ISSN: 0973-7510
E-ISSN: 2581-690X
Water is essential to life. The existence of all forms of life is dependent on an adequate water supply. The exigent need for water supply in homes prompted the construction of water sources and water storage devices in the homes. This however does not guarantee that the water is safe to drink. If the water is safe at the source, it may be contaminated during transportation storage and drawing at home. This study was carried out to determine the microbial counts, antibiotics susceptibility and plasmid profile of bacteria isolates from household water distribution tanks in the Ado-Ekiti metropolis. The total bacteria and coliform counts were determined using the pour plating technique. The antibiotic susceptibility pattern of the isolates was determined using the disc diffusion technique while the plasmid profile of the isolates was determined using the alkaline lysis method and agar gel electrophoresis. The mean total bacteria count of the water sample was 6.96 log10 CFU/ml, while the mean total of coliform count is 5.50 log10 CFU/ml. The isolates with multiple antibiotics resistance belonged to five bacteria genera namely: Escherichia, Pseudomonas, Klebsiella, Enterobacter and Proteus. The plasmid analysis showed that four of the resistant strains had multiple plasmids, Enterobacter aerogens had 3 plasmids (1kb, 1.5kb and 2kb), Pseudomonas aeruginosa and Klebsiella aerogens had two plasmids (1kb, 1.5kb) respectively while Proteus vulgaris and Escherichia coli had no plasmid.
Water sanitation, coliforms, plasmids, Antibiotic susceptibility, water storage tanks
Water is the most essential commodity for human consumption. Adequate supply of potable water is essential for the well-being of all people around the world1, 2. Human activities such as agriculture, trading, industries cannot function properly without adequate water supply.
The two most common sources of drinking water are surface water and ground water3,4. Rural communities in Nigeria usually source their water for drinking and domestic purposes from streams and well while those in urban areas source water from well, boreholes and water distribution centers. Due to the inevitable importance of water and scarcity especially during dry season, they store water in buckets, drums, basins and tanks for easy accessibility. Meanwhile, water from these sources have been reported to contain certain pathogens and other contaminants which compromise the aesthetic and microbial quality of the water5, 6, 7.
Most homes of middle income earners in Ado-Ekiti pump their water from the source (well, borehole) and store in storage tanks. The tank is piped to all sections of the house for convenience and easy accessibility to water when needed. It is assumed that over time, organic particulates and microorganisms in the water will settle via sedimentation. The pipes also can be coated with organic matter which may serve as nutrient for the growth of microorganisms.
Regular washing and disinfection of tanks, inspection of pipes for leakages, semi-annual testing of water for total coliforms and faecal coliforms which are means of accessing the quality of water are not practiced in the homes. Most of them are usually carried away by the comfort of easy and regular accessibility to water and forget to wash the tanks and take other precaution measures.
Inadequate storage conditions and vulnerable water storage containers have been documented as factors contributing to increased microbial contamination of household water8, 9. Increased risks of waterborne diseases from inadequately stored water compared to water stored in an improved vessel have also been reported10. Loss of disinfectant residual, bacteria re-growth, poor turn over and excessive detention time are the common problems in storage tanks and reservoirs11.
The deterioration of water quality as a result of anthropogenic activities, indiscriminate discharge of wastes has been reported12,13, 14. Polluted water has been identified as the major cause of water borne disease and epidemics looming the developed and developing countries15. Waterborne diseases represent major health problem in many parts of the world and reported to cause about 842,000 diarrhoea deaths per year 16. Many diseases such as Cholera, typhoid fever, bacillary dysentery, and others can be transmitted through this route10. Bacteria genera commonly isolated from water includes; Enterobacter, coliforms and Escherichia coli17. Their ability to resist the inhibitory effect of antibiotics is of great public health concern.
Microbial contamination of drinking water remains a significant threat to living organisms and therefore constant vigilance is essential because many pathogens can be transmitted through the supply of water18. Hence, this study aimed at determining the microbial quality of water from household water distribution tanks in Ado-Ekiti metropolis. This will provide baseline information on the quality of the water and create awareness for prevention of waterborne diseases.
