ISSN: 0973-7510
E-ISSN: 2581-690X
https://dx.doi.org/10.22207/JPAM.13.2.35 | © The Author(s). 2019
Bacterial biofilms gained attention in the medical field with the emergence of antibiotic-resistant strains. The potency of lactic acid bacteria (LAB), isolated from kefir, against the development of a biofilm populated by the multidrug-resistant (MDR) Klebsiella pneumoniae was accessed in this study. The ability of LAB to inhibit biofilm formation was screened using the crystal violet method, and the morphology of the biofilms was analyzed using electron microscopy. Cell viability of K. pneumoniae after administration of LAB cell free supernatant was determined by MTT assay. Lactobacillus isolates were characterized using partial sequencing against 16S rRNA. Quorum sensing inhibition of K. pneumoniae by LAB was conducted with the help of the AHL reporter strain. Result showed that addition of cell free supernatant (CFS) as well as pellet of 40 isolates LAB on to K. pneumoniae culture could decrease their culture absorbance. Among those isolates, LAB G24 and G25 from grain kefir and LAB K10 and K15 from milk kefir demonstrated the strongest effect by reducing the absorbance by eight to nine-fold. Analysis of biofilm morphology showed that the CFS of Lactobacillus could prevent the attachment between K. pneumoniae cells. In addition, cell viability of K. pneumoniae was significantly reduced after the addition of the LABs CFS. Partial sequencing of 16S rRNA identified the LAB isolate as Lactobacillus helveticus. In conclusion the L. helveticus isolated from kefir demonstrated anti-biofilm activity against MDR K. pneumoniae. L. helveticus are potential probiotic strains for therapeutic application in the clearance of MDR K. pneumoniae.
Anti-biofilm activity, lactic acid bacteria, Kefir, Klebsiella pneumoniae, Lactobacillus helveticus.
Klebsiella pneumoniae is one of the main causes of infectious diseases such as urinary tract infections (UTI), pneumonia, intra-abdominal infections, blood flow infections (BSI), meningitis, and pyogenic liver abscess (PLA)1. Infection with K. pneumoniae is becoming increasingly complicated because of multidrug-resistant (MDR) K. pneumoniae, hence, raising a serious therapeutic challenge. Bacterial biofilms have been studied by clinicians in the context of the emergence of antibiotic-resistant strains2. K. pneumoniae is a gram-negative bacteria that has been extensively studied in biofilm production3. Compared to their planktonic forms, the adherent bacteria in biofilms are more recalcitrant to antimicrobial agents such as antibiotics, leading to new problems of antibiotic resistance and more aggressive clinical progression4.
Many bacteria utilize quorum sensing (QS) to produce biofilms and virulence factors during pathogenesis. QS is an intercellular communication system involving a number of auto-inducer (AI) proteins that act via a ligand-receptor mechanism. Acylated homoserine lactone (AHL) is an AI that plays a role in K. pneumoniae biofilm formation. When the levels of AHL increase above a certain threshold, it triggers the signal transduction pathway leading to biofilm formation5. Therefore, inhibition of the QS network is an attractive approach to control bacterial infection.
Biofilm induced resistance to antibiotics compels new measures for managing the infections caused by these biofilms6. Several studies have screened beneficial bacteria for the natural competitors of pathogenic bacteria in order to prevent colonization or biofilm formation by the latter. Lactobacilli have been extensively studied in this regard because of their remarkable ability to inhibit the growth of pathogenic bacteria by producing bactericidal compounds such as lactic acid as a metabolic by-product, which also includes hydrogen peroxide, diacetyl, and bacteriocin7,8. Lactobacillus acidophilus and Lactobacillus fermentum produce antimicrobial compounds that not only inhibit bacterial growth but also inhibit biofilm formation of K. pneumoniae9. In addition, whole cell cultures and supernatants isolated from milk fermentation products of L. fermentum showed inhibitory activity on K. pneumoniae biofilm formation10. However, only limited studies have been performed to assess the potential of Lactobacillus and its compounds as natural antimicrobial agents being used to overcome the problem of antibiotic resistance in K. pneumoniae infections.
The aim of the present study was to assess the ability of lactic acid bacteria (LAB) isolated from goat milk kefir and kefir grains to inhibit biofilm formation of the MDR K. pneumoniae. The Klebsiella strain was isolated from a patient with severe pneumonia and was used to test the effect of LAB supernatant and pellet on biofilm development on the isolated bacteria.
