ISSN: 0973-7510
E-ISSN: 2581-690X
https://dx.doi.org/10.22207/JPAM.13.1.38 | © The Author(s). 2019
The number of infections caused by microorganisms is increasing significantly over the last few years. A total of 140 patients admitted to the central teaching hospital of pediatrics from the 1st of Jun 2017 to 31 October 2017. The Clinical samples was processed from culture and sensitivity testing. Antibiotic discs used for gram negative isolates. The most prevalent gram negative isolates included Escherichia coli 63 (45.0 %), Pseudomonas spp. 21 (15.0 %), Klebsiella spp. 19 (13.6 %) predominantly. Escherichia coli were the most prevalent isolates from urine 45 (71.4 %), Klebsiella spp. 11 (57.9 %) and Enterobacter spp. 11 (68.8 %) followed by Escherichia coli 10 (15.9 %) predominant from blood. 68 (48.6 %) of specimens were urine, 47 (33.6 %) were blood and 25 (17.8 %) were from other origins. Resistance to CTR was 42 (51.2 %) in Escherichia coli, 15 (18.3 %) in Pseudomonas spp. 9 (11.0 %) in Klebsiella spp. and similarly AMC showed 41 (62.1) resistance among Escherichia coli, 8 (12.1 %) among Pseudomonas spp. and Klebsiella spp. and also GEN showed 27 (57.4 %) resistance among Escherichia coli, 7 (14.9 %) among Pseudomonas spp. and 6 (12.8 %) in Klebsiella spp.
Antimicrobial Resistance, Gram Negative bacilli, Patient Pediatric, Hospital.
Number of infections caused by microorganisms is increasing significantly over the last few years; one of the reasons for this increase is development of microbial resistance to drugs used to treat these infections1. The infection due to gram negative bacteria increasing problem in recent years2. Most common reasons responsible for these infections are multidrug resistant gram negative bacilli particularly members of the family Enterobacteriaceae and non-fermenting gram negative rods3. Enterobacteriaceae family had shown high rates of antibiotic resistance4-6. Resistance gram negative bacteria are a serious global public health concern especially in developing countries7-10. Gram negative bacilli are common cause of intra- abdominal infections, urinary tract infections, nosocomial pneumonia and bacteremia11. Most infections may be related to unbalanced microbiota and host defenses mechanisms but undoubtly hospital environments are great source of potentially pathogenic microorganisms12. A number of factors have been reported that were related to infection with resistant microorganisms, including previous use of antibiotics, corticosteroid therapy, mechanical ventilation, length of hospital stay and use of invasive device such as catheters13. The prevalence of resistant gram negative bacteria can vary at local, regional, national and international levels14. The mortality is increasing every day because of global changing of antimicrobial resistance patterns16.
A total of 140 children admitted to the central teaching hospital of pediatrics from the 1 st of Jun 2017 to 31 October 2017. Gram-negative isolates from clinical samples were included in the study. The Clinical samples included urine, blood, wound secretions, pus, ear, endotracheal, doubluma, peritoneal. The sample was processed from culture and sensitivity testing. Antimicrobial susceptibility was evaluated by the Kirby- Bauer disk diffusion method in guide lines of clinical and laboratory standards Institute. The sample was processed from culture and sensitivity testing. Specimens inoculated onto agar plates according to the type of sample and incubated aerobically at 37°C over night. The cultured plates were examined after 24 hours and organisms identified by their colonial morphology, Gram staining and API 20 E kits biochemical tests using standard techniques. Antimicrobial susceptibility was evaluated by the Kirby- Bauer disk diffusion method in guide lines of clinical and laboratory standards Institute17. Antibiotic discs used for gram negative isolates [Ampicillin (AMP), Augmentin (AMC), Cefotaxime (CTX), Ceftriaxone (CTR), Cefipime (CFP), Cefixime (CFM), Cephalothin (CLT), Gentamycin (GEN), Amikacin (AK), Nitrofurantin (NIT)].
Statistical Analysis
The suitable statistical methods were used in order to analyze and assess the results, they include the followings:
Descriptive statistics: Statistical tables including observed frequencies with their percentages.
Inferential statistics: These were used to accept or reject the statistical hypotheses, Persons Chi-Square test (c2) at level of significance 0.05.
P value < 0.05 level of significance was considered statistically significant.