Collection of Samples
Water samples were collected in sterile sample bottles from household distribution tanks from ten different locations in Ado-Ekiti metropolis (7°37’16” N5°12’17”E). The samples were transported in ice packs to the laboratory for immediate microbiological analyses.
Isolation and Identification of Isolates
Total heterotrophic bacteria and coliforms were isolated from the samples using ten-fold serial dilution and pour plate method as described by Oluyege6. Pure culture was stored on nutrient agar slants and stored at 4°C. The isolates were subjected to morphological and biochemical tests such as Gram staining, motility test, catalase test, coagulase test, oxidase test, indole test, citrate test, urease test, methyl red test and their identities were determined according to Bergey’s Manual of Determinative Bacteriology19.
Antibiotic Susceptibility test of the Isolates
The antibiotics susceptibility of the isolates was determined by the disk diffusion method on Mueller-Hilton agar according to Cheesbrough20. The isolates were tested against ten ABTEK antibiotic discs which comprised of ceftazidine (CAZ) 30µg, tarivid (10µg), gentamycin (GEN) 10µg, Septrim (30µg), ofloxacin (OFL) 5µg, augmentin (AUG) 30µg, ofloxacin (OFL) 5µg, ciprofloxacin (CPR) 5µg, Perfloxacin (5µg) and Sparfloxacin (10µg). The inoculums were standardized by adjusting their densities to the turbidity of a Barium sulphate (BaSO4) (0.5 McFarland turbidity standard). One milliliter of each of the standardized broth cultures of the test isolates were swab on the Mueller Hinton agar plates, the antibiotic discs were placed firmly on solidified plates and incubated for 24 hours at 37°C. Un-inoculated agar plates with antibiotics served as the control. The diameter of the zone of growth inhibition was measured to the nearest whole millimeter and interpreted on the basis of CLSI guideline21.
Plasmid Profiling of Antibiotic-resistant Isolates
Plasmid analysis was performed on representative isolates selected on the basis of their antibiotic resistance phenotypes.
Extraction of Plasmid
Plasmid extraction was carried out using Fast and Easy Plasmid Mini-prep Kit as described by Olowomofe22,23. The extracted plasmid DNA was separated using agarose gel electrophoresis. The plasmid DNA was loaded into pre-cast wells in the gel and electric current (100V) was applied for 1 hour. The agarose gel was stained with 0.5ug/ml ethidium bromide for 20 minutes and visualized by UV-trans illumination according to Robins-Browne24.
Statistical analysis
Standard deviation of the mean of data obtained from this study were determined using 2010 Microsoft Excel.
Water storage and sanitation practices
A total of fifteen households were examined in the study. Sixty percent sourced their water from well while remaining forty percent sourced water from borehole. Forty-seven percent of the households do not engage in regular cleaning of their water tanks while fifty-three percent clean their tanks once in a year. Water guard and chlorination were the methods of disinfection used in these households. Sixty percent of the households had their septic tanks in less than 50 feets to their water source while the septic tanks and water source of the remaining forty percent were 50 to 100ft apart (Table 1).
Table (1):
Survey of water sources and sanitary practices in households in Ado-Ekiti.
Households |
Water source |
Frequency of cleaning water tanks |
Mode of disinfection |
Proximity of water source to septic tank |
|---|---|---|---|---|
N=15 |
Well (60%) Borehole (40%) |
Never (47%) Yearly (53%) |
None (47%) Water guard (33%) Chlorination (20%) |
<50ft (60%) 50-100 ft (40%) |
Mean total bacteria and total coliforms counts
The enumeration of heterotrophic and coliforms in water samples from household water distribution tank in Ado-Ekiti is shown in Table 2. Substantial count of bacteria was recovered from water samples from all the household examined. The mean total bacteria count and total coliforms were 6.96 CFU/ml and 5.50 CFU/ml respectively.
Table (2):
Total Bacteria Count and Coliform Count of Isolated Bacteria from Household Water Distribution Tanks in Ado-Ekiti.