Bacterial strains
The bacteria Klebsiella pneumoniae was provided by the Department of Microbiology, Faculty of Medicine, Brawijaya University. They were isolated from a patient with severe pneumonia, and susceptibility test against several antibiotics using Vitek@2Compact (BioMיrieux, France) indicated that the K. pneumoniae was resistant to ciprofloxacin and trimethoprim/sulfame-thoxazole, cefazolin, ceftazidime, ceftriaxone, cefepime, and aztreonam. Lactobacillus strains were isolated from goat milk kefir that was purchased from a small dairy in Malang, Indonesia. One gram of kefir grains as well as kefir was homogenized in 9 mL sterile H2O, and serially diluted from 10–1 to 10–4. Of each dilution, 100µL was spread on de Man Rogosa and Sharpe (MRS) agar (Merck Millipore) and anaerobically incubated at 37°C for 48 h11. The colonies were counted, and each colony was streaked on MRS medium. Individual colonies were inoculated into milk yeast extract (MYE) broth (10% skim milk, 15% glucose, 0.5% yeast extract; HiMedia) with 5% glycerol, and the stocks were stored at –20°C.
Biofilm assay
Lactic acid bacteria that have been successfully grown from whole kefir as well as kefir grain were sub-cultured in 15 ml tubes containing 5 ml of MRS broth and incubated for 16 hours at 37°C. After incubation, LAB cultures were centrifuged at 12,000 rpm for 15 min, and the cell free supernatants (CFS) and pellets were separated. K. pneumoniae isolates were sub-cultured in Luria Bertani (LB) broth and incubated at 37°C for 18 h. Next day, after ascertaining confluency at OD600 40.5, 200µL of the culture was dispensed per well in a 48-well plate (Corning-Costar®). This was followed by the addition of 200µL LAB CFS or pellet (diluted with MRS medium), or MRS medium as the negative control. The plates were incubated for 6 h at 37°C11.
Biofilm inhibition was tested using crystal violet11. The cultures were discarded, and the plates were washed thrice by submerging in water. To each well, 125µL of 0.1% crystal violet solution (Bioworld®) was added and incubated for 15 min at room temperature (RT). The staining solution was discarded, and the plate was washed thrice with sterile water to remove the excess stain. The resulting biofilms were diluted with 125µL 30% acetic acid per well, and after incubation for 15 min at RT, the OD of the supernatant was measured at 570nm using ELISA reader (iMark™ Microplate Absorbance Reader, BioRad). Biofilm formation (and inhibition) was quantified after removing the planktonic cells on the basis of OD570 values: the isolates with OD570 < 0.120 were considered as weak biofilm producers, those with OD570 0.120–0.240 had biofilm formation ability, and isolates with OD570 > 0.240 were classified as potent biofilm producers12. The supernatants and pellets from two LAB isolates resulted in the lowest OD570 values, indicating best inhibitory effects, and were chosen for further analysis. Each sample was analyzed in triplicate.
MTT assay of Lactobacillus cell-free supernatant against K. pneumoniae
The four LAB isolates with the lowest OD i.e. K10 and K15 (from milk kefir) and G24 and G25 (from kefir grain) were each sub-cultured in 1 mL MRS broth and incubated anaerobically for 16 h at 37°C. The cultures were centrifuged at 12,000 rpm for 15 min and the CFS and pellets were separated. The K. pneumoniae were sub-cultured in LB medium at 37°C for 18 h. After reaching confluence (OD600 0.5), 100µL of each culture was dispensed per well in a 96-well plate (Corning-Costar®), followed by addition of 100µL LAB CFS or pellet, or MRS medium (negative control). The co-cultures were incubated at 37°C for 6 h.
The antibacterial activity of LAB CFS on K. pneumoniae was determined using the MTT assay13. Briefly, 5µL MTT solution (Cell Quanti-MTTTM Cell Viability Assay Kit) was added to each well followed by 1 h incubation at 37°C away from light. The cultures were discarded, and the wells were washed with 250µL PBS each. To each well, 100µL dimethyl sulfoxide (DMSO; Merck) was added and incubated for 10 min at 37°C away from light followed by 10 min agitation. The OD of the supernatants was measured at 570 nm using an ELISA reader. The procedure was repeated three times for each sample. OD570 values were converted to percentage of cell viability (% viability) relative to the control group by using the following equation:
% activity = (Abssample/Abscontrol) x 100,
where Abs sample refers to sample treated with LAB CFS, and Abscontrol refers to the untreated control. High OD570 indicated high cell viability14.
Microscopic determination of biofilm inhibition using scanning electron microscopy (SEM)
Biofilm formation on the surface of plastic coverslips (diameter 13 mm) (Thermanox®) was monitored as per the manufacturer’s instructions. Briefly, a coverslip was placed inside each well of a 24-well plate containing 200µL K. pneumoniae in LB broth and 200µL LAB CFS or pellet from soluble MRS and incubated for 6 h. The culture media was removed, and the wells were washed several times with water. The coverslips were air dried and placed onto larger (22 x 22 mm) glass coverslips for imaging using FEI Verios 460 scanning electron microscope (FEI, Oregon, USA).
