ISSN: 0973-7510
E-ISSN: 2581-690X
Bacterial infections represent a very serious problem that threatens human health, antibiotics were designed to attack the causative agents of infectious diseases, but some bacterial pathogens became virulent and resistant to antibiotics by different mechanisms, resistance genes represented one of those mechanisms. This study attempts to screen the existence of five different resistance genes (mecA, TEM, FemA, MexD, and AmpC) among 25 bacterial isolates divided into two groups the first was non-clinical bacterial type strains including Bacillus subtilis, Escherichia coli, Klebsiella pneumonia, Pseudomonas aeruginosa, Salmonella typhi, and the other group includes some clinical bacterial isolates. Evaluation of their susceptibilities to different 12 antibiotic discs and attempting to find the relationship between genotype and phenotype assessment. Different responses were reported which varied from slightly susceptible to multidrug-resistant such as P. aeruginosa and K. pneumonia which could be considered as multidrug-resistant strains. Therefore, detection of resistance gene became crucial and critical to recognize the mechanism of resistance, five pairs of primers were included to investigate five responsible genes belonging to beta-lactamases, efflux pump, and methicillin resistance. Conclusively, the PCR technique is a very accurate tool to check the genetic resistance whether being expressed to phenotype or not. Moreover, the clinical bacterial isolates appeared more resistant that reflecting the impact of the surrounding environment on bacterial behavior.
Multidrug-resistant, mecA, femA, TEM, MexD, AmpC
The effects of pathogenic microbes extend to include several sectors such as human and animal health, agriculture, post harvested crops,1 and economics worldwide.2,3 Bacteria extensively affected public health causing infectious diseases for any part of the body thus it has a large impact on morbidity and mortality among human beings, bacteria are conveyed to human beings through the air, food, water, or other alive vectors.4 Bacterial pathogens mostly interact with humans via different thinking. Sometimes, many pathogens had been transmitted to humans directly from one individual to another, moreover, pathogens sometimes can originate from or transfer through the surrounding environment to infect the living host. Bacterial pathogens must have the ability to resist external environmental stresses outside their hosts, this resistance enables bacteria to reinfect other hosts. The external environment outside the living host is designated as the non-clinical environment.5 Persisters are typical phenotypic forms of bacteria that are resistant to antibiotics that are genetically indistinguishable from the other members of the bacterial population. The only difference is their physiological condition. The persistence arose in response to antibiotic-producing microorganisms.6 Antibiotic resistance has emerged from the overuse and misuse of antibiotics. Also, unpretentious infection control may lead to antibiotic resistance. Some precautions should be followed to minimize the phenomenon of antibacterial resistance through following the instructions of physicians, as well physicians should prescribe the suitable antibiotic after the recommendation of susceptibility
report.7 The problem of antimicrobial resistance has been recognized due to many reasons such as the misuse of the antimicrobial agents in addition to unavailability of newer drugs associated with crucial regulatory requirements and reduced financial inducements.8 Thus, there is a crucial need to synthesize new effective antibiotics (in particular those belonging to the natural origin) that improve human health to different degrees, and subsequently, the economic development is indirectly influenced in different levels.9
Lately, the bacterial RNAs have emerged as regulating keys of post-transcriptional regulators during the gene expression process, in addition, the bacterial RNAs were involved in several cellular processes, including bacterial virulence.10 MecA is a chromosomal gene on S. aureus that clarifies methicillin-resistant S. aureus (MRSA),11 while femA and femB genes encode proteins that affect the level of methicillin resistance of S. aureus.12 TEM is the major gene determinant of the resistance to amino-penicillin in E. coli, and the most common β-lactamase-encoded plasmid,13 TEM is frequently encoded by Gram-negative bacteria. As well, TEM encoded genes nearly represent 90% of the resistance against ampicillin in Gram-negative bacteria. The TEM-type ESBLs are mostly plasmid-mediated; that is resulting from the classic TEM genes mutations. Those mutations might be occurred by single or multiple amino acid substitution near the active site. The reason for calling TEM was because it had been firstly isolated from E. coli that recovered from a patient having a name; Temoneira. TEM-1 was firstly reported in 1965 in Athens, Greece.14 MexD acts as one of the efflux operons which encode an inner membrane RND transporter,15 one of the efflux pump genes affected Pseudomonas aeruginosa which is an important nosocomial pathogen of lower respiratory tract infections LRTI especially in ICUs and is often resistant to several antibiotics.16 AmpC β-lactamases are medically important cephalosporinases determinant on the chromosomes of many bacterial groups in particular; the Enterobacteriaceae.17
There is no available information referring the comparison between the genetic resistance pattern among the bacteria provided from the culture collection units and those isolated form the patients. Accordingly, the present study aimed to detect some resistant genes in different bacterial groups and attempt to find a relation between the existence of those genes and the phenotypic expression of the bacteria whether in clinical or non-clinical environments in addition to linking genotypic to phenotypic assessments to determine the exact antibiotic which reflect the presence of the interesting gene.
