ISSN: 0973-7510
E-ISSN: 2581-690X
This study was aimed to know the prevalence of biotypes, serotypes and phage types and the antibiotic susceptibility pattern of Vibrio cholerae isolates in our region.1,975 consecutive diarrheal stool samples were processed between 2009 and 2013. Standard microbiological methods and guidelines were followed to isolate, identify Vibrio cholerae and to perform antibiotic susceptibility test. Serotypes, biotypes and phage types of the isolates were determined. 106 Vibrio cholerae O1 strains were isolated from 1,975 stool samples (5.36%). All isolates were El Tor biotype. Predominant serotype was Ogawa (101/106, 95.2%). Phage types T2 (Basu & Mukerjee) and T27 (new scheme) were most common. High level of resistance was seen towards ampicillin and Co-Trimoxazole. Least resistance was towards chloramphenicol. No specific trend was shown against tetracycline. To conclude, V. cholerae O1 El Tor, biotype Ogawa was the most common serotype found in this area. Doxycycline still remains the drug of choice. Antibiotic susceptibility pattern of V. cholerae must be monitored by tertiary care centers.
Vibrio cholerae O1, cholera, diarrheal diseases, Serotyping
Cholera is a diarrheal disease caused by Vibrio cholerae. It is of high epidemiological significance because of its potential to cause epidemics having high mortality. Though maintenance of fluids and electrolyte balance is enough to manage most cases, antibiotics do play an important role in severely dehydrated patients.1,2 Antibiotic resistance among V. cholerae has been frequently reported.3–6 Injudicious use of antibiotics has led to antibiotic resistance in V. cholerae also. Since antibiogram of isolates keeps changing depending on, change in biotype and serovar, careful epidemiological monitoring of circulating strains is recommended.2,7,8 Therefore, this study was conducted with the aim of finding the biotype, serotype and antibiotic susceptibility pattern of V. cholerae isolates encountered in our area.
The study was conducted at the Department of Microbiology of a tertiary health care centre for 5 years between 2009 and 2013. A total of 1,975 consecutive stool samples from patients suffering from loose stools were processed. Specimens were inoculated on Blood agar, MacConkey’s agar, Xylose lysine deoxycholate (XLD) agar and Thiosulfate-citrate-bile salt-sucrose (TCBS) agar plates. The samples were also inoculated in alkaline peptone water and subcultures were made on MacConkey’s and TCBS plates after 8 hours of incubation. After overnight incubation the suspected colonies were identified by standard biochemical reactions.9 Antibiotic susceptibility was determined against Ampicillin (10 µg), Chloramphenicol (30 µg), Co-Trimoxazole (Trimethoprim/Sulphamethoxazole; 25 µg (1.25/23.75 µg), and Tetracycline (30 µg) according to Clinical Laboratory Standards Institute (CLSI) guidelines.10 Serotyping was done by using antisera from King’s Institute of Preventive Medicine, Guindy, Chennai. The isolates were sent to National Institute of Cholera and Enteric Diseases (NICED), Kolkata for confirmation and typing.
A total of 106 V. cholerae O1 strains were isolated from 1,975 stool samples with an isolation rate of 5.36% during the study period. The rate of isolation of V. cholerae ranged from 2.91% for the year 2011 to 6.92% for the year 2012 (Table 1, Figure 1). All the isolates belonged to El Tor biotype. Predominant serotype was Ogawa (101 of 106, 95.2%). Phage types T2 (Basu & Mukerjee) and T27 (new scheme) were the most common. Classical and O139 serotypes were not encountered during our study. 89.62% (95) and 83.01% (88) of isolates showed resistance towards Ampicillin and Co-Trimoxazole respectively. Only 3 (2.83%) isolates were resistant to Chloramphenicol. We observed that 9.43% (10) of our isolates were resistant to Tetracycline (Figure 2). No major variation in antibiotic susceptibility pattern was seen between biotypes of V. cholerae isolates.