Households |
Total Bacteria Count (Log10 CFU/ml) |
Total Coliform Count (Log10 CFU/ml) |
|---|---|---|
A |
8.17±0.05 |
6.35±0.07 |
B |
7.41±1.25 |
5.52±0.02 |
C |
7.31±0.08 |
7.60±1.21 |
D |
5.10±0.05 |
4.73±0.08 |
E |
6.71±0.06 |
4.84±0.02 |
F |
7.35±1.05 |
5.18±1.12 |
G |
6.84±0.02 |
4.02±1.05 |
I |
8.25±0.05 |
5.45±0.07 |
J |
6.16±1.08 |
4.23±0.04 |
K |
7.08±0.08 |
6.22±0.07 |
L |
5.32±0.03 |
4.14±1.21 |
M |
8.01±1.23 |
6.68±1.05 |
N |
7.79±1.06 |
7.23±1.11 |
O |
7.15±1.03 |
6.30±1.31 |
P |
5.89±0.04 |
4.10±0.04 |
Mean |
6.96 |
5.50 |
Based on the cultural and biochemical characteristics, the most frequent bacteria isolated from household water distribution tanks were Escherichia coli (20%), Pseudomonas aeruginosa (14%), Proteus vulgaris (16%), Klebsiella aerogenes (19%), Enterobacter aerogenes (9%), Serratia marcescens (8%), Bacillus sp. (8%) and Enterococcus sp. (6%) as indicated in Table 3.
Table (3):
Percentage Occurrence of Bacteria isolated from Household Water Distribution Tanks in Ado-Ekiti.
Isolates |
Percentage of occurrence (%) |
|---|---|
Escherichia coli |
20 |
Pseudomonas aeruginosa |
14 |
Proteus vulgaris |
16 |
Klebsiella aerogenes |
19 |
Enterobacter aerogenes |
9 |
Serratia marcescens |
8 |
Bacillus sp |
8 |
Enterococcus sp |
6 |
Total |
100 |
Antibiotics susceptibility pattern of the isolates
The antibiotics susceptibility test of the bacteria isolates reflects variation in their response to the different antibiotics examined as shown in Table 4. Their average percentage resistances to antibiotics are as follows: Amoxyllin (92%), Gentamycin (81%), Augmentin (80%), Sparfloxacin (78%), Chloraphenicol (75%), Perfloxacin (29%), Ofloxacin (24%), Streptomycin (53%), Cotrimoxazole (42%), and Ciprofloxacin (46%).
Table (4):
Antibiotics Resistance Pattern of Bacteria isolated from Household Water Distribution Tanks in Ado-Ekiti.
| No. | Isolates | Antibiotic resistance patterns of the bacterial isolates (%) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| AM | AU | CN | PEF | OFX | S | COT | CHL | SP | CPX | ||
| 1. | Pseudomonas aeuriginosa (n= 12) | 100 | 100 | 92 | 28 | 32 | 98 | 20 | 100 | 100 | 78 |
| 2. | Proteus vulgaris (n= 10) |
100 | 100 | 75 | 20 | 18 | 50 | 35 | 30 | 98 | 50 |
| 3. | Escherichia coli (n= 15) | 100 | 92 | 80 | 25 | 30 | 70 | 45 | 50 | 100 | 45 |
| 4. | Klebsiella aerogenes (n=10 ) | 98 | 72 | 98 | 38 | 28 | 65 | 58 | 100 | 100 | 35 |
| 5. | Enterobacter aerogenes (n= 10) |
92 | 62 | 75 | 35 | 22 | 45 | 50 | 98 | 50 | 30 |
| 6. | Serratia marcescens (n=8 ) | 90 | 75 | 80 | 23 | 15 | 35 | 50 | 58 | 65 | 35 |
| 7. | Bacillus sp (n= 5) |
75 | 68 | 72 | 30 | 25 | 28 | 45 | 100 | 65 | 50 |
| 8. | Enterococcus sp (n=7 ) | 80 | 70 | 75 | 35 | 25 | 30 | 32 | 62 | 45 | 45 |
| Average resistance | 92 | 80 | 81 | 29 | 24 | 53 | 42 | 75 | 78 | 46 | |
KEY: AMX – Amoxycilin, CPX- Ciprofloxacin, , OFL-Ofloxacin, CHL- Chloraphenicol, SP- Sparfloxacin, PEF- Perfloxacin, COT- Cotrimoxazole, S- Streptomycin, CN- Gentamycin, AU- Augmentin.