Bacterial specimens
Eleven type strains were provided by Al-Azhar center for fermentation biotechnology representing the non-clinical bacterial group that selected roughly, all data available in Table 1. Over and above that, 14 clinical bacterial isolates were recovered from infected patients; specimens were collected on a nutrient agar medium from different sites of patients’ bodies. These bacterial isolates were identified by VITEK® MS in parallel with the routine laboratory examination through investigation the bacterial isolates behavior on blood agar medium, and MacConkey agar medium as well as their microscopic characteristics by Gram’s stain.18,19 All clinical specimens were obtained by the microbiology laboratories in the intended hospitals.
Table (1):
List of provided type strains of bacteria with their ATCC codes.
Organisms |
ATCC code |
Gram reaction |
|---|---|---|
E. coli |
ATCC-8739 ATCC-25922 |
Negative |
Pseudomonas aeruginosa |
ATC C-9027 |
Negative |
K. pneumonia |
ATCC-13883 |
Negative |
Salmonella typhi |
ATCC-14028 ATCC 6539 |
Negative |
Proteus vulgaris |
ATCC-13315 |
Negative |
Staph. aureus |
ATCC-25923 ATCC-6538 |
Positive |
Bacillus cereus |
ATCC-14579 |
Positive |
Bacillus subtilis |
ATCC-6633 |
Positive |
Antimicrobial susceptibility of the clinical and non-clinical bacteria
As a phenotype marker assessment, both bacterial groups were investigated to determine their responses to different antibacterial discs using agar disc diffusion assay described by the Kirby-Bauer method,20 the disc diffusion assay was performed on Muller Hinton Agar (Hi Media India) using 12 different antibacterial discs (Bioanalyse); ciprofloxacin CIP (5 µg), ceftriaxone CRO (30 µg), clotrimazole CL (30 µg), levofloxacin LEV (5 µg), vancomycin VA (30 µg), rifampicin RF (30 µg), ceftriaxone CTX (30 µg), ceftazidime CAZ (30 µg), cefuroxime CXM (30 µg), amikacin AK (30 µg), amoxicillin/K. clavulanate AMC (20/10 µg), and ampicillin/sulbactam SAM (10/10 µg).
Genotypic detection of some resistant genes in the clinical and non-clinical bacteria
DNA of the investigated bacterial groups was extracted according to QIAGEN protocol, PCR was performed using 5 different pairs of primers listed in Table 2. according to QIAGEN PCR kit in a volume of 50 µl by (Gene Amp PCR System 2400) for 35 cycles after initial denaturation for 10 min at 95°C, the final extension was extended to 7 min at 72°C in the final stage 7.
Table (2):
List of primers used in the detection of genetic resistance to antibiotics.
| Target gene | Primer | Sequence 5′-3′ | Amplicon size | Annealing temperature | Reference |
|---|---|---|---|---|---|
| mecA | F | AAA ATC GAT GGT AAA GGT TGG C | 533 | 57 | (Kobayashi et al. 1994) |
| R | AGT TCT GCA GTA CCG GAT TTG C | ||||
| femA | F | AGA CAA ATA GGA GTA ATG AT | 509 | 57 | (Kobayashi et al. 1994) |
| R | AAA TCT AAC ACT GAG TGA TA | ||||
| TEM | F | TCA ACA TTT TCG TGT CGC CC | 766 | 56 | (Murugan et al. 2018) |
| R | AAC TAC GAT ACG GGA GGG CT | ||||
| AmpC | F | GAT GAA GGC CAA TGA CAT TCC G | 576 | 58 | (Murugan et al. 2018) |
| R | CAT GTC GCC GAC CTT GTA GTA A | ||||
| MexD | F | AGG TGA TCA ACG ACT TCA CCA A | 951 | 56 | (Murugan et al. 2018) |
| R | CAG CCA GAC GAA ACA GAT AGG T |
The final PCR products were visualized on the agarose gel stained with ethidium bromide to determine the presence of genes of interest through all investigated bacterial strains.
