley HLT. Pathogenic Escherichia coli. Nat. Rev. Microbiol., 2004; 2(2): 123-140.
- Croxen MA, Finlay BB. Molecular mechanisms of Escherichia coli pathogenicity. Nat. Rev. Microbiol., 2010; 8(1): 26-38.
- Mandal A, Sengupta A, Das S. Molecular Epidemiology of Extended-Spectrum b-Lacta-mase–Producing Escherichia coli Pathotypes in Diarrheal Children from Low Socioeconomic Status Communities in Bihar, India: Emergence of the CTX-M Type. Infect Dis. Res. Treat., 2017; 10(1): 478-483.
- Langendorf C, Le Hello S, Moumouni A. Enteric bacterial pathogens in children with diarrhea in niger: Diversity and antimicrobial resistance. Rajashekara G, ed. PLoS One, 2015; 10(3): e0120275.
- Contreras TJO and Carmen A. Enteropathogenic E. coli (EPEC) infection in children. Curr. Opin. Infect. Dis., 2011; 24(5): 478–483.
- Croxen MA, Law RJ, Scholz R, Keeney KM, Wlodarska M, Finlay BB. Recent advances in understanding enteric pathogenic Escherichia coli. Clin. Microbiol Rev., 2013; 26(4): 822-880.
- Malvi S, Appannanavar S, Mohan B. Comparative analysis of virulence determinants, antibiotic susceptibility patterns and serogrouping of atypical enteropathogenic Escherichia coli versus typical enteropathogenic E. coli in India. J Med Microbiol., 2015; 64(10): 1208-1215.
- Cabal A, Garcםa-Castillo M, Cantףn R, Gortבzar C, Domםnguez L, ֱlvarez J. Prevalence of Escherichia coli virulence genes in patients with diarrhea and a subpopulation of healthy volunteers in Madrid, Spain. Front Microbiol., 2016; 7(5): 1-6.
- Trabulsi L, Keller R, Tardelli Gomes T. Typical and atypical enteropathogenic Escherichia coli. Emerg Infect Dis., 2002; 8(5): 508-513.
- Cepeda-Molero M, Berger CN, Walsham ADS. Attaching and effacing (A/E) lesion formation by enteropathogenic E. coli on human intestinal mucosa is dependent on non-LEE effectors. Coombes BK, ed. PLOS Pathog., 2017; 13(10): e1006706.
- Usein CR, Ciontea AS, Militaru CM. Molecular characterisation of human shiga toxinproducing Escherichia coli O26 strains: Results of an outbreak investigation, Romania, february to august 2016. Eurosurveillance, 2017; 22(47): 1-8.
- Heiman KE, Mody RK, Johnson SD, Griffin PM, Gould LH. Escherichia coli O157 Outbreaks in the United States, 2003-2012. Emerg. Infect. Dis. 2015; 21(8): 1293-1301.
- Karami P, Bazmamoun H, Sedighi I, Mozaffari Nejad AS, Aslani MM, Alikhani MY. Antibacterial resistance patterns of extended spectrum b-lactamase -producing enteropathogenic Escherichia coli strains isolated from children. Arab J Gastroenterol., 2017; 18(4): 206-209.
- Kotloff KL, Platts-Mills JA, Nasrin D, Roose A, Blackwelder WC, Levine MM. Global burden of diarrheal diseases among children in developing countries: Incidence, etiology, and insights from new molecular diagnostic techniques. Vaccine, 2017; 35(49): 6783-6789.
- Shepard SM, Danzeisen JL, Isaacson RE, Seemann T, Achtman M, Johnson TJ. Genome sequences and phylogenetic analysis of K88- and F18-positive porcine enterotoxigenic Escherichia coli. J Bacteriol., 2012; 194(2): 395-405.
- Taxt A, Aasland R, Sommerfelt H, Nataro J, Puntervoll P. Heat-stable enterotoxin of entero-toxigenic Escherichia coli as a vaccine target. Infect Immun., 2010; 78(5): 1824-1831.
