ISSN: 0973-7510
E-ISSN: 2581-690X
Pseudomonas aeruginosa is an adaptable bacteria causing an extensive spectrum of infections and intrinsically resistant to many antibiotics. As antimicrobial resistance has increased due to many resistance mechanisms. This study was done to evaluate the antibiogram of Pseudomonas aeruginosa at a tertiary care centre. Thirty seven isolates were recovered from various specimens for a period of 6 months from June to December 2020 and the disc diffusion method was used for antibiotic susceptibility testing as per CLSI guidelines. Pseudomonas aeruginosa was found to be high (45.9%) in pus/wound than other samples. Antibiotic resistance rate of the isolates were 29.7% to ceftazidime, 16.2% to Piperacillin-tazobactam, 27 % to gentamicin and ciprofloxacin, 16.2% to tobramycin and imipenem, 24.3% to meropenem, 27% to ciprofloxacin, 13.5% to aztreonam, 21.6% to amikacin, 24.3% to cefepime and levofloxacin, 21.6 to tigecycline. All strains were sensitive to colistin. 27% of the organism were found to be multidrug resistance. Hence periodic susceptibility testing can curb the resurgence of these bacterial pathogens.
Pseudomonas aeruginosa, antibiotic susceptibility, carbapenem
Pseudomonas aeruginosa (P. aeruginosa) is an opportunistic bacteria which causes a extensive spectrum of infections ranging from ear infections, bacteremia, urinary tract infections, burn infections, bacteremia and respiratory tract infections.1,2 Prevalence rate of P. aeruginosa infection ranges from 10-30% in India.3 It causes both hospitalized and community acquired infections. P. aeruginosa infection pose a therapeutic challenge as the organism has both intrinsic and acquired resistance to various classes of antibiotics. As antibiotic resistance is increasing drastically among the P. aeruginosa which is a threat to the Public health. Hence monitoring these bacterial populations is necessary to formulate the antibiotic treatment policy. This analysis was done to assess the antibiotic profile of P. aeruginosa isolates from different specimens.
This retrospective analysis was done in tertiary care in Chennai during the period of June-December 2020. Thirty seven non-duplicate P. aeruginosa were recovered from samples such as urine, pus, sputum, wound swab, ear swab, blood, endotracheal aspirate from various departments of Sree Balaji Medical college and hospital (Table 1). The clinical samples were inoculated by streak plate method on nutrient agar (Himedia, Mumbai, India), Mac Conkey agar (Himedia, Mumbai, India) and blood agar (Himedia, Mumbai, India) from urine, pus, wound swab, ear swab, endotracheal secretion. Blood was inoculated in BacT/Alert 3D (Biomerieux, Marcy l’ Etoile, France) and positive cultures were plated on blood agar, Mac Conkey agar and nutrient agar. The isolated colonies on the different media were identified based on the morphology of the colony, pyocyanin production, Gram staining, oxidase test and other standard biochemical test like Triple sugar Iron agar, Indole, Citrate, Urease, Mannitol motility agar. All the media, oxidase disc, Gram staining kit purchased from Himedia, Mumbai, India.
Table (1):
Isolation of Pseudomonas aeruginosa from various samples.