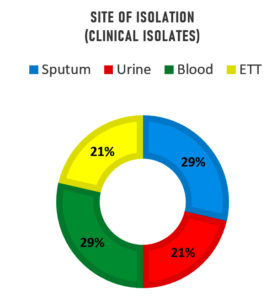
Different infected specimens were collected from different patients (see Table 3), this experiment led to the recovery of 14 bacterial strains which had been screened by Gram’s stain and fully identified by VITEK-MS (see Table 4). Fig. 1. displays those four bacterial isolates (29%) that were recovered from sputum which were Acinetobacter sp. (3 isolates) and E. coli (1 isolate). As well, four bacterial isolates (29%) were recovered from the blood which were Enterobacter sp. (1 isolate), E. coli (1 isolate), Enterococcus sp. (1 isolate), and S. aureus. Moreover, three bacterial isolates (21%) from urine were E. coli (1 isolate) and Enterococcus sp. (2 isolates). Finally, three bacterial isolates were recovered from endotracheal tube ETT (21%) which were 3 isolates of Enterobacter spp. This finding is in agreement with23 who reported that several bacteria isolates were detected in sputum specimens from asthmatic patients such as A. xylosoxidans, B. catarrhalis, Coliform, H. influenzae, K. oxytoca , P. aeruginosa, M. catarrhalis, methicillin-resistant S. aureus (MRSA), S. aureus, S. pneumoniae, and S. maltophilia. Also, bacterial growth was recovered in 47% of the TB patients Pseudomonas aeruginosa (9%), and Streptococcus spp. (9%) were the dominant bacteria.24 Regarding the bacterial isolates recovered from the blood samples; Streptococcus spp., Staphylococcus aureus, E. coli, and the Enterobacter sp. were detected as reported by.25 Referring to UTI patients, the most isolated Gram-negative strain is E. coli while, the most frequent Gram-positive strain was Enterococcus faecalis in addition to the presence of Bacillus cereus, Streptococcus gallolyticus, Streptococcus agalactiae.26 Acinetobacter baumannii, Enterobacter cloacae, Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, Serratia marcescens, Staphylococcus aureus, Stenotrophomonas maltophilia, and Group B Streptococcus were isolated from ETT.27
Table (3):
Susceptibility of the investigated bacterial strains to different antibacterial discs.
| Bacterial strain | Source | Mean Diameter of Inhibition Zone (mm) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CIP | CRO | CL | LEV | VA | RF | CTX | CAZ | CXM | AK | AMC | SAM | ||
| Non-clinical bacterial strains | |||||||||||||
| E. coli (ATCC-8739) |
ACFB | 25 | 25 | 10 | 30 | – | 9 | 30 | – | 25 | 12 | 15 | 20 |
| E. coli (ATCC-25922) |
ACFB | 25 | 20 | – | 26 | – | – | 20 | – | – | 17 | 13 | 15 |
| Pseudomonas aeruginosa (ATC C-9027) |
ACFB | 40 | – | – | 35 | – | – | 15 | – | – | 16 | – | – |
| K. pneumonia (ATCC-13883) |
ACFB | – | – | – | – | – | – | – | – | – | 12 | – | – |
| Salmonella typhi (ATCC-14028) |
ACFB | 27 | 20 | 15 | 26 | – | – | 25 | – | 22 | 15 | 23 | 23 |
| Salmonella typhi (ATCC 6539) |
ACFB | 28 | 25 | 14 | 26 | – | – | 20 | – | 15 | 15 | 13 | 15 |
| Proteus vulgaris (ATCC-13315) |
ACFB | 13 | – | 17 | – | – | 10 | 15 | 14 | – | 22 | 25 | 18 |
| Staph. aureus (ATCC-25923) |
ACFB | 21 | 9 | 9 | 24 | 15 | 17 | 12 | 16 | – | 15 | 18 | 21 |
| Staph. aureus (ATCC-6538) |
ACFB | 28 | 14 | 13 | 30 | 21 | 27 | 21 | 22 | – | 20 | 25 | 28 |
| Bacillus cereus (ATCC-14579) | ACFB | 23 | – | – | 25 | 15 | 15 | 12 | – | – | 20 | 13 | 12 |
| Bacillus subtilis (ATCC-6633) | ACFB | 32 | 28 | 30 | 32 | 25 | 20 | 38 | – | 38 | 24 | 36 | 30 |
| The clinical bacterial strains | |||||||||||||
| Acinetobacter sp. | Sputum | – | – | – | – | 15 | 13 | – | – | – | – | – | 9 |
| Acinetobacter sp. | Sputum | – | – | – | – | 12 | 13 | – | – | – | – | – | – |
| Acinetobacter sp. | Sputum | – | – | – | – | – | – | – | – | – | – | – | – |
