Pankaj Prakash Verma*, Sanjana Thakur and Mohinder Kaur
Department of Basic Science (Microbiology section), Dr. Yashwant Singh Parmar University of Horticulture and Forestry, Nauni, Solan – 173 230, India.
Received on 10 May 2016 and accepted on 27 July 2016
ABSTRACT
Plant growth promoting rhizobacteria prove a key to sustainable agriculture provided they effectively colonize roots, survive, proliferate in the rhizosphere and enhance plant growth by a variety of mechanisms. In the present study, several samplings were conducted in different apple growing regions of Himachal Pradesh in order to isolate a strain capable of showing multifarious plant growth promoting activities and antagonism against Dematophora nectarix a major apple plant pathogen. In this research five isolates of fluorescent Pseudomonas sp. were isolated from apple rhizosphere. Out of five isolates, one isolate DE-18 was selected on the bases of high production of PGP activities and was identified by 16S rRNA gene sequencing. The isolate DE-18 showed maximum phosphate solubilisation and siderophore production of 425 µg/ml and 42.18 %SU respectively. Among the five isolate, DE-18 showed production of both protease (40 mm) and chitinase (28 mm). The isolate DE-18 also showed antagonism against Dematophora nectarix (38.46%) and Phytopathora cactoram (36.18%) and can act as Bioprotectant. The 16S rDNA based phylogenetic analysis demonstrated that the isolate DE-18 belonged to the Pseudomonas putida and sequence was deposited in the GenBank nucleotide sequence databases under accession number KU139388.
Keywords: PGPR, Pseudomonas putida, Dematophora nectarix, Biocontrol efficacy.
Introduction
Plant growth-promoting rhizobacteria (PGPR) are naturally occurring soil bacteria that aggressively colonize plant roots and benefit plants by providing growth promotion1. In the last few years, the number of PGPR that have been identified has seen a great increase, mainly because the role of the rhizosphere as an ecosystem has gained importance in the functioning of the biosphere. Various species of bacteria like Pseudomonas, Azospirillum, Azotobacter, Klebsiella, Enterobacter, Alcaligenes, Arthrobacter, Burkholderia, Bacillus and Serratia have been reported to enhance the plant growth2-4. There are several PGPR inoculants currently commercialized that seem to promote growth through at least one mechanism; suppression of plant disease (termed Bioprotectants), improved nutrient acquisition (Biofertilizers), or phytohormone production (Biostimulants). Inoculant development has been most successful to deliver biological control agents of plant disease i.e. organisms capable of killing other organisms pathogenic or disease causing to crops.
Pseudomonas sp. is ubiquitous bacteria in agricultural soils and has many traits that make them well suited as PGPR. The most effective strains of Pseudomonas have been Fluorescent Pseudomonas sp. and have been studied for decades for their plant growth-promoting effects through effective suppression of soil borne plant diseases. Among various biocontrol agents, Fluorescent pseudomonads, equipped with multiple mechanisms for biocontrol of phytopathogens and plant growth promotion, are being used widely as they produce a wide variety of antibiotics, chitinolytic enzymes, growth promoting hormones, siderophores, HCN and catalase, and can solubilize phosphorous5-8. Considerable research is underway globally to exploit the potential of fluorescent Pseudomonads. Specific strains of the Pseudomonas fluorescens-putida group have recently been used as seed inoculants on crop plants to promote growth and increase yields9.
Biological control has gained considerable attention and appears to be promising alternative to chemical control. Biological control has significant potential in terms of both environmental and economic issues for incorporation into organic temperate fruit production. A number of studies have demonstrated benefits resulting from application of plant growth promoting and disease suppressive rhizobacteria to subsequent growth of apple in replant soil. Replant problem is caused by abiotic and biotic factors10. Various species of fungi like Fusarium equiseti, F. oxysporum, F. solani, Rhizoctonia sp., Cylindrocladium sp., Rosellinia necatrix, Penicillium claviforme, P. janthinellum, Phytophthora sp., Pythium sp., Cylindrocarpon sp., and of nematodes like Pratylenchus penetrans and Xiphinema sp. have been found associated with replant disease by various workers11. Root rot is a very serious soil-borne disease infecting temperate fruits especially apple. It is caused by Rosellinia necatrix Berl. ex Prill. (Anam. Dematophora necatrix Hartig). Agarwala12 (1961) observed this disease on apple trees in Himachal Pradesh. The annual losses estimated due to this disease are about Rs. 1.3 million13 which are expected to be much more as the disease is reported to occur in all apple growing regions of the country.