Identification of Lactobacillus
The LAB isolates were first identified based on colony morphology, physiology, and biochemical reactions as per the criteria of Bergey’s Manual of Systematic Bacteriology. Lactobacillus was characterized by cell morphology, Gram reaction, catalase and oxidase activity, sporulation, and cell motility. The final 4 LAB isolates with potent anti-biofilm activity were identified to the species level by using 16S ribosomal RNA gene sequencing. Total genomic DNA was isolated from 5 mL overnight cultures grown in MRS broth at 37°C using spin column method according to manufacturer’s instructions (PureLink® Genomic DNA, Life Technologies Corp., Carlsbad, USA). Targeted genes of each isolate were amplified using Lac16f (5′ -AGAGGTTTGATCCTGGCTCAG-3′) and Lac16r (5′ -CTACGGCTACCTTGTTACGA-3′) primers15. A 50µL PCR reaction mix was prepared for each sample containing 25µL 2X AmpMaster®Taq (GeneAll Biotechnology, Songpa-gu, Korea), 100 ng template DNA, and 10 pmol/µL of each primer. The PCR conditions were as follows: initial denaturation at 95°C for 2 min, followed by 35 cycles of 2 min denaturation at 95°C, 20s annealing at 49°C and 1 min extension at 72°C, and a final extension at 72°C for 5 min. The PCR products were visualized with DNA gel electrophoresis Mini-Sub® Cell GT Systems (Bio-Rad Laboratories, Singapore), and DNA fragments corresponding to the correct length of 16S rRNA were purified using Gel/PCR DNA Fragments Extraction Kit® (Geneaid Biotech, Taiwan). The purified products were sent for sequencing to Macrogen Corporation (Seoul, South Korea). The nucleotide sequences of the 16S rRNA gene were reconstructed using Unipro UGENE software version 1.24.0, and the identities of the isolates were determined on the basis of the highest BLAST score (http://www.ncbi.nlm.nih.gov.blast) against the GenBank DNA database (www.ncbi.nlm. nih.gov/Genbank/).
QS assay via inactivation of AHL by LAB
To determine whether the inhibitory effect of LAB on K. pneumoniae biofilm formation was related to the inactivation of AHL-mediated QS, AHL inactivation was qualitatively assessed with the help of a biosensor agent, Agrobacterium tumefaciens NT1 (tra::lacZ749) (ATCC®)16 (kindly provided by Dr. Ating, Faculty of Fishery, UB). The reporter strain was grown in LB medium supplemented with 25µg/mL gentamicin at 28°C until it reached an OD600 1.5. LAB isolates were inoculated in MRS broth medium and incubated overnight at 28°C under microaerophilic conditions. The cultures were centrifuged for 5 min at 13000 rpm, and then 50µL supernatant was dispensed into each well of a 96-well plate containing 50µL of 10µM 3OC8HSL per well (Sigma-Aldrich). The plates were incubated at 28°C for 5 h.
Minimal agar medium plates supplemented with 50µg/mL X-gal were prepared, and the agar was cut into 6 separate bars 1-cm wide using a sterile scalpel by removing 2-3mm strips between the bars. Starting from one end of each bar, 5µL of the LAB CFS and 3OC8HSL mix were spotted along its length, followed by 0.5µL drops of the A. tumefaciens culture; MRS broth medium and 3OC8HSL without LAB served as negative control. The plates were tightly sealed using plastic wrap and incubated for 24 h at 28°C. The biosensor spots turned to blue in the presence of detectable AHL signal.
Statistical analysis
One-way ANOVA was used to evaluate the effect of the CFS and pellet of LAB on in vitro biofilm inhibition test and viability test (optical density). Following ANOVAs, Tukey’s test was used for multiple pairwise comparisons among treatment levels. Significance level was set at P<0.05 for all comparisons. All statistical analyses were performed using SPSS version 16.
Profile of MDR K. pneumoniae isolated from a patient with pneumonia
The microorganism isolated from a patient with pneumonia was proven to be K. pneumoniae with 99% certainty. Antibiotic susceptibility test showed that the isolate was resistant to at least six antibiotics (Table 1).