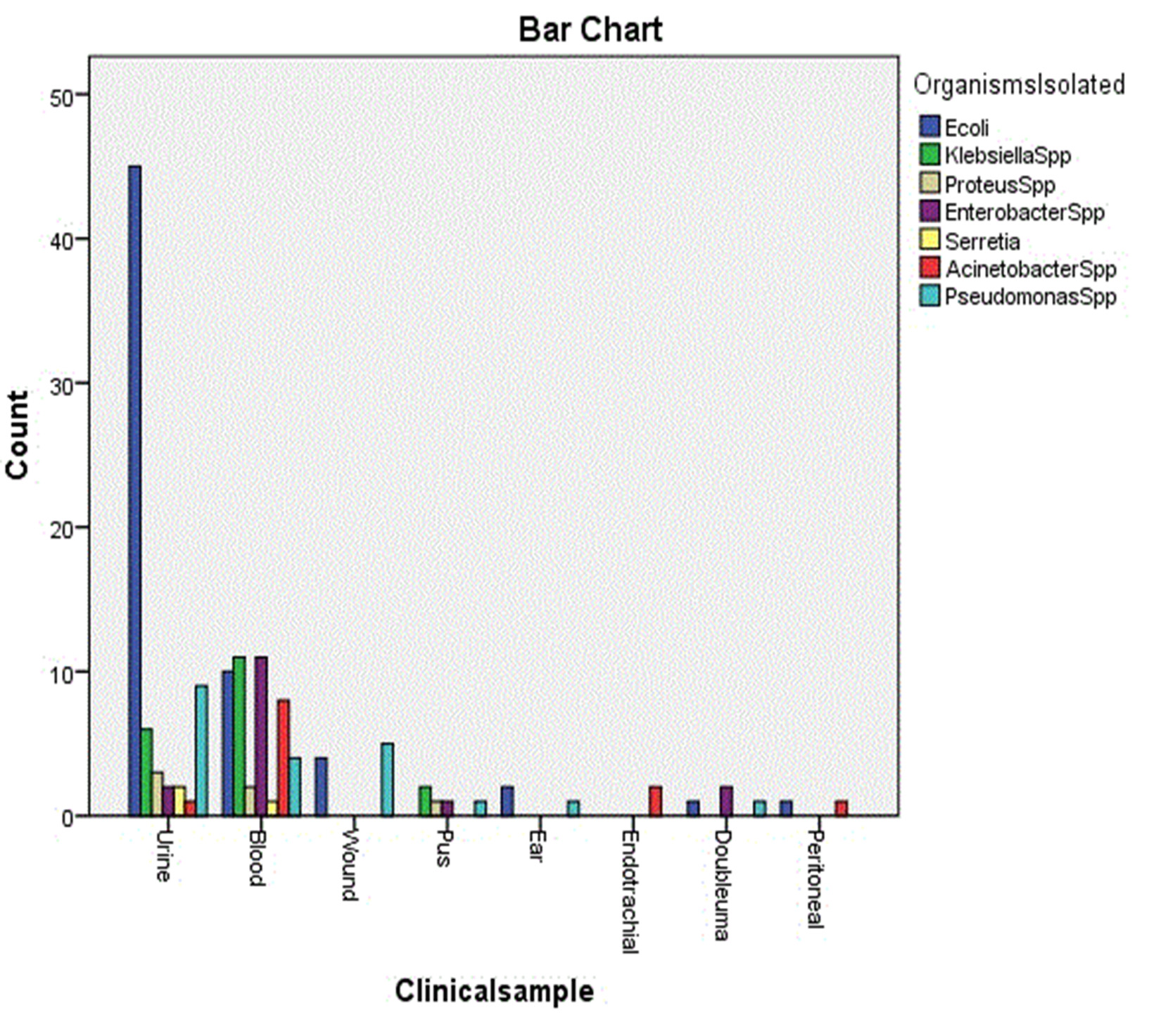
In a total of 140 different clinical samples which were processed. The most prevalent gram negative isolates included Escherichia coli 63 (45.0 %), Pseudomonas spp. 21 (15.0 %), Klebsiella spp 19 (13.6 %) predominantly. Escherichia coli were the most prevalent isolates from urine 45 (71.4 %), Klebsiella spp 11 (57.9 %) and Enterobacter spp 11 (68.8 %) followed by Escherichia coli 10 (15.9 %) predominant from blood. 68 (48.6 %) of specimens were urine, 47 (33.6 %) were blood and 25 (17.8 %) were from other origins. Resistance to CTR was 42 (51.2 %) in Escherichia coli, 15 (18.3 %) in Pseudomonas spp 9 (11.0 %) in Klebsiella spp and similarly AMC showed 41 (62.1) resistance among Escherichia coli, 8 (12.1 %) among Pseudomonas spp and Klebsiella spp and also GEN showed 27 (57.4 %) resistance among Escherichia coli, 7 (14.9 %) among Pseudomonas spp and 6 (12.8 %) in Klebsiella spp.