A diversity of bacterial species has been identified that suppress individual causal elements and enhance growth of plants in replant soil. Biological control of Phytophthora cactorum which contributes to replant disease has been reported in response to application of Enterobacter aerogenes14. Biological control of soil borne pathogens with antagonistic bacteria has gained considerable attention and appears to be promising alternative to chemical control. So the present research was undertaken to isolate and identify a PGPR inoculant that will promote growth and can act as bioprotectant, biofertilizer and a biostimulant.
MATERIALS AND METHODS
Collection of soil samples
Soil samples were collected from rhizosphere of apple plants from Shimla district which is the main apple belt of Himachal Pradesh. The randomly selected apple plants were used for collection of soil samples along with feeder roots from different replant sites of Shimla district viz., Magawta, Shrontha, Siao and Deola. The roots and soil samples from each tree basin were drawn from 15 cm soil depth with the help of soil auger. The samples were carried to the laboratory in polythene bags and were stored in refrigerator at 4° C till further analysis.
Isolation and purification of fungal pathogen
The rhizospheric soil samples were used for isolation of fungal pathogens. The media employed for the isolation of fungus was Potato Dextrose agar (PDA). The plates were incubated at 28 ± 2ºC for 3-4 days and the well isolated fungal colonies appeared on plates were purified by subsequent sub culturing. The purified colonies were grown on PDA slants and stored thereafter at 4ºC and were subcultured periodically on the same media at 28 ± 2ºC. The predominant isolates were identified on the basis of their spore arrangement and fungal isolates were finally identified from National Centre for Fungal Taxonomy, New Delhi.
Isolation of fluorescent Pseudomonas, media and growth conditions
Isolation of fluorescent Pseudomonas sp. was made from apple rhizhosphere in Shimla district of Himachal Pradesh (India). Ten grams of each rhizospheric soil sample were mixed and shaken in 90 ml sterile distilled water blank in 250 ml Erlenmeyer flask for 20-30 minutes to obtain standard soil suspension. Isolation of fluorescent Pseudomonas sp. was made by following the serial dilutions and pour plate method using the specific King’s B medium15. Plates were incubated at 28±2°C and enumerations of fluorescent Pseudomonas colonies were done after 48 h of incubation.
These were identified on the bases of morphological, biochemical and physiological tests viz., Gram’s staining, pigment production, oxidase test, catalase test, gelatine liquification, denitrification test and growth at optimum temperature i.e 4°C and 41°C.
Evaluation of PGPR traits
All the Pseudomonas isolates from apple rhizosphere were qualitatively and quantitatively characterized by standard protocols for the presence of PGPR traits viz., phosphate solubilisation, siderophore production, HCN production, ammonia production, antifungal activity and for production of hydrolytic enzymes which are known to play an essential role in growth promotion of plants.
Quantification of phosphate solubilisation
Solubilization of P by all the five Pseudomonas isolates were estimated using a known amount of inert phosphorus source (tricalcium phosphate) in Pikovskaya’s agar medium16. The composition of the medium was (g/l-1): Glucose, 10.0; Ca3(PO4), 5.0; (NH4)2SO4, 0.5; MgSO4.7H2O, 0.2; MnSO4,0.1; FeSO4,0.0001; Yeast extract, 0.5; Bromocresol purple, 0.1. Quantification of phosphate solubilization was done by spectrophotometric method17-19. Phosphate solubilizing activity was expressed in terms of tricalcium phosphate solubilization, which in turn represents µg/ml of available orthophosphate as calibrated from the standard curve of KH2PO4 (10-100 µg/ml).
Siderophores detection and quantification
Siderophore production by Pseudomonas isolates was detected by observing orange halos production around the bacterial colony on CAS agar plates20 after 72 h of growth. For quantification of siderophores, to 0.5 ml of cell free culture supernatant of each Pseudomonas isolate grown in liquid CAS medium, 0.5 ml of CAS reagent and 10µl of shuttle solution was added and absorbance was measured at 630 nm against a blank. Siderophores content was expressed as percentage siderophore units using the formula:
% Siderophore units (%SU) = (Ar-As)/Ar × 100
Where Ar = absorbance of reference at 630 nm (CAS reagent) and As = absorbance of sample at 630 nm
HCN and ammonia detection
Bacterial cultures were tested for the production of ammonia in peptone water. Freshly grown cultures were inoculated in 10 ml peptone water and incubated for 48-72 h at 28±2°C. After incubation, Nessler’s reagent was added in each tube. Development of brown to yellow colour will be a positive test for ammonia production21 and King’s B medium amended with 0.44% of Glycine was used for detection of hydrogen cyanide following the method of Bakker and Schippers22 (1987).