| E. coli | Urine | 18 | – | – | 20 | – | – | 7 | – | – | 12 | – | – |
| E. coli | Blood | – | – | – | – | – | – | – | – | – | – | – | – |
| E. coli | Sputum | – | – | – | 7 | – | – | – | – | – | 12 | – | 8 |
| Enterobacter sp. | ETT | 23 | – | – | 21 | – | – | – | – | – | 13 | 11 | 9 |
| Enterobacter sp. | Blood | – | – | – | 7 | 7 | – | – | – | – | – | – | – |
| Enterobacter sp. | ETT | 25 | – | – | 25 | – | – | – | – | – | 11 | 16 | 14 |
| Enterobacter sp. | ETT | 20 | – | – | 20 | – | – | 8 | – | – | 10 | 17 | 14 |
| S. aureus | Blood | – | – | – | 8 | – | – | – | – | – | 15 | – | – |
| Enterococcus sp. | Urine | 18 | 12 | – | 20 | 18 | 12 | 20 | – | 19 | 8 | 24 | 17 |
| Enterococcus sp. | Blood | 22 | – | – | 23 | – | – | – | – | – | 13 | – | – |
| Enterococcus sp. | Urine | 13 | 9 | – | 23 | 11 | 15 | 19 | – | – | 11 | 20 | 18 |
The negative results were expressed as a hyphen (-) while the abbreviations of antibacterial discs were expressed as follow; CIP for ciprofloxacin (5 µg), CRO for ceftriaxone (30 µg), CL for clotrimazole (30 µg), LEV for levofloxacin (5 µg), VA for vancomycin (30 µg), RF for rifampicin (30 µg), CTX for ceftriaxone (30 µg), CAZ for ceftazidime (30 µg), CXM for cefuroxime (30 µg), AK for amikacin (30 µg), AMC for amoxicillin/K. clavulanate (20/10 µg), and SAM for ampicillin/sulbactam (10/10 µg). ACFB is the abbreviation for Al-Azhar center for fermentation biotechnology, the place that provided the non-clinical bacterial strains.
Table (4):
Screening of the resistance genes distributed in the clinical and non-clinical bacteria.
| Bacterial strain | Source | Gram reaction | Resistance genes | ||||
|---|---|---|---|---|---|---|---|
| mecA | femA | TEM | AmpC | MexD | |||
| Non-clinical bacterial strains | |||||||
| E. coli (ATCC-8739) | ACFB | Negative | – | – | – | – | + |
| E. coli (ATCC-25922) | ACFB | Negative | + | + | + | – | – |
| Pseudomonas aeruginosa (ATC C-9027) | ACFB | Negative | + | + | – | – | + |
| K. pneumonia (ATCC-13883) | ACFB | Negative | + | + | – | – | + |
| Salmonella typhi (ATCC-14028) | ACFB | Negative | + | + | + | – | – |
| Salmonella typhi (ATCC 6539) | ACFB | Negative | + | + | – | – | – |
| Proteus vulgaris (ATCC-13315) | ACFB | Negative | + | + | – | – | – |
| Staph. aureus (ATCC-25923) | ACFB | Positive | + | + | – | – | – |
| Staph. aureus (ATCC-6538) | ACFB | Positive | + | + | – | – | – |
| Bacillus cereus (ATCC-14579) | ACFB | Positive | + | + | + | – | – |
| Bacillus subtilis (ATCC-6633) | ACFB | Positive | + | + | – | – | – |
| The clinical bacterial strains | |||||||
| Acinetobacter sp. | Sputum | Negative | + | + | – | – | + |
| Acinetobacter sp. | Sputum | Negative | + | + | – | – | – |
| Acinetobacter sp. | Sputum | Negative | + | + | + | + | – |
| E. coli | Urine | Negative | + | + | + | – | – |
| E. coli | Blood | Negative | + | + | + | – | – |
| E. coli | Sputum | Negative | + | + | + | – | – |
| Enterobacter sp. | ETT | Negative | + | + | + | – | – |
| Enterobacter sp. | Blood | Negative | + | + | + | – | – |
| Enterobacter sp. | ETT | Negative | + | + | + | – | – |
| Enterobacter sp. | ETT | Negative | + | + | + | – | – |
| Coagulase-negative S. aureus | Blood | Positive | + | + | + | – | – |
| Enterococcus sp. | Urine | Positive | + | + | – | – | – |
| Enterococcus sp. | Blood | Positive | + | + | + | – | – |
| Enterococcus sp. | Urine | Positive | + | + | + | – | – |
| Total | 24 | 24 | 14 | 1 | 4 | ||
| % | 96 | 96 | 56 | 4 | 16 | ||
Fig. 1. A doughnut chart represents the percentage of the clinical bacterial isolates according to the site of isolation.