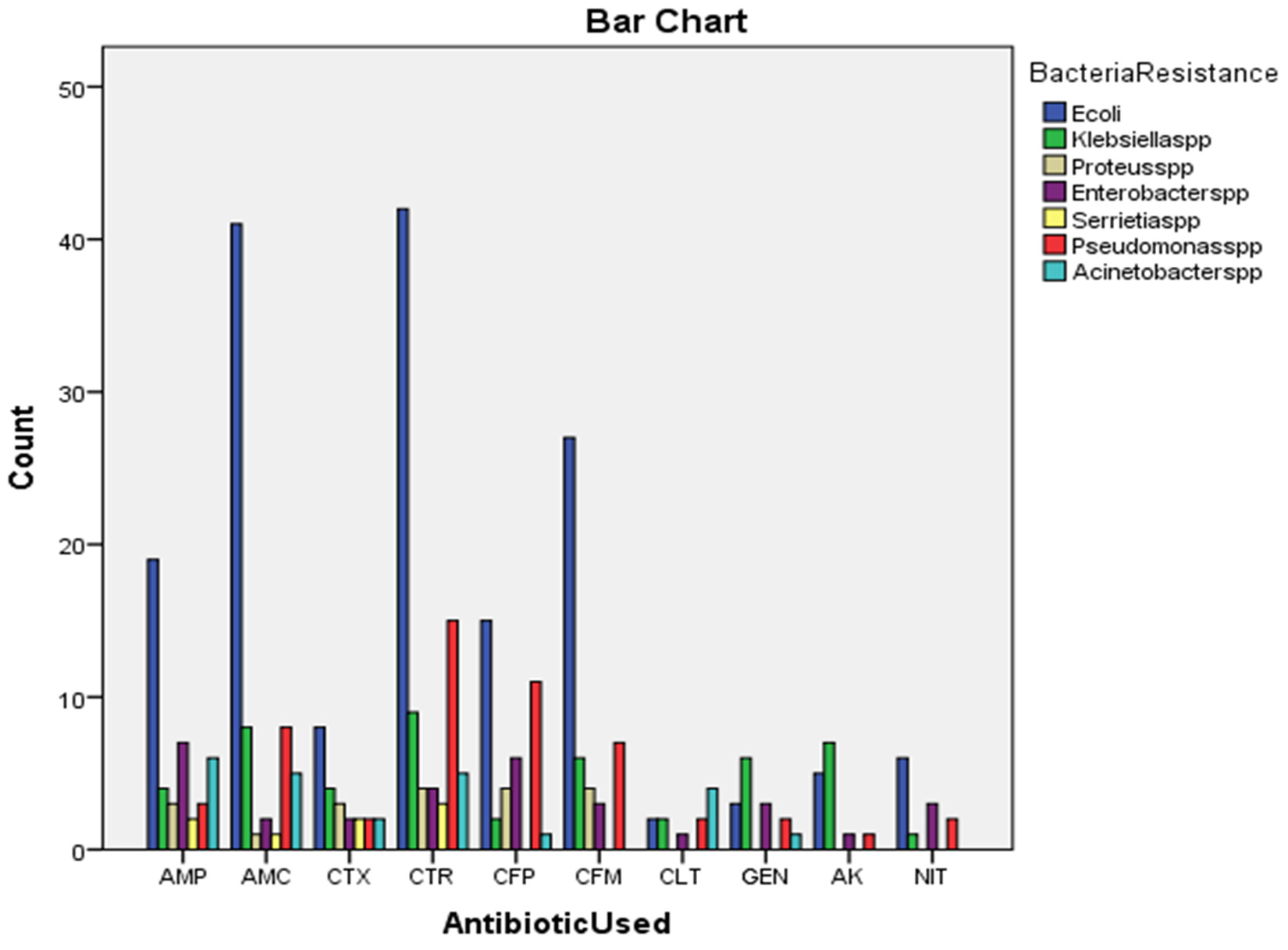
The Microbial Pathogens as well as their antibiotic sensitivity patterns may change from place to place and from time to time18. In present study show in Table (1) the most common microorganisms isolated were Escherichia coli 63 (45.0 %), Pseudomonas spp 21 (15.0 %), Klebsiella spp 19 (13.6 %). Escherichia coli found to be the most common in study which was isolated from urine and this agree with Javeed et al.19, Saravanan et al.20 and Vipin et al.21. Showed significant differences (P < 0.05) in Table (2). Urine samples were 68 (48.6 %) and blood samples were 47 (33.6 %) and other origins 25 (17.8 %) this agreement with Sharif et al. 22. Showed significant differences (P < 0.05) in Table (3). Resistance emerges from over utilization of antibiotics trying to sterilize the environment and also the inappropriate use of the antibiotics for treatment, uncontrolled antibiotics resistance surveillance23. There was 168 (47.6 %) resistance rate of Escherichia coli to [CTR 42 (51.2 %), AMC 41 (62.1 %), CFM 27 (57.4 %), AMP 19 (43.2 %)] which is the most resistance and there was 53 (15.0 %) resistance rate Pseudomonas spp to [CTR 15 (18.3 %), CFP 11 (28.2 %), AMC 8 (12.1 %), CFM 7 (14.9 %) also 49 (13.9 %) resistance rate of Klebsiella spp to [CTR 9 (11.0 %), AMC 8 (12.1 %), AK 7 (50.0 %), CFM 6 (12.8 %), GEN 6 (40.0 %), this result show in Table (3) with significant differences (P < 0.05).
Table (1):
