ISSN: 0973-7510
E-ISSN: 2581-690X
Klebsiella is a pathogen that causes a significantly high number of community-acquired and hospital-acquired infections, with infections being one of the leading causes of death in ICU patients worldwide due to increasing antibiotic-resistance and a lack of therapeutic options. A total of 230 Klebsiella spp. were collected from various clinical samples. After initial identification, the drug-resistant strain was subjected to standard Clinical Laboratory and Standards Institute methods such as Kirby–Bauer disc diffusion. All isolates were screened and confirmed for ESBL/AmpC β-lactamase/carbapenemase production. The isolated Klebsiella spp. were found to be K. pneumonia (89%), K. oxytoca (6.5%), and K. aerogenes (4.5%). Among the 230 isolates, 80 (34.7%) isolates were found to be ESBL producers via screening; of these, 53 (23.5%) were verified by a confirmatory test. Moreover, 115 isolates (50%) were screened as AmpC producers; of these, 23 isolates (10%) were verified by a confirmatory test. Carbapenemase producers accounted for 69 (30%) isolates, identified by screening; 25 (10.86%) were verified by a confirmatory test. ESBL producers accounted for the majority of Klebsiella spp. isolates, followed by carbapenem and AmpC producing strains.
Klebsiella, Antibiotic resistance, ESBL ,Amp C β-lactamase, Carbapenemase
Klebsiella is a bacterial genus in the Enterobacteriaceae family, which is gram negative, non-motile, non-sporing bacilli that normally reside in the mammalian gastrointestinal tract and infect the host opportunistically.1 These bacteria often cause life-threatening infections such as septicemia, urinary tract infections, pneumonia, and liver abscess in immuno-compromised patients.2 Klebsiella pneumoniae is the most prevalent nosocomial and opportunistic pathogen that causes infections in humans among Klebsiella spp.3
The emergence of antimicrobial resistance in Klebsiella spp. is becoming a global threat owing to increasingly ineffective treatment options. In empirical treatment protocols, resistant screening plays an important role. Bacteria produce various enzymes that promote drug resistance. Extended spectrum beta lactamases (ESBLs) are enzymes that hydrolyze most penicillins and cephalosporins, including oxyimino-β-lactam compounds (i.e., cefuroxime, third- and fourth-generation cephalosporins and aztreonam) but not cephamycins nor carbapenems. Most ESBLs belong to the Ambler class A of β-lactamases and are inhibited by β-lactamase inhibitors (clavulanic acid, sulbactam, and tazobactam) and bi-diazabicyclooctanes (avibactam).4 These enzymes are primarily responsible for resistance in Klebsiella spp. In Klebsiella pneumonia, the primary factor influencing high ESBL production is the usage of third-generation cephalosporins. Other risk factors include long-term hospitalization in the intensive care unit (ICU), catheterization, mechanical ventilation, preterm birth, and low weight at birth.5
AmpC-beta lactamases are Ambler class C β-lactamases that hydrolyze penicillins, cephalosporins (including third-generation but generally not the fourth-generation compounds) and monobactams.6 In general, AmpC-type enzymes are poorly inhibited by classical ESBL inhibitors, as clavulanic acid mediates resistance to antibiotics such as cefotetan and cefoxitin and is produced by some strains of Klebsiella spp.7-9
Carbapenemases are β-lactamases that hydrolyze penicillins, in most cases cephalosporins and, to various degrees, carbapenems and monobactams. The latter are not hydrolyzed by metallo-β-lactamases. The carbapenem resistance of K. pneumoniae is a major reason for the strain’s high morbidity and mortality.10 Hence, studying the characteristics that promote resistance in this strain will promote the effective use of antibiotics in treatment. Moreover, studying antibiotic-resistant K. pneumoniae elucidates the importance of screening and confirmatory methods for research and treatment.11
The screening test alone is insufficient for detecting antibiotic resistance. Hence, there is a need for reliable phenotypic confirmatory testing to identify resistant Klebsiella spp. in clinical samples. The present study emphasized the importance of confirmatory tests compared to screening tests and provided foundational data for the appropriate use of antibiotics.