- Wang X, Gao X, Hardwidge PR. Heat-labile enterotoxin-induced activation of NF-ךB and MAPK pathways in intestinal epithelial cells impacts enterotoxigenic Escherichia coli (ETEC) adherence. Cell Microbiol., 2012; 14(8):1231-1241.
- Bliss J, Bouhenia M, Hale P. High prevalence of shigella or enteroinvasive Escherichia coli carriage among residents of an internally displaced persons camp in South Sudan. Am. J. Trop. Med. Hyg., 2018; 98(2): 595-597.
- Hosseini Nave H, Mansouri S, Taati Moghadam M, Moradi M. Virulence Gene Profile and Multilocus Variable-Number Tandem-Repeat Analysis (MLVA) of Enteroinvasive Escherichia coli (EIEC) Isolates From Patients With Diarrhea in Kerman, Iran. Jundishapur J. Microbiol., 2016; 9(6): e33529.
- Gomes TAT, Elias WP, Scaletsky ICA. Diarrheagenic Escherichia coli. Brazilian J. Microbiol., 2016; 47(1): 3-30.
- Karambu S, Matiru V, Kiptoo M, Oundo J. Characterization and factors associated with diarrhoeal diseases caused by enteric bacterial pathogens among children aged five years and below attending Igembe District Hospital, Kenya. Pan Afr. Med. J., 2013; 16(1): 37.
- Centers for Disease Control and Prevention (CDC). Vital signs: incidence and trends of infection with pathogens transmitted commonly through food—foodborne diseases active surveillance network, 10 U.S. sites, 1996-2010. MMWR Morb Mortal Wkly Rep., 2011; 60(22): 749-755.
- CDC. Antibiotic Resistance Threats in the United States, 2013.; 2013.
- Von Baum H, Marre R. Antimicrobial resistance of Escherichia coli and therapeutic implications. Int. J. Med. Microbiol., 2005; 295(6-7): 503-511.
- Liang W juan, Liu H ying, Duan GC. Emergence and mechanism of carbapenem-resistant Escherichia coli in Henan, China, 2014. J. Infect. Public Health, 2018; 11(3): 347-351.
- Bebrone C. Metallo-b-lactamases (classification, activity, genetic organization, structure, zinc coordination) and their superfamily. Biochem Pharmacol., 2007; 74(12): 1686-1701.
- Paterson DL, Bonomo RA. Extended-spectrum beta-lactamases: a clinical update. Clin. Microbiol Rev. 2005; 18(4): 657-686.
- Drawz SM, Bonomo RA. Three decades of b-lactamase inhibitors. Clin. Microbio.l Rev., 2010; 23(1): 160-201.
- Liu X, Liu Y. Detection of plasmid-mediated AmpC b-lactamase in Escherichia coli. Biomed. Reports, 2016; 4(6): 687-690.
- Lantero M, Diego I De, Zarazaga M, Torres C. Mechanisms of resistance to expanded-spectrum cephalosporins in Escherichia coli isolates recovered in a Spanish hospital. J Antimicrob Chemother. 2005; 56 (10): 1107-1110.
- Sharma J, Sharma M, Ray P. Detection of TEM & SHV genes in Escherichia coli & Klebsiella pneumoniae isolates in a tertiary care hospital from India. Indian J. Med. Res., 2010; 132(9): 332-336.
- El-Badawy MF, Tawakol WM, Maghrabi IA, Mansy MS, Shohayeb MM, Ashour MS. Iodometric and Molecular Detection of ESBL Production Among Clinical Isolates of E. coli Fingerprinted by ERIC-PCR: The First Egyptian Report Declares the Emergence of E. coli O25b-ST131clone Harboring bla GES. Microb. Drug Resist. 2017; 23(6): 703-717.
- Trung NV, Nhung HN, Carrique-Mas JJ. Colonization of Enteroaggregative Escherichia coli and Shiga toxin-producing Escherichia coli in chickens and humans in southern Vietnam. BMC Microbiol. 2016; 16(1): 208.
- Akiyama Y, Saito E, Futai H. Comprehensive Study of Pathogenic Genes Distributed in Escherichia coli Isolated from Cattle. Food Hyg. Saf. Sci., 2014; 56(3): 118-122.