Antifungal activity
Isolated bacterial cultures were tested for growth inhibitory effect on the mycelium growth of Dematophora sp., Fusarium sp., Phytopathora sp, Pythium sp. and Rhizocotina sp, the major fungal pathogens of apple plants by well plate assay method23.
Chitinase and Protease activity
Chitinase activity was measured according to Chernin24 et at. (1995) and protease activity according to Kaur25 et at. (1988).
DNA Extraction and amplification of 16S rRNA gene fragments
The fluorescent Pseudomonas isolate DE-18 was selected on the basis of overall plant growth promoting and disease suppressing activities for their genotypic characterization by 16S rRNA gene sequencing. Genomic DNA was extracted using conventional method (Phenol: Chloroform method) and the DNA was quantified via agarose gel electrophoresis (using 1.0% agarose). 16S rRNA gene analysis was done using Pseudomonas specific oligonucleotide primer sequences viz., FP-1 (20-mer): GGTCTGAGAGGATGATCAGT and RP-1 (18-mer): TTAGCTCCACCTCGCGGC in MJ Mini BIO-RAD personal thermal cycler-100 (PTC-100) with a total of 35 cycles. These primers were designed and were used to find their taxonomic affiliation to fluorescent Pseudomonas species of group I which mainly comprise Pseudomonas aeruginosa, Pseudomonas putida and Pseudomonas fluorescence.
The PCR amplification was carried out in 0.2 ml PCR tubes with 50 µl reaction volume consisting of following components: 5.0 µl Taq buffer A, 3.0 µl dNTP Mix, 1.0 µl FP-1, 1.0 µl RP-1, 0.40 µl Taq DNA polymerase, 2.0 µl genomic DNA, 37.6 µl double distilled water. A PCR program was implemented as follows: denaturation at 94ºC for 1min, two minutes annealing at 55ºC and extension for 2 min at 72ºC. The final extension was for 10 min at 72ºC. For DNA sequencing, eluted amplified DNA product was first purified followed by sequencing in B. Genei, India pvt. Ltd. (Bangalore)
Phylogenetic analysis
The sequencing results were aligned to 16S rRNA gene sequences from GenBank database at NCBI (National Centre of Biotechnological information) through web site http://www.ncbi.nlm.nih.gov using BLAST program for screening of sequence similarity. Phylogenetic reconstruction was accomplished with the phylogeny MEGA 6.06 inference package. Phylogenetic trees were constructed from distance matrices by the neighbour-joining method and tree topology was estimated by bootstrap analysis, which includes 1000 replicate data sets.
RESULTS AND DISCUSSION
In the present research work, five isolates of fluorescent Pseudomonas sp. were isolated from Apple rhizosphere of Magawta, Shrontha, Siao and Deola (Shimla distt., Himachal Pradesh) Table 1. These isolates were further subjected to morphological, physiological and biochemical characterization. All the isolates of fluorescent Pseudomonas were studied in detail for colony, colour, growth type, fluorescence, and cell shape. It was evident from the observations that all the five isolates viz., M1, M2, SH1, SO1 and DE-18 produced round shaped colonies with an entire margin and rod shaped cells Table 2. Through morphological and biochemical identification, all the isolates were found to be gram-negative, aerobic, oxidase positive, catalase positive with a fluorescent pigmentation. However, negative responses were also identified for some Pseudomonas isolates such as for gelatin hydrolysis and their ability to grow at 4°C and 41°C. The isolate DE-18 could not hydrolyse gelatine and isolates M1, SH1 and DE-18 did not showed growth at 41°C Table 3. Reynolds26 (2004) also characterized isolates on the bases of biochemical tests including oxidase, catalase, gelatin hydrolysis, nitrate reduction and performing growth at 4 and 41°C and identified them as P. fluorescens.