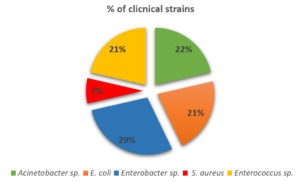
Depending upon Fig. 2.; the more dominant recovered strain is Enterobacter sp. (4 isolates represent 29%) while the rarest one is coagulase-negative S. aureus (1 isolate represents 7%). It was clearly noticed that Enterobacter spp. are important opportunistic human pathogens. 28,29 The prevalence of clinically isolated Enterococcus was reported in Europe, Asia, America and Latin America, and Africa.30 As well, Acinetobacter spp. represent a public health concern due to their antimicrobial resistance particularly in Africa and Western Pacific.31
Antimicrobial susceptibility of the clinical and non-clinical bacterial strains
Eleven type strains of non-clinical bacteria strains and 14 clinical bacterial isolates were investigated to determine their susceptibilities to 12 different antibiotic discs and the data resulted had been presented in Table 3. which clearly revealed that the clinical group is highly resistant, and the inhibition zones ranged from 9 to 40 mm in case of the non-clinical bacterial strains, while the inhibition zones ranged from 7-25 mm in case of the clinical bacterial isolates. The most susceptible bacterial strain among the non-clinical group is P. aeruginosa (ATC C-9027) towards ciprofloxacin followed by Bacillus subtilis (ATCC-6633) towards ceftriaxone and cefuroxime, while the most resistant strain is K. pneumonia (ATCC-13883) towards all tested antibacterials except weak susceptibility towards amikacin. On the other hand, the most susceptible bacterial strain among the clinical group is Enterobacter sp. (ETT) towards ciprofloxacin and levofloxacin followed by Enterococcus sp. towards ceftriaxone and cefuroxime, while the most resistant strain is E. coli (blood) followed by S. aureus (blood) towards all antibacterials included with weak activities of levofloxacin and amikacin. It was also observed that there is a noticeable difference in the same species whether clinical or non-clinical strain. This remark is in harmony with32 who reported that the susceptibility of the bacterial isolated to each respiratory quinolones varies, even within the same bacterial species, and that the expected efficacy also varies between the drugs. As well, majority of the bacterial isolates were resistant to almost all the examined antimicrobials. Particularly, Klebsiella pneumoniae, E. coli, and Pseudomonas aeruginosa exhibited highly resistant to the tested antibiotics with high rate of multiple antibiotic resistances.33
The prevalence of antimicrobial resistance among 60 isolates of Enterobacter spp. from clinical specimens of pets in Japan, also the resistance mechanisms were characterized, and the results indicate multidrug resistance for the investigated isolates and it was suggested to the presence of intra- and inter-hospital prevalence of E. cloacae clones co-docking ESBLs and PMQRs among companion animals.34
In contrary to the finding regarding susceptibility of non-clinical bacteria,35 found that Acinetobacter baumannii, Enterobacter cloacae, Escherichia coli, Klebsiella pneumoniae, and Pseudomonas aeruginosa were selected as Gram-negative nosocomial pathogenic bacteria from the Culture Collection of the Emerging Antibiotic Resistance Unit (University of Fribourg, Switzerland), all of them exhibited multidrug resistance for several antibiotics but octenidine dihydrochloride (OCT) is with unlimited efficacy against multidrug-resistant (MDR) Gram-negative bacterial pathogen within a short-term time. This contradiction might because presence/absence of extrachromosomal materials accepted/lost by accident during the storage period. This interpretation is in accordance with36 who investigated two genetically identified Lactiplantibacillus plantarum plus another type strain (ATCC-202195), and he reported that both isolated species are totally similar to each other and differ from the type strain due to their susceptibility to the most commonly prescribed antibiotics.
The negative results were expressed as a hyphen (-) while the abbreviations of antibacterial discs were expressed as follow; CIP for ciprofloxacin (5 µg), CRO for ceftriaxone (30 µg), CL for clotrimazole (30 µg), LEV for levofloxacin (5 µg), VA for vancomycin (30 µg), RF for rifampicin (30 µg), CTX for ceftriaxone (30 µg), CAZ for ceftazidime (30 µg), CXM for cefuroxime (30 µg), AK for amikacin (30 µg), AMC for amoxicillin/K. clavulanate (20/10 µg), and SAM for ampicillin/sulbactam (10/10 µg). ACFB is the abbreviation for Al-Azhar center for fermentation biotechnology, the place that provided the non-clinical bacterial strains.