Table (1):
Identification and antimicrobial susceptibility of Klebsiella pneumonia isolated from patient with pneumonia
| Antimicrobial | MIC | Interpretation | Antimicrobial | MIC | Interpretation |
|---|---|---|---|---|---|
| ESBL | POS | + | Ertapenem | ≤0.5 | S |
| Ampicillin | ≥32 | R | Meropenem | ≤0.25 | S |
| Ampicillin/Sulbactam | ≥32 | R | Amikacin | ≤2 | S |
| Piperacillin/ Tazobactam | 8 | S | Gentamicin | ≤1 | S |
| Cefazolin | ≥64 | R | Ciprofloxacin | ≥4 | R |
| Ceftazidime | 16 | R | Tigecycline | 1 | S |
| Ceftriaxone | ≥64 | R | Nitrofurantoin | 64 | S |
| Cefepime | ≥64 | R | Trimethoprim/
Sulfametoxazole |
≤20 | R |
| Aztreonam | 32 | R | |||
| Selected Organism | 99% Probability | Klebsiella pneumoniae | |||
R= resistant, S= sensitive
Screening of Lactobacillus from kefir with anti-biofilm activity against K. pneumoniae
Fifteen and 15 isolates were successfully isolated from whole kefir and from kefir grains, respectfully. The supernatants (CFS), as well as the pellets of all LAB isolates, were tested for their ability to inhibit the development of K. pneumoniae biofilms. LAB isolated from whole kefir and kefir grains showed anti-biofilm activity as determined by absorbance after crystal violet staining. K. pneumoniae cultured without LAB showed a high ability to produce a strong biofilm. Upon incubation of K. pneumoniae with LAB CFSs and pellet, two isolates (K10 and K15), out of the 15 LAB isolates from goat milk kefir, showed significant absorbance reduction, up to 70% and 80%, respectfully, indicating biofilm disruption. Similar results were obtained when K. pneumoniae was incubated with CFSs and pellets of kefir grain. Out of 25 LAB isolates, only two isolates (G24 and G25) demonstrated substantial absorbance reduction, up to 90% (Table 2). These four isolates were selected for further assays. Taken together, LAB decreased K. pneumoniae biofilm formation.
Table (2):
Measurement of OD of K. pneumonia biofilm after incubation with Lactic Acid Bacteria (p<0.001).
| Vol of Lactobacillus sp. | Average OD570 + SD | |||
|---|---|---|---|---|
| Isolate Number | Amount (µL) | K. pneumoniae + LAB CFS | K. pneumoniae + LAB Pellet | |
| Control | 200 | 0.894±0.036 | 0.894±0.036 | |
| Kefir Grain | G1 | 200 | 0.161±0.084 | 0.333±0.247 |
| G2 | 200 | 0.144±0.025 | 0.150±0.078 | |
| G3 | 200 | 0.126±0.021 | 0.307±0.019 | |
| G4 | 200 | 0.134±0.010 | 0.540±0.138 | |
| G5 | 200 | 0.133±0.080 | 0.178±0.013 | |
| G6 | 200 | 0.160±0.019 | 0.191±0.014 | |
| G7 | 200 | 0.237±0.026 | 0.264±0.031 | |
| G8 | 200 | 0.259±0.008 | 0.254±0.042 | |
| G9 | 200 | 0.205±0.019 | 0.391±0.059 | |
| G10 | 200 | 0.185±0.024 | 0.189±0.005 | |
| G11 | 200 | 0.252±0.034 | 0.317±0.051 | |
| G12 | 200 | 0.231±0.076 | 0.224±0.055 | |
| G13 | 200 | 0.179±0.029 | 0.241±0.071 | |
| G14 | 200 | 0.213±0.037 | 0.263±0.031 | |
| G15 | 200 | 0.291±0.040 | 0.226±0.073 | |
| G16 | 200 | 0.258±0.111 | 0.166±0.024 | |
| G17 | 200 | 0.243±0.039 | 0.199±0.049 | |
| G18 | 200 | 0.231±0.037 | 0.174±0.027 | |
| G19 | 200 | 0.252±0.036 | 0.208±0.062 | |
| G20 | 200 | 0.177±0.125 | 0.210±0.046 | |
| G21 | 200 | 0.249±0.028 | 0.252±0.018 | |
| G22 | 200 | 0.239±0.025 | 0.249±0.045 | |
| G23 | 200 | 0.099±0.016 | 0.098±0.012 | |
| G24 | 200 | 0.084 ±0.005* | 0.092±0.012 | |
| G25 | 200 | 0.089±0.006 | 0.087±0.016* | |
| Whole Kefir | K1 | 200 | 0.151±0.017 | 0.131±0.011 |
| K2 | 200 | 0.159±0.014 | 0.155±0.026 | |
| K3 | 200 | 0.140±0.030 | 0.147±0.018 | |
| K4 | 200 | 0.148±0.012 | 0.147±0.015 | |
| K5 | 200 | 0.139±0.009 | 0.135±0.009 | |
| K6 | 200 | 0.141±0.016 | 0.123±0.014 | |
| K7 | 200 | 0.134±0.027 | 0.140±0.016 | |
| K8 | 200 | 0.135±0.027 | 0.139±0.018 | |
| K9 | 200 | 0.140±0.009 | 0.136±0.011 | |
| K10 | 200 | 0.124±0.011* | 0.148±0.040 | |
| K11 | 200 | 0.251±0.083 | 0.243±0.051 | |
| K12 | 200 | 0.283±0.015 | 0.263±0.024 | |
| K13 | 200 | 0.417±0.183 | 0.211±0.036 | |
| K14 | 200 | 0.347±0.219 | 0.168±0.029 | |
| K15 | 200 | 0.133±0.054 | 0.097±0.008* | |
*Lowest OD570 value
Species Identification
Physiological and biochemical tests showed that all the above isolates were Gram-positive, non-motile, catalase negative and oxidase negative (Table 3). The isolates were identified to the species levels by partial 16S rRNA sequencing. The amplified region was identified by alignment with known genes using BLASTn (Table 4). All four isolates were identified as Lactobacillus helveticus.