Distribution of culture positive Bacterial Isolates.
Bacterial Type |
No. |
% |
|---|---|---|
Escherichia coli |
63 |
45.0 |
Klebsiella spp |
19 |
13.6 |
Proteus spp |
6 |
4.3 |
Enterobacter spp |
16 |
11.4 |
Serretia spp |
3 |
2.1 |
Acinetobacter spp |
12 |
8.6 |
Pseudomonas spp |
21 |
15.0 |
Total |
140 |
100.0 |
Table (2):
Distribution of Organisms Isolated from various Clinical sample.
Clinical
Sample |
Escherichia coli
N % |
Klebsiella spp
N % |
Proteus spp
N % |
Enterobacter spp
N % |
Serretia spp
N % |
Acinetobacter spp
N % |
Pseudomonas spp
N % |
Total
N % |
|---|---|---|---|---|---|---|---|---|
Urine |
45 (71.4) |
6 (31.6) |
3 (50.0) |
2 (12.5) |
2 (66.7) |
1 (8.3) |
9 (42.9) |
68 (48.6) * |
Blood |
10 (15.9) |
11 (57.9) |
2 (33.3) |
11 (68.8) |
1 (33.3) |
8 (66.7) |
4 (19.0) |
47 (33.6) * |
Wound |
4 (6.3) |
0 (0.0) |
0 (0.0) |
0 (0.0) |
0 (0.0) |
0 (0.0) |
5 (23.8) |
9 (6.4) |
Pus |
0 (0.0) |
2 (10.5) |
1 (16.7) |
1 (6.2) |
0 (0.0) |
0 (0.0) |
1 (4.8) |
5 (3.6) |
Ear |
2 (3.2) |
0 (0.0) |
0 (0.0) |
0 (0.0) |
0 (0.0) |
0 (0.0) |
1 (4.8) |
3 (2.1) |
Endo-
tracheal |
0 (0.0) |
0 (0.0) |
0 (0.0) |
0 (0.0) |
0 (0.0) |
2 (16.7) |
0 (0.0) |
2 (1.4) |
Doubluma |
1 (1.6) |
0 (0.0) |
0 (0.0) |
2 (12.5) |
0 (0.0) |
0 (0.0) |
1 (4.8) |
4 (2.9) |
Peritoneal |
1 (1.6) |
0 (0.0) |
0 (0.0) |
0 (0.0) |
0 (0.0) |
1 (8.3) |
0 (0.0) |
2 (1.4) |
Total |
63 (45.0) * |
19 (13.6)* |
6 (4.3) |
16 (11.4) |
3 (2.1) |
12 (8.6) |
21 (15.0) * |
140 (100.0) |
(P < 0.05).