Methods
Identification
Clinical samples were obtained in an aseptic manner and processed using standard microbiological techniques. Ethical approval was obtained from the Institutional Ethics Committee of SRM Medical College Hospital and Research Center (1860(A)/IEC/2019). Conventional culture techniques were used to isolate the Klebsiella spp. After obtaining the sample, direct gram stain was performed, and samples were cultured on blood agar, MacConkey agar, and chocolate agar. All suspected isolates are identified by Gram stain and hanging drop method. Klebsiella spp. were speciated using the following biochemical tests: indole, methyl red, Voges–Proskauer, triple sugar iron, urease, citrate, OF dextrose, nitrate reduction, lysine, ornithine, and arginine.12,13 Antibiotic susceptibility testing (AST) was carried out according to the Clinical and Laboratory Standards Institute (CLSI) using Kirby–Bauer disc diffusion method in Muller–Hinton agar.14
AST Procedure
AST was conducted according to drugs listed in the CLSI guidelines for Klebsiella spp. Testing was carried out using Muller–Hinton Agar (MHA) with the Kirby–Bauer disc diffusion method The following antibiotic discs were used: ampicillin (10 µg), amoxicillin–clavulanate (20 µg/ 10 µg), ceftazidime (30 µg) ceftazidime with clavulanic acid (30 µg/20 µg), cefepime (30 µg), ceftriaxone (30 µg), ceftizoxime (30 µg), ertapenem (10 µg), meropenem (10 µg), piperacillin tazobactam (100 µg/10 µg), amikacin (30 µg), chloramphenicol (30 µg), ciprofloxacin (5 µg), cefotaxime (30 µg), cefoxitin (30 µg), cefazolin (30 µg), gentamycin (10 µg), imipenem (10 mg), tetracycline (30 µg), tigecycline (15 µg), and nitrofurantoin (300 µg) (purchased from Himedia, Mumbai, India). The zone size was interpreted after 18–24 h of incubation according to CLSI guidelines.
Screening Tests for ESBL, AmpC, and Carbapenemase Producers
ESBL screening test was performed according to CLSI guidelines by a disk diffusion method in which the isolate was streaked as a lawn culture over MHA and antimicrobial concentrations of cefotaxime (30 µg) and ceftazidime (30 µg) were used. If the zone size was <22 mm and <27 mm after 18–24 h of incubation, the isolate was considered positive for ESBL production.
AmpC screening test was performed according to EUCAST guidelines15 by a disk diffusion method in which the isolate was streaked as a lawn culture over MHA and an antimicrobial concentration of cefoxitin (30 µg) was used. If the zone size was ≤14 mm after 18–24 h of incubation, the isolate was considered positive for AmpC production.
Carbapenemase screening test was performed based on CLSI guidelines using a disk diffusion method in which the isolate was streaked as a lawn culture over MHA and an antimicrobial concentration of ertapenem (10 µg) was used. If the zone size was ≤ 19 mm after 18–24 h of incubation, the isolate was considered positive for carbapenemase production.
Confirmatory Tests for ESBL, AmpC, and Carbapenemase Production
Combined testing for ESBL detection was carried out based on CLSI guidelines using a disk diffusion method in which the isolate was streaked as a lawn culture over Muller–Hinton agar and two antibiotic discs were used: ceftazidime (30 µg) and ceftazidime with clavulanic acid (CAC) (30 µg/20 µg). The CAC disc was placed in the center of the MHA plate, the and ceftazidime disc was placed approximately 1.5 cm away from the CAC disc. If the zone size difference for each exhibited a ≥5 mm increase in inhibition zone, the sample was confirmed for ESBL production.
Double Disc Synergy Test
The double disc synergy test relied on the ability of cloxacillin to inhibit AmpC enzyme function. This test was conducted based on EUCAST guidelines. Cefoxitin (30 µg) and cloxacillin (200 µg) discs were used (Himedia, Mumbai). The strains were streaked as a lawn culture on MHA with 0.5 McFarland and incubated for 18–24 h at 37°C. A zone of inhibition ≥4 mm between the cefoxitin–cloxacillin and cefoxitin zones confirmed AmpC production.
Modified Hodge Test (MHT)
MHT
To verify carbapenemase enzyme production, the MHT was conducted according to CLSI guidelines. A carbapenem-susceptible strain of E. coli ATCC 25922 is known to grow toward the carbapenem from the test isolate, which also generates the enzyme.