Table (3):
Characterization of Lactic Acid Bacteria (LAB) isolated from kefir grain and goat milk kefir
| Characteristics | Isolate Number | |||
|---|---|---|---|---|
| K10 | K15 | G24 | G25 | |
| Cell Morphology | ||||
| Gram | Pos | Pos | Pos | Pos |
| Cell shape | Bacillus | Bacillus | Bacillus | Bacillus |
| Colony Morphology | ||||
| Shape | Circular | Circular | Circular | Circular |
| Size | 1.23 mm | 1.71 mm | 2.03 mm | 1.78 mm |
| Margin | Entire | Entire | Entire | Entire |
| Elevation | Convex | Convex | Convex | Convex |
| Consistency | Dry | Dry | Dry | Dry |
| Cell Physiology | ||||
| Motility | Neg | Neg | Neg | Neg |
| Sporulation | Neg | Neg | Neg | Neg |
| Biochemistry | ||||
| Oxidase | Neg | Neg | Neg | Neg |
| Catalase | Neg | Neg | Neg | Neg |
Table (4):
Identification of LAB isolate based on homology (%) of DNA fragment sequence region 16S rRNA.
Isolate Number |
Type |
Species |
GenBank® Accession Number |
% Identity |
|---|---|---|---|---|
G24 |
Grain kefir |
Lactobacillus helveticus strain RU27-3 |
KC836674.1 |
97% |
G25 |
Grain kefir |
Lactobacillus helveticus strain TW2-2 |
KJ026564.1 |
97% |
K10 |
Whole Kefir |
Lactobacillus helveticus strain C2 |
KJ702501.1 |
95% |
K15 |
Whole Kefir |
Lactobacillus helveticus strain AJT |
JQ775393.1 |
97% |
MTT Assay
The LAB supernatant isolated from kefir grains demonstrated a stronger inhibition activity by reducing the absorbance by almost 75%, followed by the LAB pellet. In contrast, the LAB supernatant, as well as pellet isolated from whole kefir, could only reduce the cell viability by 50% (Table 5).
Table (5):
Measurement of OD of K pneumoniae cell viability after incubation with Lactic acid bacteria (p<0.001).
Isolate Number |
Amount (µL)
of CFS |
Cell viability (%) |
|---|---|---|
K. pneumoniae |
100 |
100.00 |
Kpn + G24 |
100 |
29.07 |
Kpn + G25 |
100 |
37.22 |
Kpn + K10 |
100 |
44.25 |
Kpn + K15 |
100 |
43.14 |
Kpn = Klebsiella pneumoniae
The inhibition of metabolic activity of 4 isolates against K. pneumoniae are presented in Table 5. MTT assay showed that LAB CFS from kefir grain possesses inhibitory activity that is stronger than the activity of isolates from kefir milk. This is shown by the reduction of growth activity of K. pneumonia upon being challenged with Lactobacillus spp. G24, G25, K10 and K15 by 30%, 37%, 44%, and 43%, respectively.
Analysis of K. pneumoniae biofilm inhibition by L. helveticus using SEM
Analysis of biofilm formation under SEM showed thick biofilms with a compact arrangement of interconnected K. pneumoniae cells that served as the positive control (Fig. 1A). In contrast, when K. pneumoniae was challenged with L. helveticus CFS (Fig. 1B and 1D) and pellet (Fig. 1C and 1E), the cells were not able to aggregate. The phenomenon of biofilm formation and LAB-mediated disruption was clear on coverslips (Fig. 1D and 1E).
AHL inactivation by LAB
We tested the K10, K15, G24 and G25 isolates for their ability to inhibit AHL using the biosensor agent A. tumefaciens NT1 (tra::lacZ749). The biosensor grew well on the medium that changed its color to blue indicating the production of b-galactosidase in NT1 took place normally. Hence reflecting the absence of any compound inhibiting the exogenously added AHL therefore QS was not disturbed. (Fig. 2).