Table (1):
Year-wise distribution of V. cholerae between 2009 and 2013
Year |
Stool samples Screened |
No. of isolates (%) |
|---|---|---|
2009 |
248 |
17 (6.85) |
2010 |
236 |
7 (2.96) |
2011 |
343 |
10 (2.91) |
2012 |
491 |
34 (6.92) |
2013 |
657 |
38 (5.57) |
Total |
1975 |
106 (5.36) |
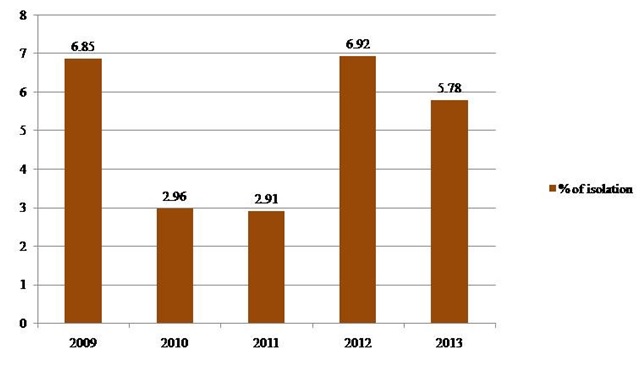
Fig. 1. Frequency of isolation of V. cholerae between 2009 and 2013
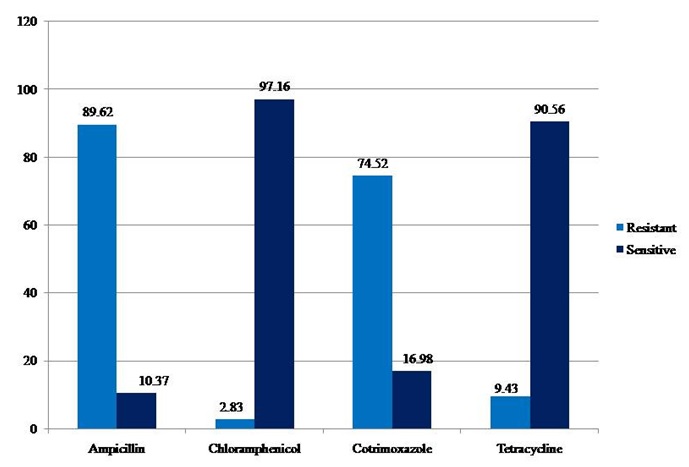
Fig. 2. Antibiogram of V. cholerae isolates during the study period (2009 – 2013)
More than 4 million people are affected by cholera and more than 1 lakh deaths occur yearly.2 India reported about 37,000 cases from 68 outbreaks between 1997 to 2006.11 The number of cases in reality are predicted to be six times higher than reported.11, 12 Large, diverse population, suboptimal water sanitation and warm climate are conducive for epidemics in India.11
Cholera is a self-limiting disease but can cause severe dehydration and death in a matter of hours.13 Rehydration is the mainstay of treatment. Antibiotics are indicated in cases of severe dehydration. Antibiotics reduce the total volume of stool passed, stopping diarrhea in 48 hours.1, 2 Hence antibiotics play a very important role in the treatment of cholera. However, emergence of drug resistance among V. cholerae has narrowed the choices of antibiotics. There has been sudden surge of multidrug resistance (MDR) in Vibrio in last 20 years. 3-5, 14-16 Besides, reports of biotype variation among V. cholerae from Inaba to Ogawa and vice versa are also known.13-15 Such variation is due to immune selection pressure in the local population. There have been reports of emergence of atypical variants of V. cholerae presenting with severe illness and MDR.4,5,16 Hence it is important to monitor biotypes and resistance pattern of the isolates.
In our study 106 V. cholerae O1 strains were isolated and all of them belonged to biotype El Tor, the predominant serotype was Ogawa. Phage typing revealed types T2 (Basu & Mukerjee) and T27 (new scheme) were most common. Workers from different parts of India during the same period also reported similar findings.7,18,19 Classical and O139 serotypes were not isolated at this centre indicating their absence in this geographical area during the study period. Over the years, frequency of Classical and O139 serotypes of V. cholerae has declined considerably throughout India.11 However a study conducted during 2000-2004 reported presence of non O1 and non O-139 strains.20
Various studies have tested a variety of antibiotics for V. cholerae including cephalosporins, fluoroquinolones and aminoglycosides. But CLSI recommends testing of only four drugs, i.e., Ampicillin, Co-Trimoxazole, Tetracycline and Chloramphenicol.10 The present study showed a high resistance to Ampicillin and Co-Trimoxazole throughout the study period. This may be linked to plasmid transfer from resistant colonic flora to V. cholerae in the gut environment.14,21 Variable resistance patterns was seen towards Tetracycline. A similar picture was observed, also among Enterobacteriaceae. Plasmid-transfer from gut flora to V. cholerae may be the reason for variable pattern of antibiotic resistance. We found that the variable resistance of our isolates to Tetracycline was not associated with the serotype and biotype of the isolate. Our isolates showed least resistance towards Chloramphenicol. However, Chloramphenicol has side effects when used in children, abridging its use as drug of choice.