Table (3):
Antibiotic Resistance in Gram Negative Bacilli.
| Antibiotic Used | Bacteria Resistance | Total | ||||||
|---|---|---|---|---|---|---|---|---|
| Escherichia coli
N % |
Klebsiella spp
N % |
Proteus spp
N % |
Enterobacter spp
N % |
Serretia spp
N % |
Acinetobacter spp
N % |
Pseudomonas spp
N % |
||
| AMP | 19 (43.2) | 4 (9.1) | 3 (6.8) | 7 (15.9) | 2 (4.5) | 6 (13.6) | 3 (6.8) | 44 (12.5) |
| AMC | 41 (62.1) | 8 (12.1) | 1 (1.5) | 2 (3.0) | 1 (1.5) | 5 (7.6) | 8 (12.1) | 66 (18.7) |
| CTX | 8 (34.8) | 4 (17.4) | 3 (13.0) | 2 (8.7) | 2 (8.7) | 2 (8.7) | 2 (8.7) | 23 (6.5) |
| CTR | 42 (51.2) | 9 (11.0) | 4 (4.9) | 4 (4.9) | 3 (3.7) | 5 (6.1) | 15 (18.3) | 82 (23.2) * |
| CFP | 15 (38.5) | 2 (5.1) | 4 (10.3) | 6 (15.4) | 0 (0.0) | 1 (2.6) | 11 (28.2) | 39 (11.0) |
| CFM | 27 (57.4) | 6 (12.8) | 4 (8.5) | 3 (6.4) | 0 (0.0) | 0 (0.0) | 7 (14.9) | 47 (13.3) |
| CLT | 2 (18.2) | 2 (18.2) | 0 (0.0) | 1 (9.1) | 0 (0.0) | 4 (36.4) | 2 (18.2) | 11 (3.1) |
| GEN | 3 (20.0) | 6 (40.0) | 0 (0.0) | 3 (20.0) | 0 (0.0) | 1 (6.7) | 2 (13.3) | 15 (4.2) |
| AK | 5 (35.7) | 7 (50.0) | 0 (0.0) | 1 (7.1) | 0 (0.0) | 0 (0.0) | 1 (7.1) | 14 (4.0) |
| NIT | 6 (50.0) | 1 (8.3) | 0 (0.0) | 3 (25.0) | 0 (0.0) | 0 (0.0) | 2 (16.7) | 12 (3.4) |
| Total | 168 (47.6) * | 49(13.9) * | 19 (5.4) | 32 (9.1) | 8 (2.3) | 24 (6.8) | 53 (15.0) * | 353 (100.0) |
(P < 0.05).
[Ampicillin (AMP), Augmentin (AMC), Cefotaxime (CTX), Ceftriaxone (CTR), Cefipime (CFP), Cefixime (CFM), Cephalothin (CLT), Gentamycin (GEN), Amikacin (AK), Nitrofurantin (NIT)].
In this study, the most common microorganisms isolated was Escherichia coli isolated from urine samples most resistance rate to Ceftriaxone 42 (51.2 %), Augmentin 41 (62.1 %), Cefixime 27 (57.4 %), Ampicillin19 (43.2 %).
The author declares that there are no conflict of interest.
- Anjum MU, Shams N, Shah SH, Mujaddad- ur-Rehman M, Hussain S. Prevalence and Antibiotic Resistance pattern of Multidrug Resistance Bacteria among Blood Isolates. Sch. J. App. Med. Sci., 2014; 2(5D): 1734-1740.
- Sankarakutty J., Kaup S., Distribution and Antibiogram of Gram Negative Isolates from Various clinical samples at a Teaching Hospital, Tunkur. Sch. J. App. Med. Sci., 2014; 2(3A): 927-931.
- Lockhart SR., Abramson MA., Beckmann SE., Gallagher G., Riedel S., Diekema DJ et al. Antimicrobial Resistance among Gram-Negative Bacilli causing infections in Intensive care unit patients in the United States between 1993 and 2004. J. Clin. Microbiol., 2007; 45(10): 3352-3359.
- Abo-state MA.,Mahday HM. Ezzat SM., Abded EH., Shakour EI and Bahnasawy EI. Antimicrobial Resistance profiles of Enteriobacteriaceae Isolated from Rosetta Branch of River Nile, Egypt. Worl. App. Sci. J., 2012; 19: 1234-1243.
- Osman KM., Ata NS.,Hedia RH., AbuElnaga ASM., El-Hariri M and Aly MAK. Emergence of an Anti-microbial Resistant Pseudomonas aeroginosa from Human and Animal Clinical samples : A zoonotic and public Health Hazard. Global Veterinaria, 2012; 19: 745-751.