No |
Age/sex |
Ward |
Sample |
Isolate |
|---|---|---|---|---|
1 |
65/M |
ICU |
Blood |
P. aeruginosa |
2 |
37/M |
GS |
Wound swab |
P. aeruginosa |
3 |
28/F |
OBG |
Urine |
P. aeruginosa |
4 |
5/M |
Pead |
Wound swab |
P. aeruginosa |
5 |
50/M |
GS |
Pus |
P. aeruginosa |
6 |
39/F |
GM |
Sputum |
P. aeruginosa |
7 |
42/M |
Ortho |
Pus |
P. aeruginosa |
8 |
33/M |
GS |
Pus |
P. aeruginosa |
9 |
30/F |
URO |
Urine |
P. aeruginosa |
10 |
45/M |
ICU |
Sputum |
P. aeruginosa |
11 |
43/F |
OBG |
Blood |
P. aeruginosa |
12 |
31/F |
OBG |
pus |
P. aeruginosa |
13 |
6/F |
Pead |
Urine |
P. aeruginosa |
14 |
52/M |
GS |
Pus |
P. aeruginosa |
15 |
44/M |
GM |
Sputum |
P. aeruginosa |
16 |
43/M |
URO |
Urine |
P. aeruginosa |
17 |
57/M |
ICU |
Blood |
P. aeruginosa |
18 |
41/F |
GS |
Wound swab |
P. aeruginosa |
19 |
33/F |
URO |
Urine |
P. aeruginosa |
20 |
36/M |
GS |
Wound swab |
P. aeruginosa |
21 |
48/F |
GS |
Wound swab |
P. aeruginosa |
22 |
63/M |
Ortho |
Pus |
P. aeruginosa |
23 |
34/F |
ENT |
Ear swab |
P. aeruginosa |
24 |
53/M |
ICU |
Blood |
P. aeruginosa |
25 |
62/M |
GS |
Sputum |
P. aeruginosa |
26 |
44/F |
OBG |
Urine |
P. aeruginosa |
27 |
34/M |
ENT |
Wound swab |
P. aeruginosa |
28 |
21/M |
GM |
Pus |
P. aeruginosa |
29 |
38/M |
GS |
Pus |
P. aeruginosa |
30 |
45/F |
ICU |
Blood |
P. aeruginosa |
31 |
38/F |
OBG |
Pus |
P. aeruginosa |
32 |
66/M |
Res.Med |
Sputum |
P. aeruginosa |
33 |
45/F |
GS |
Pus |
P. aeruginosa |
34 |
59/M |
GM |
Blood |
P. aeruginosa |
35 |
61/F |
ICU |
ET secretion |
P. aeruginosa |
36 |
42/F |
GS |
Pus |
P. aeruginosa |
37 |
62/F |
Res.Med |
Sputum |
P. aeruginosa |
GM – General Medicine, GS – General surgery, ICU – Intensive care unit, OBG – Obstetrics and gynecology, Res.Med – Respiratory medicine, ENT- Ear, nose throat, Uro – urology, Pead – Paediatrics, Ortho – Orthopedics.
Antibiotic susceptibility testing was done by disk diffusion technique on Mueller-Hinton agar medium (MHA) (Himedia, Mumbai, India). Antibiotics impregnated paper discs (Himedia, Mumbai, India): imipenam (10 µg) (Himedia, Mumbai, India), piperacillin-tazobactam (100/10 µg ) (Himedia, Mumbai, India), meropenem (10 µg) (Himedia, Mumbai, India), Ceftazidime (30 µg) (Himedia, Mumbai, India), ciprofloxacin (5 µg) (Himedia, Mumbai, India), amikacin (30 µg) (Himedia, Mumbai, India), colistin (10 µg) (Himedia, Mumbai, India), Tigecycline (10 µg) (Himedia, Mumbai, India), cefepime (30 µg) (Himedia, Mumbai, India), aztreonem (30 µg) (Himedia, Mumbai, India), levofloxacin (5 µg) (Himedia, Mumbai, India), tobramycin (10 µg) (Himedia, Mumbai, India) and gentamicin (10 µg) (Himedia, Mumbai, India) were placed on lawn cultures of MHA (Himedia, Mumbai, India) and incubated overnight at 37°C. Measurement of the inhibition zone was taken and interpreted as Susceptible, intermediate and resistant based on CLSI guidelines 2020.4
P. aeruginosa ATCC 27853 (Himedia, Mumbai, India) was used for control strains for culture and susceptibility testing.
Thirty seven P. aeruginosa (Fig. 1) were isolated from 676 different samples. The prevalence rate of the organism was found to be 5.5%.
Among 37 P. aeruginosa isolates, 20 (54.05%) were from males and 17(45.9%) from females. P. aeruginosa was isolated from 17 pus/wound samples, 6 urine, 6 sputum, 1 ET secretions, 1 ear swab and 6 blood samples. 45.9% of the isolates were from Pus or wound sample (Table 2). 29.7% of strains were isolated from the samples sent from General surgery followed by 16.2% from ICU shown in Table 3. The isolation of P. aeruginosa was more from inpatients than out patients.
Table (2):
Frequency of Pseudomonas aeruginosa isolates in different samples.