Apart from plasmid transfer of antibiotic resistance, there are other reasons for emergence of resistance among V. cholerae. Extensive antibiotic usage in veterinary and poultry industry, in order to increase the diary and meat production, is one of the important reasons.22 Pressure exerted by the antibiotics lead to selection of resistant mutants. Studies have also shown that during an outbreak, if antibiotics are used for treatment, there is rapid development of resistance.23 Biotype switching is another cause of shift in drug resistance.7,24 Presence of a single biotype of V. cholerae in a geographical area leads to development of increased immunity levels in the community and in such situations, V. cholerae are known to exhibit a revolutionary strategy by changing the biotype from Ogawa to Inaba and vice versa. Such biotype variation is linked to variation in antimicrobial resistance in some studies.14,17
Since antibiotics play important role in limiting the disease severity, they should be wisely used. Clinical microbiology laboratories should adhere to the CLSI guidelines of testing only four antibiotics for V. cholerae.10 Testing and reporting sensitivity to antibiotics, not recommended by CLSI, would lead to their injudicious use adding to the menace of drug resistance. Many studies have also documented reversal of resistance with judicious use of antibiotics.7, 24
Clinical laboratories do not test antibiotic susceptibility of V. cholerae routinely. Therefore, unlike the other gram negative bacteria, only a little data on sensitivity pattern of this epidemiologically important pathogen exists.
The risk of cholera to community can be prevented by availability of good quality potable water and sanitation measures. Use of vaccines also is highly effective in halting the spread of cholera. Vaccination is an effective measure to prevent drug resistance in pathogens.25
Our study had a few limitations. Only those cases presenting to our hospital were included in this study and the data does not include isolates from cases treated at other government or private health care centers in this area. And also we could not perform the minimum inhibitory concentration (MIC) of Tetracycline for resistant isolates. Inclusion of most isolates from a region would provide a comprehensive perspective of the prevalence of a pathogen.
V. cholerae El Tor biotype Ogawa is the most common serotype found in this area. Doxycycline remains the drug of choice for therapy. Resistance to Ampicillin and Co-Trimoxazole is common and this study also showed the presence of Tetracycline resistant isolates. Regular monitoring of antibiotic susceptibility profile of epidemiologically relevant bacterial pathogens must be carried out by tertiary health care centers.
- World Health Organization, Department of Child and Adolescent Health and Development. The treatment of diarrhoea: a manual for physicians and other senior health workers. Geneva: Dept. of Child and Adolescent Health and Development, World Health Organization; 2005.
- WHO | Cholera [Internet]. WHO. [cited 2016 Jan 18]. Available from: http://www.who.int/mediacentre/factsheets/fs107/en/
- Wang R, Lou J, Liu J, Zhang L, Li J, Kan B. Antibiotic resistance of Vibrio cholerae O1 El Tor strains from the seventh pandemic in China, 1961–2010. Int J Antimicrob Agents. 2012; 40(4):361–4.
- Shrestha SD, Malla S, Adhikari BR, Shakya G, Basnyat SR, Sharma S. Antibiotic susceptibility patterns of Vibrio cholerae isolates. JNMA J Nepal Med Assoc. 2010; 49(179):232–6.
- Miwanda B, Moore S, Muyembe J-J, Nguefack-Tsague G, Kabangwa IK, Ndjakani DY, et al. Antimicrobial Drug Resistance of Vibrio cholerae , Democratic Republic of the Congo. Emerg Infect Dis. 2015; 21(5):847–51.
- Kar SK, Pal BB, Khuntia HK, Achary KG, Khuntia CP. Emergence and Spread of Tetracycline resistant Vibrio cholerae O1 El Tor variant during 2010 cholera epidemic in the tribal areas of Odisha, India. Int J Infect Dis. 2015; 33: 45–9.