- Ahmed HA, El Hofy FI, Hefny AA, Ammar AM, Abd El Tawab AA. ERIC-PCR Genotyping of Some Campylobacter jejuni Isolates of Chicken and Human Origin in Egypt. Vector-Borne Zoonotic Dis., 2015; 15(12): 713-717.
- Ateba CN, Mbewe M. Genotypic characterization of Escherichia coli O157:H7 isolates from different sources in the north-west province, South Africa, using enterobacterial repetitive intergenic consensus PCR analysis. Int. J. Mol. Sci., 2014; 15(6): 9735-9747.
- Ranjbar R, Tabatabaee A, Behzadi P, Kheiri R. Enterobacterial repetitive intergenic consensus polymerase chain reaction (ERIC-PCR) genotyping of Escherichia coli strains isolated from different animal stool specimens. Iran J. Pathol., 2017; 12(1): 25-34.
- Blanco AE, Barz M, Cavero D. Characterization of Enterococcus faecalis isolates by chicken embryo lethality assay and ERIC-PCR. Avian Pathol., 2018; 47(1): 23-32.
- Abia ALK, Ubomba-Jaswa E, Momba MNB. High prevalence of multiple-antibiotic-resistant (MAR) Escherichia coli in river bed sediments of the Apies River, South Africa. Environ. Monit. Assess., 2015; 187(10): 652.
- Bester LA, Essack SY. Observational Study of the Prevalence and Antibiotic Resistance of & lt;I> Campylobacter </I> spp. from Different Poultry Production Systems in KwaZulu-Natal, South Africa. J Food Prot. 2012; 75(1): 154-159.
- CLSI M100-S24. M45. Methods for Antimicrobial Dilution and Disk Susceptibility Testing of Infrequently Isolated or Fastidious Bacteria/ ; Proposed Guideline. Vol 35.; 2015.
- Ranjbar R, Pezeshknejad P, Khamesipour F, Amini K, Kheiri R. Genomic fingerprints of Escherichia coli strains isolated from surface water in Alborz province, Iran. BMC Res. Notes. 2017; 10(1):1-6.
- Heras J, Domםnguez C, Mata E. GelJ – a tool for analyzing DNA fingerprint gel images. BMC Bioinformatics. 2015; 16(1): 1-8.
- Canizalez-Roman A, Flores-Villaseסor HM, Gonzalez-Nuסez E. Surveillance of diarrheagenic escherichia coli strains isolated from diarrhea cases from children, adults and elderly at Northwest of Mexico. Front Microbiol., 2016; 7(11): 1924.
- Malema MS, Luther A, Abia K. Antibiotic-Resistant Pathogenic Escherichia coli Isolated from Rooftop Rainwater-Harvesting Tanks in the Eastern Cape, South Africa. Int. J. Environ. Res. Public Health. 2018; 15(1):1-14.
- Chigor VN, Umoh VJ, Smith SI, Igbinosa EO, Okoh AI. Multidrug resistance and plasmid patterns of Escherichia coli o157 and other E. coli isolated from diarrhoeal stools and surface waters from some selected sources in Zaria, Nigeria. Int. J. Environ. Res. Public Health. 2010; 7(10): 3831-3841.
- Laaveri T, Pakkanen SH, Antikainen J. High number of diarrhoeal co-infections in travellers to Benin, West Africa. BMC Infect. Dis., 2014; 14(1): 81-92.
- Denno DM, Shaikh N, Stapp JR. Diarrhea etiology in a pediatric emergency department: A case control study. Clin. Infect. Dis. 2012; 55(7): 897-904.
- Puentes SS, Dunstan M. Escherichia coli Complications in Pediatric Critical Care. Crit Care Nurs Clin North Am., 2018; 30(1): 149-156.
- Tobias J, Kassem E, Rubinstein U. Involvement of main diarrheagenic Escherichia coli, with emphasis on enteroaggregative E. coli, in severe non-epidemic pediatric diarrhea in a high-income country. BMC Infect. Dis., 2015; 15(1): 79-89.