The multiple antibiotics resistant pattern of the bacteria isolates is shown in Table 5. The isolates displayed different resistance pattern to the antibiotics, Pseudomonas aeruginosa, Escherichia coli and Klebsiella aerogenes were resistant to 70 % of the antibiotics examined, while Enterobacter aerogenes, Serratia marcescens and Proteus vulgaris showed resistance to 60 % of the antibiotics and Bacillus sp. was resistant to 40 %.
Table (5):
Multiple antibiotic resistance patterns of bacteria isolates from Household Water Distribution Tanks in Ado-Ekiti.
No. |
Isolates |
Resistotype |
|---|---|---|
1. |
Pseudomonas aeuriginosa |
AM-AU-CN-S-CH-SP-CPX |
2. |
Proteus vulgaris |
AM-AU-CN-S -SP-CPX |
3. |
Escherichia coli |
AM –AU-CN -S -CHL –SP-CPX |
4. |
Klebsiella aerogens |
AM -AU -CN -S -COT –CHL-SP |
5. |
Enterobacter aerogens |
AM-AU-CN-COT -CHL -SP |
6. |
Serratia marcescens |
AM-AU-CN-COT –CHL-SP |
7. |
Bacillus sp |
AM- AU-CN -CHL |
KEY: AMX – Amoxyllin, CPX- CiprofloXacin, , OFL-Ofloxacin,CHL-Chloraphenicol, SP- Sparfloxacin, PEF- Perfloxacin, COT- Cotrimoxazole, S- Streptomycin, CN- Gentamycin, AU- Augmentin
Plasmid profiling of the isolates
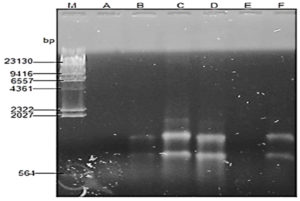
The plasmid profile of the isolates with multiple antibiotic resistance is shown in Figure 1. The result showed that four out of the resistant strains harbored multiple plasmids, Escherichia coli had three plasmids (1kb, 1.5kb and 2kb), Pseudomonas aeruginosa, Klebsiella aerogenes and Enterobacter aerogenes had two plasmids with 1kb, 1.5kb each while Proteus vulgaris and Serratia marcescens had no plasmid.
Table (6):
Plasmid profiling of the multiple resistant Bacteria isolated from Household water Distribution Tanks in Ado-Ekiti.
Isolates |
Number of Plasmid |
Molecular Weight |
|---|---|---|
Proteus vulgaris |
Nil |
Nil |
Pseudomonas aeruginosa |
2 |
1kb, 1.5kb |
Escherichia coli |
3 |
1kb, 1.5kb, 2kb |
Enterobacter aerogens |
2 |
1kb, 1.5kb, |
Serratia marcescens |
Nil |
Nil |
Klebsiella aerogens |
2 |
1kb1.5kb. |
Key: A: Proteus vulgaris, B: Pseudomonas aeruginosa, C: Escherichia coli, D: Enterobacter aerogenes, E: Serratia marcescens, F: Klebsiella aerogenes, bp – represents molecular sizes
Fig. 1. Plasmid Profile of Antibiotics resistant Bacteria isolated from household water distribution tanks in Ado-Ekiti
The numerous reports about the occurrence of pathogenic microorganisms in drinking water, their ability to resist antibiotics and associated diseases prompted this study, to access the microbial quality of water in storage tanks which serve as drinking and other domestic purpose for majority of homes in Ado-Ekiti.