Genotypic detection of multidrug resistance pattern of clinical and non-clinical bacterial strains
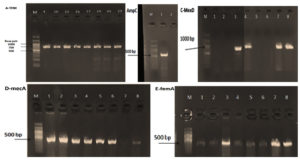
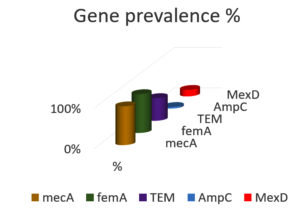
Five different genes responsible for bacterial resistance were determined by PCR (Fig. 3); mecA gene (533 bp) and femA gene (509) were detected in 24 out of 25 strains (96%), AmpC gene (576 bp) was only detected in one strain (4%), MexD gene (951 bp) was detected in 4 strains (16%), and finally, TEM gene (799 bp) was detected in 14 strains (56%) as reported in Table 4.
Fig. 3. Agarose gel of the investigated bacterial PCR products of the selected genes; A. TEM (799 bp), B. AmpC (576 bp), C. MexD (951 bp), D. mecA (533 bp), and E. femA (509). The bands that appeared on the agarose gel expressed only the positive results` so, the negative results are not represented. M is the DNA ladder (gene ruler 100 bp) purchased from QIAGEN.
It had been reported that PCR analysis showed that the mecA gene was present only in 43 (22.2%) of 194 S. aureus isolates.37 As well, it was reported that the mecA gene was determined in 60 (90.9%) out of 66 S. aureus isolates.7 Regarding the femA gene, it was reported that resistance property could be determined rapidly by a microfluidic device based on loop-mediated isothermal amplification (LAMP).38 MexD gene is widely detected in a large number of P. aeruginosa which demonstrated the presence of efflux pump overexpression, mandating for large multicentric studies.39
Regarding mecA and femA genes, there is no significant difference in their existence among both clinical and non-clinical bacterial strains. In the case of the TEM gene, the probability of the existence of this gene in the non-clinical isolates (3 out of 11) is less than in the clinical strains (11 out of 14). This finding is in a harmony with40 who reported that 41.6% (5/12) tested positive for the blaTEM gene among ESBL producer E. coli isolate.
Fig. 4. A column chart represents the percentage of the presence of the resistance genes in all tested bacterial strains.
Whilst MexD gene was detected in 3 out of 11 non-clinical bacterial strains and it had been detected once in the case of clinical isolates. Lastly, AmpC was detected once in the clinical bacterial isolates (Table 4 and Fig. 4). It was noticeably observed that each of mecA, femA, TEM, and AmpC is more dominant in the clinical bacterial isolates than the non-clinical bacterial strains as evidence for emerging the bacterial resistance, diverse reasons in the hospital contribute to the dissemination of the bacterial resistance counter to the wild strains which never face the same critical conditions in the hospitals. Unlikely, the MexD gene is more dominant in the wild bacterial strains than the clinical group, and this may due to the nature of the gene itself because it was commonly associated with P. aeruginosa clinical infection and this type of infection was not reported in our study. The mexD gene is the second least detected genes among the investigated antibiotics resistance genes ARGs, and this finding is in a harmony with 41 who stated that tetB, mefA and mexB were the three least abundant ARGs with the average abundance lower than 1.67 × 10-3 copies/16 S copies, otrA and mexD even were not detected in E2 effluent.
Our finding is very important because it demonstrated the reasons associated with hospitals such as arbitrary use of antibiotics, use of antiseptics and disinfectants, etc. which leads the bacteria to find a defense mechanism to protect itself. Also, it was markedly observed the emerging of bacterial resistance to several types of antibiotics which called multidrug resistance MDR. Both phenotypic and genotypic are successful tools to recognize bacterial resistance with remarkable superiority for the genotypic tool because it can predict the existence of bacterial resistance in the future.
ACKNOWLEDGMENTS
The authors thank the Director of Al-Azhar Center for Fermentation Biotechnology for providing the type strains of bacteria.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
ETHICS STATEMENT
Not applicable.
AVAILABILITY OF DATA
All datasets generated or analyzed during this study are included in the manuscript.
- Suleiman WB, El Bous M, Ibrahim M, El Baz H. In vitro evaluation of Syzygium aromaticum L. ethanol extract as biocontrol agent against postharvest tomato and potato diseases. Egyptian Journal of Botany. 2019;59(1):81-94.
- Geddes-McAlister J. Pathogenesis of Fungal and Bacterial Microbes. Pathogens. 2020;9(8):602.
Crossref - Gad A, Suleiman WB, Beltagy EA, El-Sheikh H, Ibrahim HA. Characterization and screening of marine-derived fungi along the coastline of Alexandria, Mediterranean Sea, Egypt. Egyptian Journal of Aquatic Biology & Fisheries. 2021;25(5):215-239.
Crossref - Doron S, Gorbach S. Bacterial Infections: Overview. International Encyclopedia of Public Health. 2008:273-282.