Fig. 1. Structure of K. pneumoniae biofilms observed by scanning electron microscopy (SEM): K. pneumoniae (A), K. pneumoniae exposed with: kefir supernatant (B), kefir pellet (C), grain kefir supernatant (D) and grain kefir pellet (E). Magnification: 10.000x (first row) and 20.000x (second row)
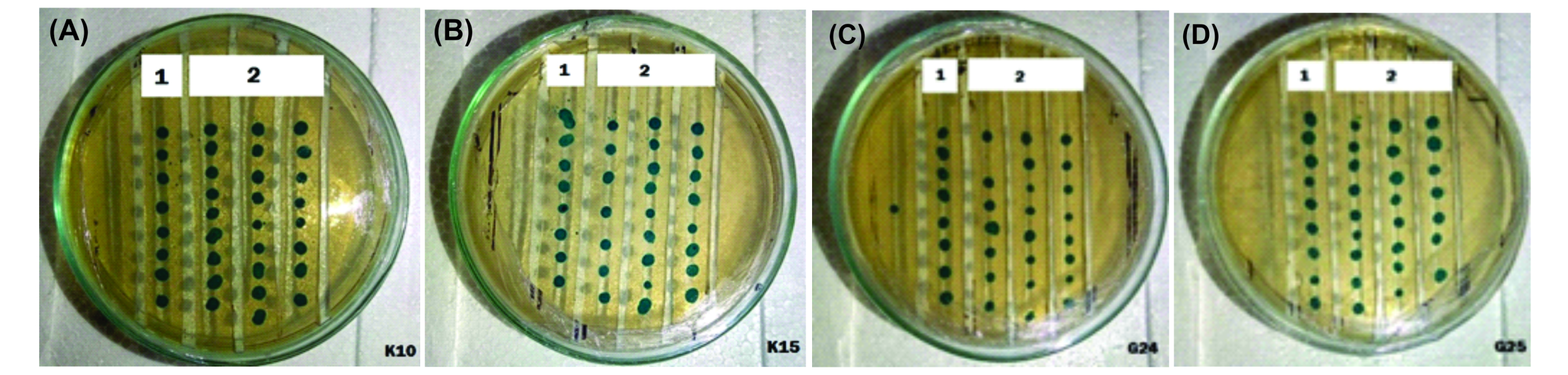
Fig. 2. AHL inactivation test using biosensor agent Agrobacterium tumefaciens NT1 (tra::lacZ749) towards Lactic Acid Bacteria isolates from kefir and kefir grain : K10 (A), K15 (B) G24 (C), and G25 (D),. There is no colour change in A. tumefaciens colonies supplemented with LAB (1) as well as without LAB (2).
Infections by several pathogens are complicated owing to the formation of a biofilm17. Biofilms render the bacteria resistant to antibiotics and serve as a barrier upon clearance by the host immune defense mechanisms. In the last decade, microbiologists have been evaluating bacteria isolated from probiotics for their anti-biofilm activity to control infections due to antibiotic-resistant pathogens18. In the present study, we assessed the ability of LAB isolated from kefir to inhibit biofilm formation of MDR K. pneumoniae.
Screening of LAB showed potent anti-biofilm activity against MDR K. pneumoniae. In general, LAB isolated from kefir grains possessed a stronger anti-biofilm capacity compared to that isolated from milk kefir. Among the 40 isolates tested, four isolates showed the strongest anti-biofilm activity—K10 and K24 from kefir milk, and G24 and G25 from kefir grain. G24 exhibited the strongest anti-biofilm activity (90%). Furthermore, both CFS and pellets of LAB isolated from the kefir grains demonstrated comparable anti-biofilm capacity. This could be due to the presence of anti-biofilm compounds and functional antimicrobial metabolites in CFS such as bacteriocins, diacetyl, and acetoin that were secreted by the LAB6,7. The pellet of LAB inhibits biofilm development through the synthesis of capsular materials or protective polymeric substances surrounding the cell19.
In addition to biofilm inhibition, LAB CFS also demonstrated antimicrobial activity, which was shown by the reduced viability of K. pneumoniae cells. The Lactobacillus strain of goat milk origin significantly suppressed the metabolic activity of several pathogens, such as Pseudomonas aeruginosa and Staphylococcus aureus20. Apparently, probiotics exert beneficial effects through synthesis of organic acids (acetic and lactic acids), bioactive peptides, peroxides, exopolysaccharides and other metabolites21.