Total bacteria and total coliform count of all the water samples analyzed in this study revealed high microbial contamination of the water (Table 2). The limit of <500 CFU/ml of heterotrophic bacteria and zero coliform or E. coli per 100ml of water as stipulated by WHO, USEPA, ISI 25,18 was exceeded in all the samples. Non-conformity of these water samples to the WHO standard decreased the water quality and renders them unfit for human consumption26. Previous researches on microbial assessment of drinking water sources have also reported high heterotrophic bacteria and coliform counts in different water sources and many of these water sources exceeded the permissible limits for quality water27,28,6. Eight bacteria genera recovered from the water samples: Escherichia coli, Pseudomonas aeruginosa, Proteus vulgaris, Klebsiella aerogenes, Enterobacter aerogenes, Serratia marcescens, Bacillus sp. and Enterococcus sp. have been isolated from different water sources29. The presence of Escherichia coli in the water is an indication that the household water has been faecally polluted. It also correlates with past studies which reported Escherichia coli as an organism that is commonly encountered in different water sources such as rivers, streams, rain water, well water, underground water and even pipe borne water30. Sixty percent of the households examined in this study sourced their water from well while the remaining 40% source their water from borehole (Table 1). Prevalence of bacteria from these genera have been reported in underground water31. High frequency of Pseudomonas species in household water also corroborates the report of Kawther and
Suaad32. Factors such as, their raw water source, treatment process employed and hygienic practices observed could influence microbial contaminations of household water33. The possibility of cross contamination of sewage and water in these household cannot be ruled out because larger percentage (60%) of them had their wells in close proximity (<50ft) to the septic tank (Table 1). Movement of water underground can lead to the sewer contaminating the water. Hence, the source of these bacteria is therefore linked to the water source rather than the storage tank.
Fifty-three percent of the households claimed they treated their water yearly with water guard or chlorination while others did not treat theirs. However, the effect of the treatment of the water by some of these households was not reflected as they also had high bacteria counts as the untreated water. This could be due to the loss of residual concentration of disinfectant used. Also, accumulation of organic matter in the tanks and pipes over time could enhance the growth of these bacteria since the households do not practice regular washing and disinfection of the water and the tanks which could have removed these contaminants. Once the water in the tank is exhausted, they immediately pump another water and overtime, the tank become heavily contaminated with organic and microbial contaminants.
The presence of these microbes in the water can present serious wellbeing dangers to consumers in general especially the immunocompromised individuals when the water is distributed.
Water borne diseases are usually combated with antibiotics. Due to their potency, they have gained global recognition as agents for treating infections. However, some bacteria have developed mechanisms of resisting the inhibitory effect of this group of antimicrobial agents. Some bacteria build living wall in response to exposure to antibiotics creating a physical barrier that shield them from and contribute further to the growing problems of drug resistant infection34. Bacteria isolated in this study exhibited similar characteristics of resisting multiple antibiotics examined. Previous studies accounted for the prevalence of antibiotic-resistant bacteria in surface and ground waters35,36.
High resistance was exhibited by the bacteria to Amoxyllin (92%), Gentamycin (81%), Augmentin (80%), Sparfloxacin (78%), and Chloraphenicol (75%). These antibiotics are the commonly prescribed drugs for people diagnosed with water related or other infections. Resistance of these bacteria to the antibiotics calls for public health concern. Mulamattathil37 likewise revealed that all bacteria from surface and drinking water in Mafikeng, South Africa analyzed were resistant to Erythromycin, Trimethoprim, and Amoxicillin. The isolates also displayed resistance to multiple antibiotics (4 to 7). Bacteria from these genera have been reported to possess multidrug resistance38, 39.
Six (6) multi-drug resistant bacteria isolated from household water samples were analyzed for plasmid out of which 4 harbored more than one plasmid. This conforms with the findings of Atuanya40 who isolated 45 antibiotics resistant bacteria from water samples and detected 31 plasmids in 14 of 45 antibiotics resistant strains with 10 carrying multiple plasmids. All the four (4) isolates containing the plasmids were resistant to Amoxyllin. The result of this study was in accordance with Ash41 who studied antibiotics resistance of gram negative bacteria from ground water in the United States and showed that their resistance to Augmentin, Amoxyllin and Erythromycin were plasmid mediated. The plasmids isolated were between the ranges of 1kb-2kb which was similar to the observation of Smith42.