Testing procedure
An overnight culture of E.coli ATCC 25922 (indicator) yielded a 0.5 McFarland standard suspension, which was diluted 1:10 in saline or broth. The suspension was inoculated onto the MHA plate using lawn culture and left to dry for 3 to 10 min. In the center, a 10 g ertapenem disc (Himedia) was placed. The test isolate and positive and negative controls were streaked from the edge of the disc to the plate’s periphery and incubated at 37°C for 24 h. Streaks were 20 to 25 mm in length. If the test strain showed growth near the inhibition zone, carbapenemase production was assumed. If the test strain did not grow near the inhibition zone, carbapenemase was not produced.16,17
Isolate identification
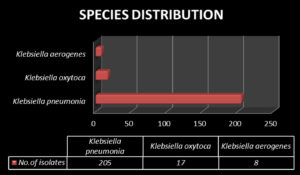
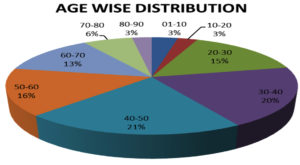
In this study, 230 isolates of Klebsiella were collected and subjected to identification by microbiological culture, and biochemical tests and AST were performed for verification. Microbiological culturing was performed using MacConkey agar, blood agar, and biochemical solutions to validate Klebsiella spp. identification. Of the 230 isolates, the following strains were identified: K. pneumonia (89%), K. oxytoca (6.5%), and K. aerogenes (4.5%) (Figure 1). Nearly 65% of the Klebsiella spp. were isolated from male patients, and 35% of Klebsiella spp. were isolated from female patients. Of the 230 isolates, 36.5% were obtained from patients in the age group 40–60 years of age, followed by 35.6% from patients in the age group of 20–40 years, 30.19% in the age group of 60–80 years, followed by (2.6%) followed by (6%) followed by 1-20 years (Figure 2).
Sample Distribution
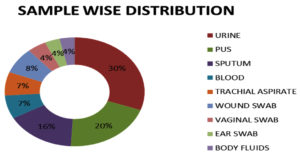
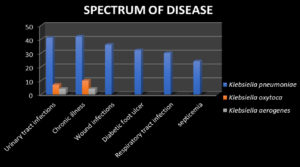
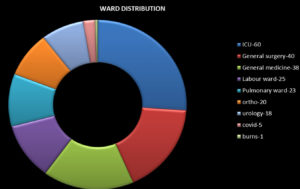
The sources of infection in the collected Klebsiella samples were found to be urine (30.43%), sputum (20.43%), pus (16.08%), tracheal aspirate (6.95%), wound swab (6.95%), vaginal blood swab (4.34%), ear swab (3.47%) and body fluids (3.47%), (Figure 3) respectively. The spectrum of disease distribution of studied isolates was as follows: urinary tract infections, chronic illness, wound infections, diabetic foot ulcer, Respiratory tract infection and septicaemia (Figure 4) The ward distribution of studied isolates was as follows: ICU (26.08%), general surgery (17.39%), general medicine (17.39%), labour ward (10.86), pulmonary ward (10%), orthopaedic ward (20%), urology ward (7.82%), COVID-19 ward (2.17%), and burn ward (0.43%) (Figure 5).
AST Results
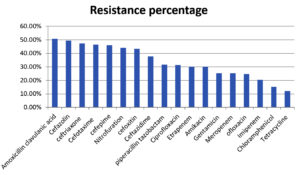
AST and ESBL, AmpC β-lactamase, and carbapenemase production analyses were carried out according to CLSI guidelines. The ESBL producers yielded zone size differences of ≥5 mm between ceftazidime and ceftazidime with clavulanic acid. Cefoxitin resistance was considered to indicate that the isolate was a AmpC producer. Imipenem, meropenem, and ertapenem resistance indicated that the isolate was a carbapenemase producer. The distribution of resistances among all isolates were determined as follows: amoxicillin-clavulanic acid (20/10 µg) (50.86%), cefazolin (30 µg) (49.56%), ceftriaxone (30 µg) (47.39%), cefotaxime (30 µg) (46.52%), cefepime (30 µg) (46%), nitrofurantoin (300 µg) (44%), cefoxitin (30 µg) (43.47%), ceftazidime (30 µg) (37.82%), piperacillin–tazobactam (100/10 µg) (31.73%), ciprofloxacin (5 µg) (31.30%), ertapenem (10 µg) (30%), amikacin (30 µg) (30%), gentamicin (10 µg) (29.13%), meropenem (10 µg) (25.21%), ofloxacin (5 µg) (24.78%), imipenem (10 µg) (20.43%), chloramphenicol (30 µg) (15.21%), and tetracycline (10 µg) (12.06%) (Figure 9). Klebsiella spp. strains were the most resistant to amoxicillin and clavulanic acid.