Characterization of the four LAB isolates with the highest anti-biofilm activity using biochemical, microbiological, and genetic analyses indicated that the LAB isolates belonged to L. helveticus, which likely secreted bacteriocin into the cell-free supernatants. L. helveticus seems to impair biofilm formation of K. pneumoniae by inhibiting cellular aggregation, since the administration of the L. helveticus CFS onto K. pneumoniae cultures lead to dispersion of the aggregates, and the cells failed to aggregate further. In addition, biofilm development was inhibited only during bacterial cell attachment to each other. Interfering with the attachment of cells to one another and to the plate is one of the mechanisms of early inhibition of biofilm development22. Our hypothesis was supported by SEM analysis, which showed that LAB loosened K. pneumoniae aggregation, resulting in the pellicle form. Failure of bacterial cells to attach will halt the maturation process, and consequently, matrix synthesis. In addition to inhibiting bacterial aggregation, biofilm formation can also be blocked by degrading biofilm polysaccharides or inhibiting the biofilm developmental process23. However, these possibilities were not tested in this study.
The antimicrobial activity of Lactobacillus can generally be classified into three mechanisms—the secretion of organic acids (acetic acid and lactic acid), antimicrobial products, e.g., bacteriocin accumulation and secretion in LAB culture supernatants, and QS24. Quorum sensing plays an important role in regulating the initiation of biofilm formation in some pathogenic bacteria25. We hypothesized that K. pneumoniae biofilm formation was inhibited by LAB through repression of AHL expression, a QS inducer type 1. However, the biosensor agent that we used in this study did not show any repression of AHL molecules by L. helveticus. Other studies have shown that Lactobacillus paracasei produced soluble molecules that accumulated in the supernatant and inhibited QS gene expression26. The discrepancy could be due to the different techniques that were used to measure the effect of LAB to QS. In the above study, QS gene expression was measured using real-time- PCR, which yields a much more accurate result than that by using a biosensor.
The present study indicates that Lactobacillus helveticus isolates G24 and G25 can be considered as potential probiotic strains owing to their anti-biofilm formation capacity as well as growth inhibition against multidrug-resistant Klebsiella pneumoniae.
Acknowledgements
We would like to express our heartfelt thanks to DR. Ating Yuniarti, Brawijaya University for providing Agrobacterium tumefaciens. The authors are grateful to Dept. of Microbiology Faculty of Medicine, Brawijaya University for providing MDR Klebsiella pneunomiae . We would like to thank Prof Lilik Eka for providing kefir grain.
Conflict Of Interest
The authors declares that there is no conflict of interest.
Authors’ Contribution
TR, AF, IK and NN designed the experiments. AF, IK and TR performed the experiments. AF, IK , TR and NH analyzed the data. AF, IK and TR wrote the manuscript. TR, AF, IK and NN read and approved the manuscript.
Funding
This study was supported by PNBP grants from Faculty of Medicine, Brawijaya University. The funders had no role on study design, data collection and analysis, decision to publish or preparation of the manuscript.
Data Availability
All datasets generated or analyzed during this study are included in the manuscript and/or the Supplementary Files.
Ethics Statement
This article does not contain any studies with human participants or animals performed by any of the authors.
- Vading M., Naucle´r P., Kalin M., Giske C.G. Invasive infection caused by Klebsiella with high comorbidity and associated with high long-term mortality. PLOS ONE, 2018; 6:1-13.
- Wu X., Wang Y., Tao L. Sulfhydryl compounds reduce Staphylococcus aureus biofilm by inhibiting PIA biosynthesis. FEMS Microbiol. Lett., 2011; 316: 44-50.
- Rosenblueth M., Martinez L., Silva J., Martinez-Romero E. Klebsiella variicola, a novel species with clinical and plant-associted isolates. System Appl. Microbiol., 2004; 27: 27-35.
- Tsering D.C., Das S., Singh T.S. Extended spectrum Beta-lactamase detection in Gram negative bacilli of nosocomial origin. J. Glob. Infect. Dis., 2008; 1(2): 87-92.
- Raffa R.B., Iannuzzo J.R., Levine D.R., Saeid K.K., Schwartz R.C., Sucic N.T., Terleckyj O.D., Young J.M. Bacterial communication “quorum sensing” via ligands and receptors: a novel pharmacologic targets for the design of antibiotic drugs. J. Pharmacol. Exp. Ther., 2004; 312(2): 417-423.
- Miquel S., Lagrafeuille R., Souweine B., Forestier C. Anti-biofilm activity as a health issue. Front Microbiol., 2016; 592(7): 1-14.
- Khiralla G., Mohamed E., Farag A., El Hariry H. Antibiofilm effect of Lactobacillus pentosus and Lactobacillus plantarum cell free supernatants against some bacterial pathogens. J. Biotechnol. Res., 2015; 6: 86-95.