Plasmids are double stranded extra-chromosomal genetic elements which reproduce autonomously. They have been recognized in numerous microbes however, they are some of the time found in eukaryotic cells43. It is notable that plasmids are quite possibly one of the most important facilitating agents in the fast spreading of antibiotics resistance among bacteria44. The microbial resistance genes frequently carried on plasmids have the ability to replicate and possibly the potential for self-transmission.
The incidence of plasmids among bacteria with multiple resistance to antibiotics in this study is alarming because plasmids have been identified as one of such movable elements through which resistance and foreign genes are being transmitted in niches45, 46. Genes that influence bacterial virulence are also frequently found on plasmids. Consequently, non-pathogenic and antibiotic susceptible bacteria can become pathogenic and resistant to antibiotics over time as a result of transmission of plasmids. This pose a health threat to the consumers because pathogenic bacteria from water sources have been identified as etiological agents of water borne diseases.
This study revealed water from household distribution tanks analyzed exceeded the permissible limits for coliforms and heterotrophic bacteria counts. Bacteria from the water samples were resistant to multiple antibiotics and the resistant strains had plasmids which could spread the resistance ability to non-resistant strains. The findings showed the water were contaminated and unfit for consumption. There is therefore need for regular washing of water storage tanks and routine disinfection of stored water to avert outbreak of waterborne diseases.
ACKNOWLEDGMENTS
None
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made substantial, direct and intellectual contributions to the work and approved it for publication.
FUNDING
None.
ETHICS STATEMENT
Not applicable.
AVAILABILITY OF DATA
All datasets generated or analyzed during this study are included in the manuscript.
- Pund DA, Aanorkar RP. Study of some physicochemical parameters of drinking water sources in Tem bhurkheda and Jarud Region Dist. Amravati, MS, India. Int. Res. J. Environment Sci. 2013; 2(10):93-95.
- Alhassan H, Kwakwa PA. When water is scarce: the perception of water quality and effects on the vulnerable. Journal of Water, Sanitation and Hygiene for Development, 2014; 4: 43-50.
Crossref - Getso BU, Mustapha A, Abubakar MM, Tijjani A. Assessment of Borehole Water Quality for Domestic Use in Three Selected Wards in Wudil Local Government Area, Kano State. Journal of Environmental Science Studies, 2018;1(1):1-5.
Crossref - The World Water, International World Water Day, United Nations Conference on Environment and Development (UNCED). Washington DC, USA: Island Press. 2009.
- Benjamin L, Atwill ER, Jay-Russell M, et al. Occurrence of generic Escherichia coli, E. coli O157 and Salmonella spp. in water and sediment from leafy green produce farms and streams on the Central California coast. International Journal of Food Microbiology, 2013 ;165.1: 65-76.
Crossref - Oluyege JO, Dada O, Oluyege AO, Olowomofe TO. Multiple antibiotic resistance index of Escherichia coli isolated from drinking water sources in Ado-Ekiti, Ekiti State. The Experiment, 2014;28(1):1896-1905.
Crossref - Onuoha C. Antibiotics Susceptibility Pattern of Escherichia coli Isolated from Well Water in Afikpo, South Eastern Nigeria. AASCIT Journal of Biology, 2015 ;3:38-42.
- Lehloesa LJ, Muyima NYO. Evaluation of impact of household treatment procedures on the quality of groundwater supplies in the rural community of the Victoria District, Eastern Cape. Water SA, 2000 ;26(2):285–290.
- Akoto O, Adiyiah J. Chemical analysis of drinking water from some communities in the Brong Ahafo region. Int. J. Environmental Science Technology, 2007;4(2): 211-214.
- Cabral JPS. Water microbiology. Bacterial pathogens and water. International Journal of Environmental and Public Health, 2010;7(10): 3657-3703.
Crossref - Duer. The science of mixing and improving water quality in water storage tanks; 2006. http://waterworld.com/content/ww/en/whitepapers/. Accessed Mar, 2015.