Crossref - Faucher SP, Charette SJ. Editorial on: Bacterial pathogens in the non-clinical environment. Frontiers in microbiology. 2015;6:331.
Crossref - Amato SM, Fazen CH, Henry TC, et al. The role of metabolism in bacterial persistence. Frontiers in microbiology. 2014;5:70.
Crossref - Suleiman WB. Detection of mecA gene in multidrug resistant Staphylococcus aureus isolated from Egyptian patients. Al-Azhar Bull Sci. 2017;9:357-63.
- Aslam B, Wang W, Arshad MI, et al. Antibiotic resistance: a rundown of a global crisis. Infection and drug resistance. 2018;11:1645.
Crossref - Suleiman WB. In vitro estimation of superfluid critical extracts of some plants for their antimicrobial potential, phytochemistry, and GC–MS analyses. Annals of Clinical Microbiology and Antimicrobials. 2020;19(1):1-12.
Crossref - Pita T, Feliciano JR, Leitão JH. Small noncoding regulatory RNAs from Pseudomonas aeruginosa and Burkholderia cepacia complex. International journal of molecular sciences. 2018;19(12):3759.
Crossref - Zuniga E, Benites NR, da Hora AS, et al. Expression of genes encoding resistance in Staphylococcus spp. isolated from bovine subclinical mastitis in Brazil. The Journal of Infection in Developing Countries. 2020;14(07):772-780.
Crossref - Maidhof H, Reinicke B, Blümel P, Berger-Bächi B, Labischinski H. femA, which encodes a factor essential for expression of methicillin resistance, affects glycine content of peptidoglycan in methicillin-resistant and methicillin-susceptible Staphylococcus aureus strains. Journal of bacteriology. 1991;173(11):3507-3513.
Crossref - Adamus-Białek W, Baraniak A, Wawszczak M, et al. The genetic background of antibiotic resistance among clinical uropathogenic Escherichia coli strains. Molecular biology reports. 2018;45(5):1055-1065.
Crossref - Rahman S, Ali T, Ali I, Khan NA, Han B, Gao J. The growing genetic and functional diversity of extended spectrum beta-lactamases. BioMed research international. 2018;2018.
- Depardieu F, Podglajen I, Leclercq R, Collatz E, Courvalin P. Modes and modulations of antibiotic resistance gene expression. Clinical microbiology reviews. 2007;20(1):79-114.
Crossref - Ozer B, Duran N, Onlen Y, Savas L. Efflux pump genes and antimicrobial resistance of Pseudomonas aeruginosa strains isolated from lower respiratory tract infections acquired in an intensive care unit. The Journal of antibiotics. 2012;65(1):9-13.
Crossref - Jacoby GA. AmpC β-lactamases. Clinical microbiology reviews. 2009;22(1):161-182.
Crossref - Abdel-Razek A, El-Sheikh H, Suleiman W, Taha TH, Mohamed M. Bioelimination of phenanthrene using degrading bacteria isolated from petroleum soil: safe approach. DESALINATION AND WATER TREATMENT. 2020;181:131-140.
Crossref - Kamel A, Suleiman WB, El-Fiky A, El-Sherbiny G, Elhaw M. Characterization of Bee Venom and Its Synergistic Effect Combating Antibiotic Resistance of Pseudomonas aeruginosa. Original article. Egyptian Journal of Chemistry. 2022;65.
- Hudzicki J. Kirby-Bauer disk diffusion susceptibility test protocol. 2009. https://asm.org/getattachment/2594ce26-bd44-47f6-8287-0657aa9185ad/Kirby-Bauer-Disk-Diffusion-Susceptibility-Test-Protocol-pdf.pdf
- Kobayashi N, Wu H, Kojima K, et al. Detection of mecA, femA, and femB genes in clinical strains of staphylococci using polymerase chain reaction. Epidemiology & Infection. 1994;113(2):259-266.
Crossref - Murugan N, Malathi J, Therese KL, Madhavan HN. Application of six multiplex PCR’s among 200 clinical isolates of Pseudomonas aeruginosa for the detection of 20 drug resistance encoding genes. The Kaohsiung Journal of Medical Sciences. 2018;34(2):79-88.
Crossref - Zhang Q, Illing R, Hui CK, et al. Bacteria in sputum of stable severe asthma and increased airway wall thickness. Respiratory research. 2012;13(1):35.
Crossref - Regmi RS, Khadka S, Sapkota S, et al. Bacterial etiology of sputum from tuberculosis suspected patients and antibiogram of the isolates. BMC research notes. 2020;13(1):1-6.