ESBL, AmpC, and Carbapenemase Producer Confirmatory Test Results
a. Combined disc diffusion test for ESBL production (Figure 6)
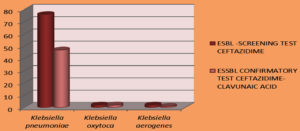
Of the 230 isolates, 80 (34.7%) isolates were found to be ESBL producers by the screening test; of these, 53 (23.5%) were verified as ESBL producers by the confirmatory test.
b. Double disc synergy test for AmpC production (Figure 7)
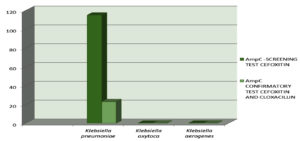
Of the 230 isolates, 115 isolates (50%) were found to be AmpC producers by the screening test; of these, 23 isolates (10%) were verified as AmpC producers by the confirmatory test.
c. MHT for carbapenemase production (Figure 8)
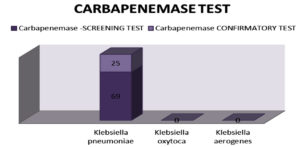
Of 230 isolates, 69 (30%) were found to be carbapenemase producers by the screening test; of these, 25 (10.86%) were verified as carbapenemase producers by the confirmatory test.
Figure 6. Among 230 isolates,80 (34.7%)isolates were found to be ESBL producer by screening test out of which 53 (23.5%) were ESBLproducer by confirmatory test
Figure 7. 115isolates(50%) were found to be AmpC producer by screening test out of which 23 isolates (10%) was only positive in confirmatory test
Figure 8. On analysis for Carbapenemase producer 69(30%) was isolated by screening test out of which 25(10.86%) isolates in confirmatory test were recorded
Among 230 Klebsiella spp. isolates, we identified the following strains: K. pneumonia (89%), K. oxytoca (6.5%), and K. aerogenes (4.5%). Similar results were suggested by Sandeep Vasikar et al.18 Male predominance is more nearly 65% of the Klebsiella spp. was isolated from male patients and 35% of Klebsiella spp. is isolated from female patients. Men are more commonly affected by Klebsiella infections than women due to the increased prevalence of alcoholism and smoking among men. This distribution is similar to that identified by Sunilkumarbiradar et al.19
In this study, isolate samples were predominantly obtained from patients in the age groups of 40–60 years (36.5%) and 20–40 years (35.6%). The next-most prevalent age group was 60–80 years (30.19%) followed by (2.6%) followed by (6%) followed by 1–20 years, which correlates with data reported by Virawan et al.20 Infection is considered to be more common around 40–60 years of age because the immune declines as individuals age and are associated with various co-morbidities. The highest number of Klebsiella spp. samples were isolated from urine followed by sputum, which reflects the findings reported by Hamida et al.21
Klebsiella spp. infections are the second-most common cause of urinary tract infection, which may be due to expression of type 1 fimbriae present in the urinary tract. Nearly one third (26.08%) of Klebsiella spp. samples are isolated from ICU patients because ICU patients are a tremendously vulnerable group and highly prone to infection, as reported by Aliyu Aminu et al.22 Carbapenem-producing Enterobacteriaceae pose a significant threat to patients in the ICU, and Klebsiella spp. are the most common bacterial species in this group, causing life threatening infections. Klebsiella spp. were primarily resistant to amoxicillin and clavulanic acid, followed by cefazolin and ceftriaxone. The lowest resistance was to tetracycline, followed by chloramphenicol and imipenem. Using similar methods as those employed in this study, Mwangi Joseph Kibuchi et al.23 reported that the highest drug sensitivities among Klebsiella spp. included that to meropenem, followed by amikacin and chloramphenicol. These discrepancies were expected because antibiotic susceptibility patterns change according to geographical location.
Among the 230 isolates, 34.7% were found to be ESBL producers by the screening method, and 23.5% were verified for ESBL production by the confirmatory method. The lower number of confirmed producers implies the importance of employing confirmatory tests to reduce false positive reporting, as this can lead to the increased unnecessary administration of antibiotics, which is known to promote drug resistance. Detecting ESBL producers among Klebsiella spp. is crucial as resistant strains are typically resistant to third-generation cephalosporins and aztreonam. Treatment failure occurs when the appropriate antibiotic is not used. Moreover, ESBL producers often contain plasmids that carry genes encoding co-resistance to other antibiotics. Iran Hadi Mehrgan et al.24 reported that ESBL producers exhibit around 70% now higher antibiotics like carbapenems are used as the drug against them.