- Shokri D., Khorasgani M., Mohkam M., Fatemi S., Ghasemi Y., Taheri-Kafrani A. The inhibition effect of Lactobacilli against growth and biofilm formation of Pseudomonasaeruginosa. Probiotics Antimicrob Proteins, 2018; 10(1): 34-42.
- Al-Matkhury H.J.F., Assal S.D.A. Inhibitory effect of Lactobacilli filtrate on Klebsiella pneumoniae biofilm. IASJ, 2012; 11(2): 168-179.
- Deshmukh P.V., Thorat P.R. Detection of antimicrobial efficacy of bacteriocin produced by lactic acid bacteria isolaed raw milk samples. ISRJ, 2013; 1(2): 987-995.
- O’Toole, G.A. Microtiter dish biofilm formation assay. J. Vis. Exp., 2011; 47: 1-2.
- Maldonado N.C., de Ruiz C.S., Cecilia M., Nader-Macias M.E.F. A simple technique to detect Klebsiella biofilm-forming-strains. Inhibitory potential of Lactobacillus fermentum CRL 1058 whole cells and products. Commun. Curr. Res. Educ. Top Trends Appl Microbiol, 2007: 52-59.
- Paula-Ramos L., Santos C., Mello D., Theodoro L., De Oliveira F., Brito G., Junqueira J.C., Jorge A.O.C., de Oliveira L.D. Klebsiella pneumoniae planktonic and biofilm reduction by different plant extract: In vitro study. Scientific World Journal, 2016; 3521413: 1-5.
- Kumar N., Attri P., Yadav D.K., Choi J., Choi E.H., Uhm H.S. Induced melanocytes cancer cell and oxidation biomolecules through deuterium oxide generated from atmosheric pressure non-termal plasma jet. Sci. Rep., 2014; 4(7589): 1-15.
- Liu W., Bao Q., Jirimutu, Qing M., Siriguleng, Chen X., Sun T., Li M., Zhang J., Yu J., Bilige M., Sun T., Zhang H. Isolation and identification of LAB from Tarag in Eastern Mongolia of China by 16s rRNA sequences and DGGA analysis . Microbiol. Res., 2012; 167(2): 110-115.
- Zhang H.B., Wang L.H., Zhang L.H. Detection and analysis of quorum quenching enzyme against Acyl Homoserine Lactone quorum-sensing signals. Curr. Protoc. Microbiol., 2007; IC 3.1-IC3.15.
- Jeong D., Kim D.H., Song K.Y., Seo K.H. Antimicrobial and anti-biofilm activities of Lactobacillus kefiranofaciens DD2 against oral pathogens. J. Oral Microbiol., 2018; 10: 1-9.
- Lagrafeuille R., Miquel S., Balestrino D., Vareille-Delarbre M., Chain F., Langella P., Forestier C. Opposing effect of Lactobacillus on in vitro Klebsiella pneumoniae in biofilm and in an in vivo intestinal colonisation model. Benef Microbes, 2018; 9(1): 87-100.
- Jeong D., Kim D.H., Song K.Y., Seo K.H. Anti-microbial and anti-biofilm activities of Lactobacillus kefiranofaciens DD2 against oral pathogens. J. Oral Microbiol., 2018; 10: 1-9.
- Singh N., Sharma C., Gulhane R.D., Rokana N., Singh B.P., Puniya A.K., Attri S., Goel G., Panwar H. Inhibitory effect of Lactobacilli of goats milk origin against growth and biofilm formation against pathogen: An in vitro study. Food Biosci., 2018; 22: 129-138.
- Cotar AI. Quorum sensing inhibitors as antipathogenic drug in the fight agains Pseudomonas aeruginosa infections. Clin Microbial., 2013; 2(4): 111-119.
- Ghigo J.M. Natural conjugative plasmids induce bacterial biofilm development. Nature, 2001; 412: 442-445.
- O’Toole G.A., Kaplan H.B., Kolter R. Biofilm formation as microbial development. Annu. Rev. Microbiol., 2000; 54: 49-79.
- McDougald D., Rice C., Barraud N., Steinberg P., Kjelleberg S. Should we stay or should we go: mechanisms and consequences for biofilm dispersal. Nat. Rev. Microbiol., 2011; 10: 39-50.
- Goldstone R., Popat R., Fletcher M., Crusz S., Diggle S. Quorum sensing and social interactions in microbial biofilms, 2012, pp. 1-24. In: Lear G, Lewis G, (eds.), Microbial biofilms. Caister Academic Press, Norfolk.
- Cotar A., Chifiriuc M., Dinu S., Pelinescu D., Banu O., Laza V. Quantitative PCR study of the influence of probiotic culture soluble fraction on the expression of Pseudomonas aeruginosa quorum sensing genes. Roum. Arch. Microbiol. Immunol., 2010; 69: 213-223.
© The Author(s) 2019. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.