Crossref - Han Y-Y, Lin Y-C, Cheng W-C, et al. Rapid antibiotic susceptibility testing of bacteria from patients’ blood via assaying bacterial metabolic response with surface-enhanced Raman spectroscopy. Scientific reports. 2020;10(1):1-18.
Crossref - Folliero V, Caputo P, Della Rocca MT, et al. Prevalence and antimicrobial susceptibility patterns of bacterial pathogens in urinary tract infections in University Hospital of Campania “Luigi Vanvitelli” between 2017 and 2018. Antibiotics. 2020;9(5):215.
Crossref - Antoine J, Inglis GD, Way M, O’Rourke P, Davies MW. Bacterial colonisation of the endotracheal tube in ventilated very preterm neonates: A retrospective cohort study. Journal of Paediatrics and Child Health. 2020;56(10):1607-1612.
Crossref - Zhu X, Li P, Qian C, et al. Prevalence of Aminoglycoside Resistance Genes and Molecular Characterization of a Novel Gene, aac (3)-IIg, among Clinical Isolates of the Enterobacter cloacae Complex from a Chinese Teaching Hospital. Antimicrobial agents and chemotherapy. 2020;64(9):e00852-20.
Crossref - De Gheldre Y, Maes N, Rost F, et al. Molecular epidemiology of an outbreak of multidrug-resistant Enterobacter aerogenes infections and in vivo emergence of imipenem resistance. Journal of clinical microbiology. 1997;35(1):152-160.
Crossref - Toru M, Beyene G, Kassa T, Gizachew Z, Howe R, Yeshitila B. Prevalence and phenotypic characterization of Enterococcus species isolated from clinical samples of pediatric patients in Jimma University Specialized Hospital, south west Ethiopia. BMC research notes. 2018;11(1):281.
Crossref - Pormohammad A, Mehdinejadiani K, Gholizadeh P, et al. Global prevalence of colistin resistance in clinical isolates of Acinetobacter baumannii: A systematic review and meta-analysis. Microbial pathogenesis. 2020;139:103887.
Crossref - Kosaka T, Yamada Y, Kimura T, et al. Susceptibility of clinically-isolated bacteria strains to respiratory quinolones and evaluation of antimicrobial agent efficacy by Monte Carlo simulation. The Japanese journal of antibiotics. 2016;69(1):27-40.
- Manikandan C, Amsath A. Antibiotic susceptibility of bacterial strains isolated from wound infection patients in Pattukkottai, Tamilnadu, India. Int J Curr Microbiol App Sci. 2013;2(6):195-203.
- Harada K, Shimizu T, Mukai Y, et al. Phenotypic and molecular characterization of antimicrobial resistance in Enterobacter spp. isolates from companion animals in Japan. PLoS One. 2017;12(3):e0174178.
Crossref - Alvarez-Marin R, Aires-de-Sousa M, Nordmann P, Kieffer N, Poirel L. Antimicrobial activity of octenidine against multidrug-resistant Gram-negative pathogens. European Journal of Clinical Microbiology & Infectious Diseases. 2017;36(12):2379-2383.
Crossref - Pell LG, Horne RG, Huntley S, et al. Antimicrobial susceptibilities and comparative whole genome analysis of two isolates of the probiotic bacterium Lactiplantibacillus plantarum, strain ATCC 202195. Scientific reports. 2021;11(1):15893.
Crossref - Alli OT, Ogbolu D, Akorede E, Onemu O, Okanlawon B. Distribution of mecA gene amongst Staphylococcus aureus isolates from Southwestern Nigeria. African Journal of Biomedical Research. 2011;14(1):9-16.
- Meng X, Zhang G, Sun B, et al. Rapid Detection of mecA and femA Genes by Loop-Mediated Isothermal Amplification in a Microfluidic System for Discrimination of Different Staphylococcal Species and Prediction of Methicillin Resistance. Frontiers in microbiology. 2020;11;1487.
Crossref - Al Rashed N, Joji RM, Saeed NK, Bindayna KM. Detection of overexpression of efflux pump expression in fluoroquinolone-resistant Pseudomonas aeruginosa isolates. International Journal of Applied and Basic Medical Research. 2020;10(1):37-42.
Crossref - Sah RSP, Dhungel B, Yadav BK, et al. Detection of TEM and CTX-M Genes in Escherichia coli isolated from clinical specimens at tertiary care heart hospital, Kathmandu, Nepal. Diseases. 2021;9(1):15.
Crossref - Chen H-Y, Li X-K, Meng L, et al. The fate and behavior mechanism of antibiotic resistance genes and microbial communities in anaerobic reactors treating oxytetracycline manufacturing wastewater. Journal of Hazardous Materials. 2022;424 Part C:127352.
Crossref
© The Author(s) 2021. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.