Among the 230 isolates, 50% were found to be AmpC producers by the screening test, and 10% were verified as AmpC producers by the confirmatory test. These findings were similar to those reported by Donati et al.25 This difference in initial and final identification further supports the necessity of confirmatory testing.
Nearly 30% of all isolates were determined to be carbapenemase producers by the screening test, and 10.86% were verified by the confirmatory test. The level of carbapenemase producers among our isolate samples was relatively low when compared with the findings of Shao et al.26 who reported 23.3% of evaluated Klebsiella samples as carbapenemase producers. Carbapenems are widely administered to various treat-life threatening infections, and the development of resistance to this drug class will increase mortality, hospitalization length, and healthcare costs. Hence, the proper detection and administration of this antibiotic is critical for limiting the development of carbapenem resistance in major bacterial species.
The widespread and non-specific use of antibiotics results in an increasing number of multidrug-resistant Klebsiella spp. This study elucidated the enzymes responsible for drug resistance in Klebsiella spp. such as ESBL, AmpC βlactamase, and carbapenemases. Among 230 Klebsiella spp. isolates evaluated in this study, ESBL producers were most encountered, followed by carbapenemase and AmpC producers. Infection control practices along with antibiotic control policies are necessary for controlling increasing resistance trends. Moreover, employing confirmatory tests such as combined disc diffusion (ESBL detection), double disk synergy (AmpC detection), and Modified Hodge (Carbapenemase detection) tests in addition to screening tests is a reliable method for identifying drug resistance and can be carried out easily as a routine microbiology technique. Implementing confirmatory tests will promote the identification of antibiotic resistance to inform the careful and specific use of antibiotics in treating high-risk infections.
ACKNOWLEDGMENTS
The authors would like to thank SRM Medical College Hospital and Research Centre, Kattankulathur, Kanchipuram district, India for providing all the equipments and facilities for performing this research work.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analysed during this study are included in the manuscript.
ETHICS STATEMENT
This study was approved by the Institutional Ethics Committee, SRM Medical College Hospital and Research Centre, Tamilnadu, India (1860(A)IEC/2019).
- Mackie MC, McCartney JE. Practical Medical Microbiology: Eds. Collee JG, Fraser AG, Marnion BP Simmons A. Tests for identification of bacteria. 14th Edition. Churchill living stone publishers, UK. 1999.
- Ramakrishnan K, Kali A, Sah S, Pagal S, Kenchappa P, Seetha KS. Molecular Profile of Emerging Multidrug Resistant Klebsiella pneumoniae Clinical Isolates from Southern India. J Clin Diagn Res. 2019;13(3):1-6.
Crossref - Orhue PO, Aliu FR. Antibiogram and susceptibility of Klebsiella Spp isolated from different clinical specimens in health care centers in Etsako West local government area of EDO state, Nigeria. American Journal of Current Microbiology. 2015;3(1):60-72.
- Bush K, Jacoby GA, Medeiros AA. A functional classification scheme for β -lactamases and its correlation with molecular structure. Antimicrob Agents Chemother. 1995;39(6):1211-1233.
Crossref - Gupta A, Ampofo K, Rubenstein D, Saiman L. Extended spectrum β lactamase-producing Klebsiella pneumoniae infections: a review of the literature. J Perinatol. 2003;23(6):439-443.
Crossref - Jacoby GA. AmpC β-lactamases. Clin Microbiol Rev. 2009;22:161-182.
Crossref - Hemalatha V, Padma M, Sekar U, Vinodh TM, Arunkumar AS. Detection of AmpC beta lactamase production in Escherichia coli and Klebsiella by an inhibitor-based methods. Indian J Med Res. 2007;126(3):220-223. PMID: 18037717
- Ejikeugwu C, Duru C, Eluu S, et al. Isolation and Phenotypic Detection of Metallo-Beta Lactamase (MBL)- Producing Klebsiella spp. from Cow Anal Swabs. Global J Pharma Sci. 2017;2(3):555586.
Crossref - Papanicolaou GA, Medeiros AA, Jacoby GA. Novel plasmid mediated β-lactamases (MIR-1) conferring resistance to oxyimino and a methoxy β-lactams in clinical isolates of Klebsiella pneumoniae. Antimicrob Agents Chemother. 1990;34(11):2200-2209.