- Obi CL, Potgieter N, Bessong PO, Matsaung G. Assessment of the microbial quality of river water sources in rural Venda communities in South Africa, Water SA, 2002; 28(3):287–292.
Crossref - Sharma A, Dubey N, Sharan B. Characterization of Aeromonads isolated from the river Narmada, India, International Journal of Hygiene and Environmental Health, 2005;208(5): 425–433.
Crossref - US Environmental Protection Agency (EPA). Quick guide to drinking water sample collection, Region 8 Laboratory 16194 W. 45th Dr. Golden, CO 80403, 2016
- Onyenekenwa CE. Effects of Water and Sanitation Crisis on Infants and Under-five Children in Africa. Journal of Environmental Science and Technology, 2011;4(2):103-111.
Crossref - WHO. Water-related diseases. Environmental Management for Vector Control. World Health Organization 2014, (http://www.euro.who.int/pubrequest)
- Calva JJ, Sifuentes-Osornio J, Ceron C. Antimicrobial resistance in faecal flora: Longitudinal community-based surveillance of children from urban Mexico. Antimicrobial Agents Chemotherapy. 1996 ;40(7): 1699–1702.
Crossref - World Health Organization. Heterotrophic plate counts and drinking water safety. The significance of HPCs for water quality and human health. 2003; Edited by Bartram J, Cotruvo J, Exner M.
- Holt GJ, Krieg NR. Sneath PHA, Stanley JT, Williams ST. Bergey’s manual of determinative bacteriology. 9th Ed; Baltimore md; Williams and wikins. Pub.co, Maryland, 1994;786.
- Cheesbrough M. District Laboratory Practice in Tropical Countries (Part ii). Cambridge University Press, 50-150. https://www.medbox.org/preview/5255d6e1-05d4-41a9-beb2-02b60e695ecc/doc.pdf
- CLSI. Performance standards for antimicrobial susceptibility testing; fifteenth informational supplement, Clinical and Laboratory Standard Institute Wayne, Pa. M100- S15, 2012;25: 1.
- Olowomofe TO, Babalola TF, Oluyide OO, Adedayo O. Microbial Assessment of In-door Air and Equipment Used in Banks within Ekiti State University, Ado-Ekiti, Ekiti State, Nigeria. Annual Research & Review in Biology, 2019; 33(5):1-13
Crossref - Baserisalehi M and Bahador N. A study on relationship of plasmid with Antibiotic Resistance in Thermophilic Campylobacter spp. Isolates from Environmental Samples Biotechnology, 2008;7(4): 813-817.
Crossref - Robins-Browne RM, Bordun AM, Tauschek M, et al. Escherichia coli and community-acquired gastroenteritis: Melbourne, Australia. Emerg Infect Dis. 2004;10(10):1797-1805.
Crossref - US Environmental Protection Agency (EPA). “Quality drinking water. 2015. http://www.epa.gov/sites/default/files/201511/documents/drinking_water_sample_collection.pdf
- World Health Organization, 2006. Protecting Groundwater for Health: Managing the Quality of Drinking Water Sources. IWA Publishing, London. https://www.who.int/water_sanitation_health/publications/PGWsection1.pdf?ua=1
- Onwa NC, Uzomaka IC, Maduako AL, Elom EE, Ikeanumba MO, Nwode VF. Antibiotic Susceptibility of Bacterial Species Isolated from Underground Waters in Abakaliki Metropolis of Ebonyi State, Nigeria. International Journal of Pharmaceutical Science Invention(IJPSI), 2019; 8(2):55-65.
- Ekhosuehi A, Akharame MO, Obayuwana P. Bacteriological quality and antibiogram of isolates from potable water sources in Ekosodin community, Benin City, Nigeria. Journal of Applied Science and Environmental Management,2018;22(1) 129-133.