Table 1. Details of soil samples and location used for isolation of fluorescent Pseudomonas sp.
fluorescent Pseudomonas isolates |
Location |
District |
State |
M1 |
Magawta |
Shimla |
Himachal Pradesh |
M2 |
Magawta |
Shimla |
Himachal Pradesh |
SH1 |
Shrontha |
Shimla |
Himachal Pradesh |
SO1 |
Siao |
Shimla |
Himachal Pradesh |
DE-18 |
Deola |
Shimla |
Himachal Pradesh |
Table 2. Morphological characterization of fluorescent Pseudomonas sp. isolated from apple rhizosphere
| fluorescent Pseudomonas isolates | Colony morphology | Gram reaction | Cell shape | |||
| Form | Elevation | Margin | Opacity | |||
| M1 | Circular | Raised | Entire | Translucent | – | Rod |
| M2 | Circular | Raised | Entire | Translucent | – | Rod |
| SH1 | Circular | Flat | Entire | Transparent | – | Rod |
| SO1 | Circular | Flat | Entire | Transparent | – | Rod |
| DE-18 | Circular | Raised | Entire | Transparent | – | Rod |
Table 3. Physiological and biochemical characterization of fluorescent Pseudomonas sp. isolated from apple rhizosphere
| Parameters | Fluorescent Pseudomonas isolates | ||||
| M1 | M2 | SH1 | SO1 | DE-18 | |
| Catalase test | + | + | + | + | + |
| Gelatin liquification | + | + | + | + | – |
| Lecithinase activity | + | + | – | – | – |
| Oxidase test | + | + | + | + | + |
| Spore staining | – | – | – | – | – |
| Starh hydrolysis | – | – | – | – | – |
| Tween 80 hydrolysis | – | – | – | – | – |
| Denitrification | + | + | + | + | + |
| Growth at 4°C | + | + | + | – | + |
| Growth at 41°C | – | + | – | + | – |
(-) Indicates negativity of test (+) indicates positivity of test
The fungal cultures from rhizospheric soil were purified by single spore and hyphal tip isolation technique. The identification of the isolated fungus was done by studying the morphological characters and by microscopic examination of each isolate. The cultures were sent to National Centre for Fungal Taxonomy, Inderpuri, New Delhi for identification to species level and the fungi were identified as Fusarium solani and Nigrospora oryzae Table 4 Figure 1.
Table 4. Identification of fungal pathogens isolated from apple rhizosphere
Sr. no |
Host |
Identification number |
Final identification |
Replant site |
1 |
Apple rhizosphere |
6219.15 |
Fusarium solani |
Magawta (Shimla distt.) |
2 |
Apple rhizosphere |
6220.15 |
Nigrospora oryzae |
Deola (Shimla distt.) |
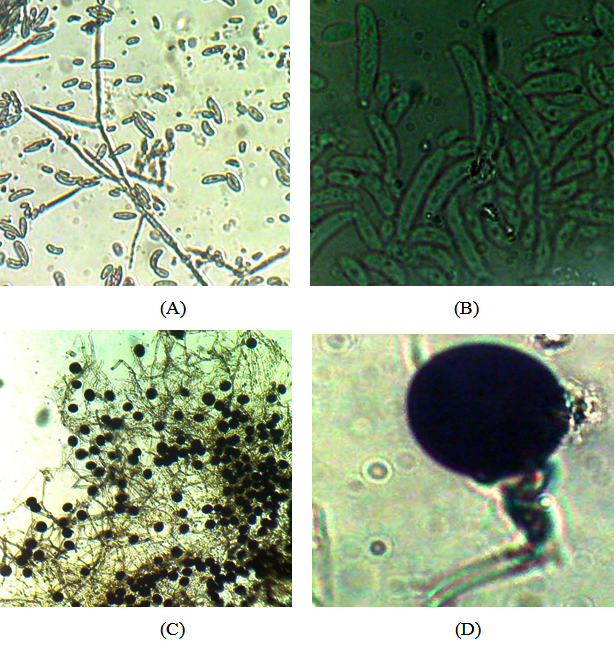 Fig. 1. Microphotographs of Fusarium solani (A, B), Nigrospora oryzae (C, D)
Fig. 1. Microphotographs of Fusarium solani (A, B), Nigrospora oryzae (C, D)
All the fluorescent Pseudomonas isolates were identified as potential phosphate solubilizers based on their capacity to solubilize tricalcium phosphate [Ca3(PO4)2] by the formation of halo zone on Pikovskaya’s agar medium. The maximum P solubilisation was recorded by DE-18 (425 µgml-1) compared to other isolates M1, M2, SH1 and SO1 (362.5, 350, 162.5 and 225 respectively) Table 5. Verma27 et al. (2014) also recorded phosphate solubilizing activity of Pseudomonas isolates in the range of 155µg/ml to 410 µg/ml. Most of phosphorus in soil is present in the form of insoluble phosphates and cannot be utilized by the plants. PGPR have been shown to solubilize precipitated phosphates and enhance phosphate availability to plant that represent a possible mechanism of plant growth promotion under field conditions28.