Sample |
Number (%) |
|---|---|
Urine |
6 (16.2) |
Pus/wound |
17 (45.9) |
Sputum |
6 (16.2) |
Blood |
6 (16.2) |
Ear swab |
1 (2.7) |
ET Secretion |
1 (2.7) |
Total |
37 |
Table (3):
Frequency of isolates from different departments.
Clinical Department |
Number (%) |
|---|---|
General surgery |
11 (29.7) |
General medicine |
4 (10.8) |
Paediatrics |
2 (5.4) |
Obstetrics and gynecology |
5 (13.5) |
Urology |
3 (8.1) |
ICU |
6 (16.2) |
ENT |
2 (5.4) |
Orthopedics |
2 (5.4) |
Respiratory medicine |
2 (5.4) |
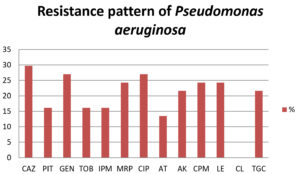
Antibiotic resistance rate of the isolates were 29.7% to ceftazidime, 16.2% to Piperacillin-tazobactam, 27% to gentamicin and ciprofloxacin, 16.2% to tobramycin and imipenem, 24.3% to meropenem, 27% to ciprofloxacin, 13.5% to aztreonam, 21.6% to amikacin, 24.3% to cefepime and levofloxacin, 21.6% to tigecycline as shown in Fig. 2. All the isolates were sensitive to colistin. 27% isolates were found to be multidrug resistance.
P. aeruginosa is the prime cause of healthcare associated infections among the Gram negative bacterial pathogens. Wider use of the antibiotics has resulted in the emergence of the multidrug resistant isolates among these organisms. Multiple drug resistance (MDR) is the resistance exhibited by an microgranism to alteast one antibiotic in three or more antibiotic categories. It has been found to cause infections in wider clinical settings especially in surgical wards and ICUs and the resistance patterns in different geographical regions. Hence antibiotic surveillance is important to the policy makers to frame the empirical treatment regime for these bacterial infections.
Our study results shows that P. aeruginosa infections was more in males than females. It was higher in inpatients than outdoor patients similar to the study by Anupurba.5 The prevalence of isolation was higher from surgical ward samples. This might be due to prolonged hospital stay after surgery resulting in colonization and infection.5 When considering the factor like sex, P. aeruginosa occurrence was predominant in males (54.05%) in our study similar to other studies.6,7
In this study, the frequency of P. aeruginosa was predominant in pus than other specimens which were similar to studies of Siguan SS et al., Masaadeh HA et al and Ranjan et al.6,8,9 In contrast in certain studies blood has been found to be the predominant sample followed by pus.10-12 In this study, most of the isolates were from surgical wards similar to a study by Ramakrishnan.13
In the present study, Prevalence rate of P. aeruginosa was 5.5% which is similar to the reports in India by Ramakrishnan et al.13 and Sorabh Singh Sambyal14 were 6.8 and 4.8%.
In this study, all our isolates were sensitive to colistin like the study by Mastoraki et al.15 In contrast few studies have shown resistance to colistin.13 Aztreonam and tobramycin, imipenem showed 16.2% of resistance. But meropenem has slightly higher rate of resistance than imipenem in contrast to study by Ahmad.16 25-30% of isolates were resistant to other aminoglycosides, cephalosporins, ciprofloxacin, levofloxacin and tigecycline. 27% of the isolates are found to be multi-drug resistant lower than the study by Doi et al.17
The susceptibility pattern is found to vary from time to time and differs in different geographical regions. Colistin, tobramycin, imipenem has been the promising antimicrobial agents to treat the P. aeruginosa infections from our study. P. aeruginosa infections were common among the inpatients and in surgical wards. Hence continuous monitoring of the antibiotic susceptibility and following the infection control can greatly help us to treat and reduce the infections.
ACKNOWLEDGMENTS
The authors are thankful to the faculty and technical assistance staff of Central laboratory, Department of Microbiology, SBMCH, Chennai, India for their support.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
Both the authors have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
ETHICS STATEMENT
This study was approved by the Institutional Ethics Committee, Sree Balaji Medical College, Chrompet, Tamilnadu, India.
AVAILABILITY OF DATA
All the data sets generated and analyzed during this study are included in the manuscript.