- Aslani MM, Alikhani MY. Molecular and phenotypic characterization of atypical enteropathogenic Escherichia coli serotypes isolated from children with and without diarrhea. J. Microbiol. Immunol. Infect., 2011; 44(1): 27-32.
- Vygen-Bonnet S, Rosner B, Wilking H. Ongoing haemolytic uraemic syndrome (HUS) outbreak caused by sorbitol-fermenting (SF) shiga toxin-producing Escherichia coli (STEC) O157, Germany, December 2016 to May 2017. Eurosurveillance, 2017; 22(21): 1-5.
- Sudershan R V, Kumar RN, Bharathi K, Kashinath L, Bhaskar V, Polasa K. E. coli pathotypes and their antibiotic resistance in young children with diarrhea in Hyderabad, India. Int. J. Curr. Microbiol. Appl. Sci., 2014; 3(9): 647-654.
- Foster MA, Iqbal J, Zhang C. Enteropathogenic and enteroaggregative E. coli in stools of children with acute gastroenteritis in Davidson County, Tennessee. Diagn. Microbiol. Infect Dis., 2015; 83(3): 319-324.
- Pasqua M, Michelacci V, Di Martino ML. The intriguing evolutionary journey of enteroinvasive E. coli (EIEC) toward pathogenicity. Front Microbiol., 2017;8(12): 2390-2399.
- Bhavnani D, De Los ֱngeles Bayas R, Lopez VK. Distribution of enteroinvasive and entero-toxigenic Escherichia coli across space and time in northwestern Ecuador. Am. J. Trop. Med. Hyg., 2016; 94(2): 276-284.
- Bliss J, Bouhenia M, Hale P. High prevalence of shigella or enteroinvasive Escherichia coli carriage among residents of an internally displaced persons camp in South Sudan. Am. J. Trop. Med. Hyg., 2018; 98(2): 595-597.
- Singh T, Das S, Ramachandran VG. Spectrum of diarrhoeagenic Escherichia coli in paediatric population suffering from diarrhoea and as commensals in healthy children. Indian J. Med. Microbiol., 2017; 35(2): 204-210.
- Escher M, Scavia G, Morabito S. A severe foodborne outbreak of diarrhoea linked to a canteen in Italy caused by enteroinvasive Escherichia coli, an uncommon agent. 2014; 142 (12): 2559-2566.
- Osiסska A, Korzeniewska E, Harnisz M, Niestךpski S. The prevalence and characterization of antibiotic-resistant and virulent Escherichia coli strains in the municipal wastewater system and their environmental fate. Sci. Total Environ., 2017; 577 (1):367-375.
- Jenniskens P, Blake DF, Kouchi A. Amorphous Water Ice. Sol. Sys.t Ices, Astrophys Sp. Sci. Libr. 1998; 101(8): 139-155.
- Ibrahim DR, Dodd CER, Stekel DJ, Ramsden SJ, Hobman JL. Multidrug resistant, extended spectrum b-lactamase (ESBL)-producing Escherichia coli isolated from a dairy farm. Simonet P, ed. FEMS Microbiol. Ecol., 2016; 92(4): 1-13.
- Eiamphungporn W, Schaduangrat N, Malik AA, Nantasenamat C. Tackling the Antibiotic Resistance Caused by Class A b-Lactamases through the Use of b-Lactamase Inhibitory Protein. Int. J. Mol. Sci., 2018; 19(8): 2222.
- Ateba C, Mbewe M, Ateba CN, Mbewe M. Genotypic Characterization of Escherichia coli O157:H7 Isolates from Different Sources in the North-West Province, South Africa, Using Enterobacterial Repetitive Intergenic Consensus PCR Analysis. Int. J. Mol. Sci., 2014; 15(6): 9735-9747.
- Nielsen EM, Scheutz F. Characterisation of Escherichia coli O157 isolates from Danish cattle and human patients by genotyping and presence and variants of virulence genes. Vet. Microbiol., 2002; 88(3): 259-273.
© The Author(s) 2019. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.