All the tested isolates of fluorescent Pseudomonads were positive for the production of siderophores, HCN, ammonia, lytic enzymes and plant growth-promoting hormones viz., auxins, gibberellins and cytokinins. The maximum siderophore production was showed by isolate DE-18 (42.18 %SU) and minimum by SO1 (4.68 %SU) (Table 5). In the development of the antagonism the siderophore production of the bacteria has an important role and such bacteria may function as stress factors including local and systematic host resistance29. Iron competition in Pseudomonads has been intensively studied and the role of the pyoverdine siderophore produced by many Pseudomonas sp. has been clearly demonstrated in the control of Pythium and Fusarium species30. Different types of siderophores in Pseudomonas species have antagonistic effect against F. graminearum31. Microbial production of HCN has been suggested as an important antifungal feature to control root fungi pathogen. Cyanide acts as a general metabolic inhibitor to avoid predation or competition. The host plants are generally not harmfully affected by inoculation with HCN producing bacteria and rhizobacteria can operate as biological control agents32.
Table 5. Characterization of indigenous fluorescent Pseudomonas sp. for plant growth promoting traits
*Phosphate solubilizing activity expressed in terms of tricalcium phosphate solubilization which in turn represents µg/ml of soluble inorganic phosphate(Pi) in supernatant as calibrated from the standard curve of KH2PO4 (10-100 µg/ml).
**The siderophore unit (% SU) expressed as percent reduction in blue color of chrome azurol-S as compared to reference i.e.
% SU= (Ar-As)/Ar×100
where, Ar = Absorbance of reference solution at 630 nm; As = Absorbance of test solution at 630 nm
Table 6. Characterization of fluorescent Pseudomonas sp. for antifungal activity against different apple plant pathogens
| fluorescent Pseudomonas isolates | Dematophora nectarix (Control=65 mm) |
Phytopathora cactoram (Control=47 mm) |
Lytic enzymes | |||
| **Protease | **Chitinase | |||||
| mm dia | %I* | mm dia | %I* | (mm dia) | (mm dia) | |
| M1 | 46 | 29.23 | – | – | 12 | – |
| M2 | – | – | 37 | 21.27 | 27 | – |
| SH1 | – | – | – | – | 30 | – |
| SO1 | – | – | – | – | – | – |
| DE-18 | 40 | 38.46 | 30 | 36.18 | 40 | 28 |
The production of IAA by PGPR generally affects the root system, increasing the size and number of adventitious roots and also the root subdivision, enabling a bigger soil amount to be exploited by the roots, thus providing large amounts of nutrients accessible to the plant33. The isolates DE-18 and M2 produced 14.0 and 10.0 µgml-1 of auxins respectively, whereas all the other isolates produced auxins between 2.0 and 8.0 µgml-1 (Table 5).Similar results were also observed by Sharma34 et al. (2014) in which all the screened fluorescent Pseudomonas sp. produced auxin in concentration of 5.1-16.9 µgml-1 .
The production of indole acetic acid by the strains of Pseudomonas sp. responsible for increasing root elongation was also reported by O’ Sullivan and O’ Gara35, (1992). However, IAA production by PGPR can vary among different species and strains, and it is also influenced by culture condition, growth stage and substrate availability36. All the isolates also produced gibberellins and cytokinins in the range of 100-
250 µgml-1 and 30-600 µgml-1 respectively (Table 5). Thakur37 et al. (2013) also observed the production of gibberellins and cytokinins by fluorescent Pseudomonas isolates in the range 15.20 to 179.48 µg/ml and 51.20 to 179.48 µg/ml respectively.