- Aloush V, Navon-Venezia S, Seigman-Igra Y, Cabili S, Carmeli Y. Multidrug-resistant Pseudomonas aeruginosa: risk factors and clinical impact. Antimicrob Agents Chemother. 2006;50(1):43-48.
Crossref - Janner D. A Clinicl Guide to Pediatric Infectious Disease. Lippincott Williams & Wilkins; Philadelphia, PA, USA: 2005.
- Senthamarai S, Reddy ASK, Sivasankari S, et al. Resistance Pattern of Pseudomonas aeruginosa in a Tertiary Care Hospital of Kanchipuram, Tamilnadu, India. J Clin Diagn Res. 2014;8(5):DC30-DC32.
Crossref - Clinical Laboratory Standards Institute . Performance Standards for Antimicrobial Susceptibility Testing: Twenty-Fifth Informational Supplement M100-S25. CLSI; Wayne, PA, USA: 2020. https://www.nih.org.pk/wp-content/uploads/2021/02/CLSI-2020.pdf
- Anupurba S, Bhattacharjee A, Garg A, Sen MR. Antimicrobial susceptibility of Pseudomonas aeruginosa isolated from wound infections. Indian J Dermatol. 2006;51(4):286-288.
Crossref - Ranjan KP, Ranjan N, Bansal SK, Arora DR. Prevalence of Pseudomonas aeruginosa in post-operative wound infection in a referral hospital in Haryana, India. J Lab Physicians. 2010;2(2):74-77.
Crossref - Siguan SS, Ang BS, Pala IM, Baclig RM. Aerobic Surgical Infection: surveillance on microbiological etiology and antimicrobial susceptibility pattern of commonly used antibiotics. Phil J Microbiol Infect Dis. 1990;19(1):27-33.
- Masaadeh HA, Jaran AS. Incident of Pseudomonas aeruginosa in post-operative wound infection. Am J Infect Dis. 2009;5(1):1-6.
Crossref - Oguntibeju OO, Nwobu RAU. Occurrence of Pseudomonas aeruginosa in post-operative wound infection. Pak J Med Sci. 2004;20(3):187-192.
- Mohanasoundaram KM. The antimicrobial resistance pattern in the clinical isolates of Pseudomonas aeruginosa in a tertairy care hospital; 2008-2010 (A 3 year study). J Clin Diagn Res. 2011;5(3):491-494.
- Arora D, Jindal N, Kumar R, Romit. Emerging antibiotic resistance in Pseudomonas aeruginosa. Int J Pharm Pharm Sci. 2011;3(2):82-84.
- Mastoraki A, Douka E, Kriaras I, Stravopodis G, Manoli H, Geroulanos S. Pseudomonas aeruginosa susceptible only to colistin in intensive care units. Surgical Infections. 2008;9(2)153-160.
Crossref - Tadvi J, Javadekar TB, Bhavsar R, Garala N. Prevalence and antibiogram of Pseudomonas aeruginosa at S.S.G. Hospital, Baroda, Gujarat, India. J Res Med Den Sci. 2015;3(3):204-207.
Crossref - Ramalakshmi K, Apparao P, Kamala P, Himabindu V. Study of colistin sensitivity pattern of Pseudomonas aeruginosa in a tertiary care hospital. International J Contemp Med Sci. 2019;6(5):E5-E8.
Crossref - Sambyal SS, Kaur A, Soodan PS, Mahajan B. Changing Antibiotic sensitivity pattern in Gram Negative Non fermenting Isolates: a Study in a Tertiary care Hospital. IOSR Journal of Dental and Medical Sciences. 2015;14(5):129-133.
- Ahmad S, Alotaibi M, Alamri MS. Antibiotic Sensitivity Pattern of Clinical Isolates of Pseudomonas aeruginosa at a Tertiary Care Hospital in Saudi Arabia. Dhaka University Journal of Pharmaceutical Sciences. 2020;19(1):77-82.
Crossref - Dou Y, Huan J, Guo F, Zhou Z, Shi Y. Pseudomonas aeruginosa prevalence, antibiotic resistance and antimicrobial use in Chinese burn wards from 2007 to 2014. J Int Med Res. 2017; 45:1124-1137.
Crossref
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.