- Adedeji BAM and Abdulkadir OA-Etiology and Antimicrobial Resistance Pattern of Bacterial Agents of Urinary Tract infections in students of Tertiary Institutions in Yola Metropolis. Advan. Biol. Resea., 2009; 3: 67-70.
- Alizargar J., Sharif MR., Sharif A. Risk factors of Methicillin-Resistant Staphylococcus aureus Colonization in Diabetic outpatients. A prospective Cohort study. Internat. J. Microbiol. Resea., 2013; 4: 147-151.
- Okonko To., Soleye FA., Amusam TA., Ogun AA., Ogunnusi TA and Ejembi J. Incidence of Multi-Drug Resistance (MDR) Organisms in Abeokuta southwest Nigeria. Glo. J. Pharma., 2009; 3: 69-80.
- Aly MEA, Essam TM and Amin MA. Antibiotic Resistance profile of E.coli strains Isolated from Clinical Specimens and Food samples in Egypt. Internation. J. Microbiol. Resea., 2012; 3: 176-182.
- Sharif MR, Alizagar J. and Sharif A. Antibiotic susceptibility of Staphylococcus aureus in Isolates of the patients with Osteomyelitis. Worl. J. Med. Sci., 2013; 9: 180-183.
- Sievert DV., Ricks P., Edwards JR., Schneider A., Patel J., Srinivasam A., Kallen A., Limbago B., Fridkin S. and National Health care safety Network (NHSN) Team and Partivcipating NHSN Facilities. Antimicrobial resistant pathogens associated with health care-associated infections: summary of Data reported to the national Health care safety Network at the center for disease control and prevention 2009-2010 infect control Hospital. Epidermiol., 2013; 34: 1-14.
- Bryce EA.,Scharf S., Walker M., Walsh A., The infection control audit as a tool for change. Am. J. infect. Control., 2007; 35: 271-283.
- Khalili H., Soltani R., Afhami S., Dashti-Khavidaki S., Alijani B., Antimicrobial resistance pattern of Gram-negative bacteria of nosocomial origin at a teaching hospital in the Islamic Republic of Iran. East. Medi. Hea. J., 2012; 18(2): 172-177.
- Livermore DM., Current epidemiology and growing resistance of Gram-negative pathogens. Korean J. Intern. Med., 2012; 27: 128-142.
- Okesola AO. And Oni AA. Antimicrobial resistance Among Common Bacterial pathogens in south western Nigeria. Ameri. Euras. J. Agr. Envie. Sci., 2009; 5: 327-330.
- Okonko TO., Soleye FA., Amusam TA., Ogun AA., Ogunnusi TA. And Ejembi J., Incidence of Multi-Drug Resistance (MDR) oragnisms in Abeokuta South Western Nigeria. Glob. J. Pharma., 2009; 3: 69-80.
- Clinical and Laboratory Standards Institutes Performance Standards for antimicrobial susceptibility testing. Eighteenth informational Supplement document M100e S18. Wayne, PA: Clinical and Laboratory Standards Institutes (CLSI), 2008.
- Pakyz AL., The Utility of Hospital Antibiograms as tool for Guiding Empiric Therapy and Tracking Resistance; Insight from the society of infectious diseases pharmacists. Pharmacotherapy, 2007; 27(9): 1306-1312.
- Javeed I., Hafeez R. and Anwar MS. Antibiotic Susceptibility patteren of Bacterial Isolates from patients Admitted to Atertiary care Hospital in Lahore. Biomedica., 2011; 27: 19-23.
- Saravanan R., Raveendaran V. Antimicrobial resistance patteren in a tertiary care hospital : An observational study. J Bas. Clin. Pharm., 2013; 4(3): 56-63.
- Vipin K., Rohit KM., Avantika C., Pramila G., Incidence of B-Lactamase producing gram-negative clinical isolates and their antibiotic susceptibility patterns A case study in Allahabad. Internati. J. Resea. Pure and Appli. Microbiol., 2011; 1(3): 36-39.
- Sharif MR., Alizargar J., Sharif A. Antimicrobial Resistance among gram-negative Bacteria Isolated from Different samples of patients Admitted to A University Hospital in Kashan, Iran. Advances. Biolog. Resear., 2013; 7(5): 199-202.
- Kritu P., Prakash G., Shiba KR., Reene KM., Ram NS., Ganesh R., Antibiograms-typing of gram negative isolates in different clinical samples of tertiary hospital. Asian J. Pharmaceu. Clinicl. Resea., 2013; 6(1): 153-156.
© The Author(s) 2019. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.