The results of inhibition of mycelium growth of the assayed phytopathogenic fungi showed inhibition against only two fungal pathogens among the Fusarium oxysporium, Pythium ultimum, Dematophora nectarix, Phytopathora cactoram and Rhizocotina sp. From the assayed five bacterial cultures, the maximum inhibition of mycelium growth of 38.46 % and 36.18 % was found by isolate DE-18 against Dematophora nectarix and Phytopathora cactoram Table 6 and Figure 2. Pseudomonas putida and Pseudomonas fluorescens, are frequently isolated from soil and plant tissue and reported as potential biocontrol agents of phytopathogens38-40. Based on the antagonistic potential and other multifarious plant growth promoting characteristics isolate DE-18 was genotypically characterized by 16S rDNA gene sequencing.
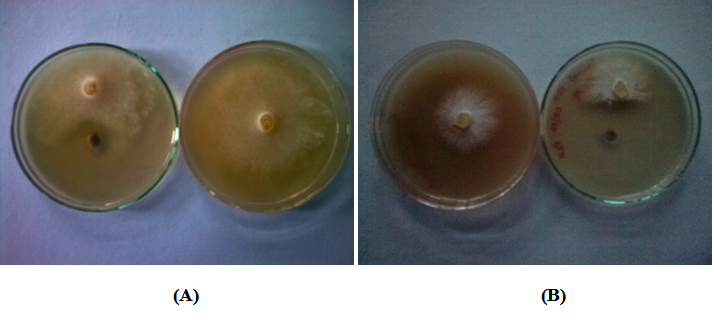
Fig. 2. Antifungal activity of Pseudomonas putida DE-18 against (A) Phytopathora cactoram and (B) Dematophora nectarix
The potential isolate DE-18 was identified based on their 16S rRNA gene using the universal primer set and BLAST analysis. The 16S rDNA based phylogenetic analysis demonstrated that the isolate DE-18 belonged to the genus Pseudomonas sp. Figure 3 described the relationship between the isolated strain and the nearest phylogenetic relatives. Similarity calculations after neighbor-joining analysis indicated that the closest relative of isolate DE-18 was Pseudomonas putida (90 %). The determined 16S rDNA sequence of strain Pseudomonas putida DE-18 was deposited in the GenBank nucleotide sequence databases under accession number KU139388.
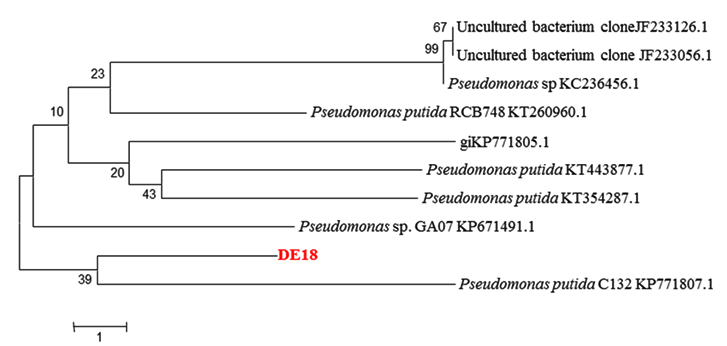 Fig. 3. Phylogenetic tree of Pseudomonas sp. strain DE-18 and related species constructed on the basis of 16S rDNA gene sequence using the neighbor-joining method
Fig. 3. Phylogenetic tree of Pseudomonas sp. strain DE-18 and related species constructed on the basis of 16S rDNA gene sequence using the neighbor-joining method
This study is assumed to be important as the agriculturally beneficial antibiotic-producing P. putida could be one of the potential candidates in the development of microbial pesticides for sustained crop productivity. Hence this P. putida DE-18 strain have a potential of being developed as a bio-inoculant for application in a agriculture and horticulture crops.
REFERENCES
- 1. Kloepper, J.W., Leong, J., Teintze, M. and Schroth, M.N. Enhanced plant growth by siderophores produced by plant growth promoting rhizobacteria. Nature, 1980; 286: 885–886.
2. Okon, Y. and Labandera-Gonzalez, C.A. Agronomic applications of Azospirillum. In: Improving Plant Productivity with Rhizosphere Bacteria (Ryder MH, Stephens PM, Bowen GD, eds). Australia: Commonwealth Scientific and Industrial Research Organization, 1994; pp 274–278.