Crossref - Olorunjuwon B, Adeleke O, Temitope O. Microbial Quality and Antibiotic Susceptibility Profile of Bacterial Isolates from borehole Water Used by Some Schools in Ijebu-Ode, Southwestern Nigeria. Scholars Acad. J. Biosci., 2013;1(1):4-13
- Environment Protection Agency, EPA. US Environment protection agency: Safe drinking water act amendment, 2002. P. EPA 816 – F – 03 –016.
- Edema MO, Atayese AO. Pure water syndrome: bacteriological quality of sachet- packed drinking water sold in Nigeria. African Journal of Food, Agriculture, Nutrition and Development.2011;11(1):4595-4609.
Crossref - Kawther FA, Suaad SA. Mineral and microbial contents of bottled water and tap water in Riyadh, Saudi Arabia. Middle-East Journal of Scientific Research. 2007; 2(3): 151-156.
- Geldreich EE. Microbial quality of water supply in distribution system. Published, 2019 by CRC Press.
- Matic L, Taddei F, Radman M. Genetics barriers among bacteria. Trends in Microbiology,1996;4(2):69-73.
Crossref - O’Dwyer J, Hynds P, Pot M, Adley C, Ryan MP. Evaluation of Levels of antibiotic Resistance in Groundwater Derived E. coli Isolates in the Mid-West of Ireland and Elucidation of Potential Predictors of Resistance. Hydrogeol. J. 2017;25:939-951.
Crossref - Efuntoye O, Apanpa O. Status of Contamination and Antibiotic Resistance of Bacteria from well water in Ago-Iwoye, Nigeria. J. Appl. Biosci. 2010;35:2244-2250.
- Mulamattathil GS, Bezuidehout C, Mbewe M, Ateba CN. Isolation of Environmental Bacteria from Surface and Drinking Water in Mafikeng, South Africa and Characterization Using Their Antibiotic Resistance Profiles. J. Pathog. 2014 ;371208.
Crossref - Losch LS, Alonso JM, Merino LA. Occurrence of Antimicrobial Resistant Enetrobacteriacea in water from Different Sources in a Sub- Tropical Region of Argentina. Ambi Agua,Taubate, 2008;3:28-36.
Crossref - Lynch JP, Clark NM, Zhanel GG. Evolution of antimicrobial resistance among Enterobacteriaceae (focus on extended spectrum b-lactamases and carbapenemases). Expert Opin. Pharmacother. 2013 ; 14: 199-210.
Crossref - Atuanya EI, Nwogu NA, Orah CU. Antibiotic resistance and plasmid profile of bacterial pathogens isolated from hair-dressing saloon effluents in Benin City, Nigeria. Nig. Jour. Life Sci. 2018;22(11):1749-1755.
Crossref - Ash RJ, Mauk B, Morgan M. Antibiotics resistance of Gram negative bacteria in wells in United States. Emerg Infect Dis., 2002;8(7):713-716.
Crossref - Smith S, Aboaba OO, Odeigha P, Shodipo K, Adeyeye NN. Plasmid profile of Escherichia coli from water samples. African Journal of Biotechnology, 2003;2 (9): 322-324. Available online at http://www.academicjournals.org/AJB
- Dale JW, Park S. Molecular genetics of bacteria 4th Ed. John Wiley and sons Inc., Chichester, UK, 2004. https://www.academia.edu/31746711/Dale_Molecular_Genetics_of_Bacteria_4th_ed
- Zhang R, Wang Y, Gu JD. Identification of environmental plasmid-bearing Vibrio species isolated from polluted and pristine marine reserves of Hong Kong, and resistance to antibiotics and mercury, Antonie van Leeuwenhoek, 2006; 89(3-4):307-15
Crossref - Bennett PM. Plasmid encoded antibiotic resistance: acquisition and transfer of antibiotic resistance genes in bacteria, Br J Pharmacol, 2008; 153(Suppl. 1): S347-S357.
Crossref - Atuanya EI, Nwogu NA, Orah CU. Antibiotic Resistance and Plasmid Profiles of Bacteria Isolated from Abattoir Effluents around Ikpoba River in Benin City, Nigeria J. Appl. Sci. Environ. Manage. 2018;22 (11) 1749–1755.
Crossref
© The Author(s) 2021. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.