- Das S, Choudhry S, Saha R, Ramachandran VG, Kaur K, Sarkar BL. Emergence of multiple drug resistance Vibrio cholerae O1 in East Delhi. J Infect Dev Ctries. 2011; 5(04):294–298.
- Safa A, Nair GB, Kong RYC. Evolution of new variants of Vibrio cholerae O1. Trends Microbiol. 2010; 18(1):46–54.
- Tille PM. Bailey & Scott’s Diagnostic Microbiology. Thirteenth edition. St. Louis, Missouri: Elsevier; 2014. pp 1038.
- CLSI (2007). Performance Standards for Antimicrobial Disk Susceptibility Tests; Approved Standard M2-A10. Wayne, PA: Clinical and Laboratory Standards Institute.
- Kanungo S, Sah B, Lopez A, Sung J, Paisley A, Sur D, et al. Cholera in India: an analysis of reports, 1997–2006. Bull World Health Organ. 2010; 88(3):185–91.
- Sarkar BL, Kanungo S, Nair GB. How endemic is cholera in India? Indian J Med Res. 2012; 135(2):246–8.
- Mahon CR, Lehman DC, Manuselis G Jr. Textbook of Diagnostic Microbiology. London: Elsevier Health Sciences;2014.
- Kitaoka M, Miyata ST, Unterweger D, Pukatzki S. Antibiotic resistance mechanisms of Vibrio cholerae. J Med Microbiol. 2011; 60 (4):397–407.
- Nair GB, Qadri F, Holmgren J, Svennerholm A-M, Safa A, Bhuiyan NA, et al. Cholera Due to Altered El Tor Strains of Vibrio cholerae O1 in Bangladesh. J Clin Microbiol. 2006; 44(11):4211–3.
- Marin MA, Thompson CC, Freitas FS, Fonseca EL, Aboderin AO, Zailani SB, et al. Cholera Outbreaks in Nigeria Are Associated with Multidrug Resistant Atypical El Tor and Non-O1/Non-O139 Vibrio cholerae. Vinetz JM, editor. PLoS Negl Trop Dis. 2013; 7(2):e2049.
- Garg P, Nandy RK, Chaudhury P, Chowdhury NR, De K, Ramamurthy T, et al. Emergence of Vibrio cholerae O1 biotype El Tor serotype Inaba from the prevailing O1 Ogawa serotype strains in India. J Clin Microbiol. 2000; 38(11):4249–53.
- Kulkarni S, Chillarge C. Antibiotic Susceptibility Pattern of Vibrio cholerae Causing Diarrohea Outbreaks in Bidar, North Karnataka, India. Int J Curr Microbiol App Sci. 2015; 4(9):957–961.
- Mandal J, Dinoop KP, Parija SC. Increasing Antimicrobial Resistance of Vibrio cholerae OI Biotype EI Tor Strains Isolated in a Tertiary-care Centre in India. J Health Popul Nutr. 2012; 30(1):12.
- Chandrasekhar MR, Krishna BVS, Patil AB. Changing characteristics of Vibrio cholerae: emergence of multidrug resistance and non-O1, non-O139 serogroups. Southeast Asian J Trop Med Public Health. 2008; 39(6):1092–7.
- Martinez JL. Antibiotics and Antibiotic Resistance Genes in Natural Environments. Science. 2008: 321(5887):365–7.
- Teuber M. Veterinary use and antibiotic resistance. Curr Opin Microbiol. 2001; 4(5):493–9.
- Mhalu FS, Mmari PW, Ijumba J. Rapid emergence of El Tor Vibrio cholerae resistant to antimicrobial agents during first six months of fourth cholera epidemic in Tanzania. Lancet. 1979; 1(8112):345–7.
- Faruque AS, Alam K, Malek MA, Khan MG, Ahmed S, Saha D, et al. Emergence of multidrug-resistant strain of Vibrio cholerae O1 in Bangladesh and reversal of their susceptibility to tetracycline after two years. J Health Popul Nutr. 2007; 25(2):241.
- Mishra RP, Oviedo-Orta E, Prachi P, Rappuoli R, Bagnoli F. Vaccines and antibiotic resistance. Curr Opin Microbiol. 2012; 15(5):596–602.
© The Author(s) 2017. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.