3. Glick, B.R. The enhancement of plant growth by free living bacteria. Canadian Journal of Microbiology, 1995; 41(2): 109–114.
4. Joseph, B., Patra, R.R. and Lawrence, R. Characterization of plant growth promoting rhizobacteria associated with chickpea (Cicer arietinum L). International Journal of Plant Production, 2007; 1(2): 141-152.
5. Banasco, P., Fuente, L., DeLa, Gaultieri, G., Noya, F. and Arias, A. Fluorescent Pseudomonas sp. as biocontrol agents against forage legume root pathogenic fungi. Soil Biology and Biochemistry, 1998; 10 (10-11): 1317–1323.
6. Kraus, J. and Loper, J. Characterization of genomic region required for production of antibiotic pyoluteorin by the biological control agent Pseudomonas fluorescens Pf-5. Applied and Environmental Microbiology, 1995; 61(3): 849–854.
7. Rodriguez, H., Fraga, R. Phosphate solubilizing bacteria and their role in plant growth promotion. Biotechnology Advances, 1999; 17(4-5): 319–339.
8. Seong, K.Y. and Shin, P.G. Effect of siderophore on biological control of plant pathogens and promotion of plant growth by Pseudomonas fluorescens ps88. Agricultural Chemistry and Biotechnology, 1996; 39: 20–24. - 9. Kloepper, J.W., Reddy, M.S., Rodríguez-Kabana, R., Kenney, D.S., Kokalis-Burelle, N., Martinez-Ochoa, N. and Vavrina, C.S. Application of rhizobacteria in transplant production and yield enhancement. Acta Horticulturae, 2004; 631: 217-229.
10. Bharat, N.K. Biological control of diseases of temperate fruit crops – a review. International Journal of Science and Nature, 2011; 2(3): 422 -431.
11. Utkhede, R. S. Replant disease and soil sickness. In: Management of soil-borne diseases (Utkhede RS and Gupta VK, eds). Ludhiana: Kalyani Publishers, 1996; pp 21-39.
12. Agarwala, R. K. Problems of root rot of apple in Himachal Pradesh and Prospectus of its control with antibiotics. Himachal Hortic., 1961; 2: 171-178.
13. Agarwala, R. K. and Sharma, V. C. White root rot disease of apple in Himachal Pradesh. Indian Phytopath., 1966; 19: 82-86.
14. Utkhede, R. S. and Smith, E. M. Phytophthora and Pythium sp. associated with root rot of young apple trees and their control. Soil Biol. Biochem., 1991; 23: 1059-1063.
15. King, E.O., Ward, M.K. and Raney, D.E. Two simple media for the demonstration of pyocyanin and fluoresccein. Journal of Laboratory and Clinical Medicine, 1954; 44: 301-307.
16. Pikovsakaya, R.E. Mobilization of phosphorus in soil in connection with vital activity of some microbial species. Microbiologia, 1948; 17: 362-370.
17. Dickman, S.R. and Bray, R.H. Colorimetric determination of phosphate. Industrial and Engineering Chemistry Analytical Edition, 1940; 12: 660-665.
18. Bray, R.H. and Kurtz, L.T. Determination of total organic available forms of phosphorus in soil. Soil Science, 1945; 23: 343-353.
19. Olsen, S.R., Cole, C.V., Whatanable, F.S. and Dean, L.A. Estimation of available phosphorus by extraction with sodium bicarbonate. Circu. US Department of Agriculture, Washington, DC. 1954; 939: 171-179.
20. Schwyan, B. and Neilands, J.B. Universal chemical assay for the detection and determination of siderophore. Analytic Biochemistry, 1987; 28(8): 751-759.
21. Lata and Saxena, A.K. Characterization of plant growth promoting rhizobacteria. In: Training manual on biofertilizer technology (Saxena AK, ed). Delhi: IARI, 2003; pp. 24-25.
22. Bakker, A.W. and Schippers, B. Microbial cyanide production in the rhizosphere in relation to total yield reduction and Pseudomonas sp. mediated plant growth stimulation. Soil Biology and Biochemistry, 1987; 19: 451-457.
23. Vincent, J.M. Distribution of fungal hyphae in presence of certain inhibitors. Nature, 1947; 150: 158-850.
24. Chernin, L., Ismailov, Z., Haran, S. and Chet, I. Chitinolytic Enterobacter agglomerans antagonistic to fungal plant pathogens. Applied and Environmental Microbiology, 1995; 61(5): 1720-1726.