Crossref - Kaur J, Sheemar S, Chand K, Chopra S, Mahajan G. Outbreak of Carbapenemase-Producing Klebsiella pneumoniae Blood Stream Infections in Neonatal Intensive Care Unit. Int J Curr Microbiol App Sci. 2016;5(1):727-733.
Crossref - Ravichitra KN, Prakash PH, Subbarayudu S, Rao US. Isolation and antibiotic sensitivity of Klebsiella pneumoniae from pus, sputum and urine samples. Int J Curr Microbiol App Sci. 2014;3(3):115-119.
- Winn Washington C, and Elmer W. Koneman. Koneman’s Color Atlas and Textbook of Diagnostic Microbiology. Philadelphia:Lippincott Williams & Wilkins, 2006;7.
- CLSI. [Performance standard of antimicrobial susceptibility]. [30th]. CLSI guideline [M100]. Wayne, PA: Clinical and Laboratory Standards Institute; [2020].
- EUCAST[detection of resistance mechanisms and specific resistances of clinical and/or epidemiological importance]Version2.1 European committee on antimicrobial susceptibility testing.
- Amjad A, Mirza Ia, Abbasi S, Farwa U, Malik N, Zia F. Modified Hodge test: A simple and effective test for detection of carbapenemase production. Iran J Microbiol. 2011;3(4):189-193.
- Cury A, Andreazzi D, Maffucci M, Caiaffa-Junior H, Rossi F. The modified Hodge test is a useful tool for ruling out Klebsiella pneumoniae carbapenemase. Clinics. 2012;67(12):1427-1431.
Crossref - Vasaikar S, Obi L, Morobe I, Bisi-Johnson M. Molecular Characteristics and Antibiotic Resistance Profiles of Klebsiella Isolates in Mthatha, Eastern Cape Province, South Africa. Int J Microbiol. 2017;2017:8486742.
Crossref - Sunilkumar Biradar and Roopa . C. Isolation and Antibiogram of Klebsiella species from Various Clinical Specimens. Int J Curr Microbiol Appl Sci. 2015;4(9):991-995.
- Virawan H, Nuryastuti T, Nirwati H. Multidrugresistant Klebsiella pneumoniae from clinical isolates at dr. Soeradji Tirtonegoro central hospital Klaten. Jurnal Kedokteran dan Kesehatan Indonesia. 2020;11(2):109-20.
Crossref - Hamida RS, Ali MA, Goda DA, Khalil MI, Redhwan A. Cytotoxic effect of green silver nanoparticles against ampicillin-resistant Klebsiella pneumoniae. RSC Adv. 2020;10(36):21136-211346.
Crossref - Aminu A, Daneji IM, Yusuf MA, et al. Carbapenem-resistant Enterobacteriaceae infections among patients admitted to intensive care units in Kano, Nigeria. Sahel Medical Journal. 2021;24(1):1-9.
- Kibuchi MJ, Mathenge S, Njoroge W. Antibiotic susceptibility profile of the Klebsiella pneumoniae isolated from Africa Inland Church Hospital Kijabe, Kenya. Int J Adv Multidiscip Res. 2019;6(2):74-86.
- Donati V, Feltrin F, Hendriksen RS, et al. Extended-spectrum-beta-lactamases, AmpC beta-lactamases and plasmid mediated quinolone resistance in klebsiella spp. from companion animals in Italy. PLoS One. 2014;4;9(3):90564.
Crossref - Mehrgan H, Rahbar M, Arab-Halvaii Z. High prevalence of extended spectrum beta lactamase producing klebsiella pneumoniae in a teritary care hospitalin Tehran Iran. J Infect Dev Ctries. 2010;4(3):132-138.
Crossref - Champs, DC, Sauvant, MP, Chanal, C, et al. Prospective survey of colonization and infection caused by expended spectrum beta lactamase- producing members of the family Enterobacteriaceae in an intensive care unit. J C Microbiol. 1989;27(12):2887-2890.
Crossref - Shao C, Jin Y, Liu S, Jiang M, Zhao S. An Outbreak of Carbapenem-Resistant Klebsiella Pneumoniae of K57 Capsular Serotype in an Emergency Intensive Care Unit of a Teaching Hospital in China. Res Sq. 2020.
Crossref
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.