25. Kaur, M., Gupta, M., Tripathi, K. K. and Gupta, K.G. Lytic effect of Pseudomonas aeruginosa elastase on Gram positive and Gram negative bacteria. Zentrablatt Beckteriologie Internatinal Journal of Microbiology, 1988; 271(2): 153-157.
26. Reynolds, J. Lab procedures manual: Biochemical tests. Richland College. http://www.rlc. dcccd.edu/mathsci/Reynolds/micro/lab_manual/TOC.html. 2004
27. Verma, P.P., Sharma, S. and Kaur, M. Effect of indigenous strains of fluorescent Pseudomonas sp. on growth of apple plants in replant site of Himachal Pradesh. Indian Journal of Applied Research, 2014; 4(7): 432-437.
28. Ashrafuzzaman, M., Hossen, F.A., Ismail, M.R., Hoque, M.A. and Islam, M.Z. Efficiency of plant growth-promoting rhizobacteria (PGPR) for the enhancement of rice growth. Afr J Biotechnol., 2009; 8: 1247-1252.
29. Kumari, B., Pragash, M.G., Cletus, J., Raman, G. and Sakthivel, N. Simultaneous phosphate solubilization potential and antifungal activity of new fluorescent pseudomonad strains, Pseudomona aeruginosa, P. plecoglossicida and P. mosselii. World J. Microbiol. Biotechnol., 2009; 25: 573-581.
30. Duijff, B.J., Meijer, J.W., Bakker, P.A.H.M. and Schippers, B. Siderophores mediated competition for iron and induced resistance in the suppression of Fusarium wilt of carnation by fluorescent Pseudomonas sp. Netherlands Journal of Plant Pathology, 1993; 99: 277-289.
31. Quan, C.S., Wang, X. and Fan, S.D. Antifungal compounds of plant growth promoting rhizobacteria and its action mode. In: Plant Growth and Health Promoting Bacteria (Maheshwari DK, ed). Berlin-Heidelberg: Springer Verlag, 2010; pp 117-156.
32. Ramette, A., Moënne Loccoz, Y. and Défago, G. Prevalence of fluorescent pseudomonads producing antifungal phloroglucinols and/or hydrogen cyanide in soils naturally suppressive or conducive to tobacco black root rot. FEMS Microbiol Ecol., 2003; 44: 35-43.
33. Ribeiro, C.M. and Cardoso, E.J. Isolation, selection and characterization of root associated growth promoting bacteria in Brazil Pine (Araucaria angustifolia). Microbiol Res., 2011; 167: 69-78.
34. Sharma, S., Verma, P.P. and Kaur, M. Isolation, purification and estimation of IAA from Pseudomonas sp. using HPLC. Journal of pure and applied microbiology, 2014; 8(4): 3203-3208.
35. O’Sullivan, D.J. and O’Gara, F. Traits of fluorescent Pseudomonas sp. involved in suppression of plant root pathogen. Microbiological Reviews, 1992; 56(2): 662-676.
36. Sajjad Mirza, M., Ahmad, W., Latif, F., Haurat, J. and Bally, R. Isolation, partial characterization, and the effect of plant growth-promoting bacteria (PGPB) on micro-propagated sugarcane in vitro. Plant Soil, 2001; 237: 47-54.
37. Thakur, D., Kaur, M. and Shyam, V. Management of replant problem by the production of plant growth regulators and phosphate solubilising potential of fluorescent Pseudomonas sp. isolated from the normal and replant sites of apple and pear. Indian Streams Research Journal, 2013; 3(4): 1-8.
38. Colyer, P.D. and Mount, M.S. Bacterization of potatoes with Pseudomonas putida and its influence on postharvest soft rot diseases. Plant Dis., 1984; 68:703-706.
39. Xu, G.W. and Gross, D.C. Selection of fluorescent pseudomonads antagonistic to Erwinia carotovora and suppressive of potato seed piece decay. Phytopathology , 1986a; 76: 414-422.
40. Xu, G.W. and Gross, D.C. 1986b. Field evaluations of the interactions among fluorescent pseudomonads, Erwinia carotovora and potato yield. Phytopathology, 1986b; 76: 423-430.

