ISSN: 0973-7510
E-ISSN: 2581-690X
Dengue virus (DENV) is still global problem and infecting millions of people a year. This virus belongs to Flavivirus and consists of the structural and non-structural proteins including envelop (E), capsid (C), NS2B/NS3, and NS5. Garcinia atroviridis Griff. ex T. Anders is traditional plant that has broad potential as antioxidant, antibacterial, and anti-cancer activities. However, the anti-DENV potential of this plant is uncertain. The objective of this research is to find out the potential of the phytochemical compounds of G. atroviridis as DENV antiviral drugs targeting E, C, NSB2/NS3, and NS5 proteins using molecular simulation approach. Sample retrieval was obtained from PubChem and RCSB PDB. Drug-likeness analysis has been assessed with Swiss ADME based on the pharmacology and pharmacokinetics aspects. Toxicity prediction was done by pkCSM webserver. PyRx was carried out to screen ligand-protein interaction virtually. Visualization of the best interaction was displayed by BIOVIA Discovery Studio. CABS-flex 2.0 version webserver was performed to predict stability interaction. Atroviridin was determined as the most promising as DENV antiviral to be tested by the wet laboratory approach.
DENV, Garcinia atroviridis, Structural Proteins, Non-structural Proteins, Bioinformatics
Dengue virus (DENV) is the frequent arthropod-borne infection that infecting human worldwide.1 It is reported infects over 400 million people each year. DENV is capped-single stranded RNA, positive sense virus that classified into the Flavivirus.2 This virus has 5 serotypes, the most recent of which was found.3,4 That condition complicated dengue control particularly in tropical climate countries where epidemic occurs regularly. DENV infects human via Aedes sp. mosquito’s transmission.5
DENV has proteins act significant roles from viral entry to viral release in human cell. Proteins are classified into two types: structural and non-structural proteins. Structural and non-structural proteins have various roles in the virion’s existence. Envelop (E), capsid (C), NS2B/NS3, and NS5 are parts of DENV proteins that play important roles in viral entrance, immune system recognition, growth and maturation, genome synthesis, and viral release.6-11
Garcinia atroviridis Griff ex T. Anders. is native to and widely dispersed throughout South and Southeast Asia regions including India, Thailand, Myanmar, and Indonesia.12 This plant belongs to Guttiferae family. This plant is known as asamgelugur in Indonesia and commonly utilized as cooking spices especially in Aceh and other Sumatra regions.13 Previous researches have shown that the extract has antioxidant, antibacterial, and anti-tumour activities.14,15,16,17 There have been no researches regarding the potential of G. atroviridis as DENV antiviral drugs. The objective of this research is to find out the potential of the phytochemical compounds of G. atroviridisas DENV antiviral drugs targeting E, C, NSB2/NS3, and NS5 proteins using molecular simulation approach.
Sample Retrieval
Various phytochemical compounds derived from G. atroviridis and specific synthetic drugs were yielded from PubChem (https://pubchem.ncbi.nlm.nih.gov/).
The compound structures have been compiled in .sdf file as ligands.18 Therefore, conversion to be protein data bank (PDB) was done to produce flexible 3D structure using PyRx version 0.9 software.19 The target proteins used were obtained from RCSB PDB (https://rcsb.org/) consists envelop (PDB ID 3UZV), capsid (PDB ID 6VSO), NSB/NS3 (PDB ID 2FOM), and NS5 (PDB ID 2J7U). Removal of water and native ligands was conducted using BIOVIA Discovery Studio 2016 16.1.0 ׳ 64 (Dassault Systems France).20 Synthetic drugs were added to the later step as a control for each target protein.21
Drug-likeness Analysis
Drug-likeness analysis was done to analyze pharmacological and pharmacokinetics similarities in each drug using SwissADME (https://www.swissadme.ch/index.php). The Lipinski, Ghose, Veber, Egan, Muege, bioavailability score (BA), and gastrointestinal absorption (GI abs) were included in this analysis. Drug-likeness analysis results must be no violation and ≥0.50 as well as high score for pharmacokinetics.22 Positive predictions are distinguished to fulfill the criteria of each category with check mark (√) and will be continued to the next step.21
Toxicity prediction
The phytochemical compounds were filtered on the drug-likeness properties basis with toxicity prediction using Predicting Small-Molecule Pharmacokinetic Properties using Graph-Based Signature (pkCSM) (https://biosig.lab.uq.edu.au/pkcsm/).23 Toxicity predictions are vital because their relations with pharmacokinetics on prospective drug analysis.24 Several endpoint parameters were applied including Ames test, maximum recommended tolerated dose (MRTD), hERG I/II inhibitors, lethal dose 50 (LD50), and hepatotoxicity. The results of toxicity prediction should have one and/or no violation on Ames test, hERG I/II inhibitors, and hepatotoxicity categories.25 For MRTD and LD50 are quantitative parameter with MRTD endpoint for human is 0.477 log(mg/kg/day) and LD50 is stated in mol/kg.26 Positive predictors would mark as check mark (√) before they continue in docking analysis.
Docking analysis and interaction visualization
Docking analysis was carried out to investigate the ligand-protein interaction with computational screening. In recent years, this approach has been significant technique apart from in vitro and in vivo for computer-aided drug development.27 This study step was carried out by PyRx version 0.9 due to the precision.28 Specific docking was modified to cover only the active sides of target protein (Table 1). Favoured interactions were indicated to the most negative binding affinity then compared to the native ligands and synthesis drugs. After docking analysis, visualization of selected ligand-protein interactions was displayed using BIOVIA Discovery Studio. 2D and 3D interactions will be visualized to examine the interaction groups.21
Table (1):
References of DENV target proteins
Protein |
Control |
Active sites |
Center |
Dimension |
Ref. |
|---|---|---|---|---|---|
Envelop (E) |
NITD448 (CID1390310) |
A: Ile308, Val309, Gln325. B: Thr33, Gly100, Trp101, Glu102 |
X: -8.701 Y: 6.916 Z: 21.773 |
X: 43.458 Y: 27.357 Z: 28.788 |
31 |
Capsid (C) |
ST-148 (CID2909914) |
A: Arg32, Phe33, Gly36, Met37, Leu38, Glu39, Leu44, Thr62, Ala63, Gly64, Arg68, Ile72, Arg82, Arg85, Leu92. B: Val26, Arg41, Leu57, Arg82, Arg85, Lys86, Leu92, Asn96 C: Arg32, Gln39, Pro43, Leu44, Leu46, Arg68, Ile72, Lys73, Arg82, Arg85, Leu92, Asn96. D: Gln39, Arg41, Leu44, Arg68, Ile72, Lys73, Arg82, Arg85, Leu92, Asn93, Asn96. E: Arg32, Phe33, Gly36, Met37, Leu38, Gln39, Lys45, Thr62, Ala63, Gly64, Arg68, Lys73, Arg82, Arg83, Lys86, Gly89, Leu92, Asn96. F: Val26, Gln39, Arg41, Leu44, Arg68, Ile72, Lys73, Arg82, Arg85, Lys86, Gly89, Leu92, Asn93, Asn96, Asn97. His51, Asp75, Ser135 |
X: 32.720 Y: 77.020 Z: 1.427 |
X: 55.925 Y: 74.438 Z: 79.230 |
32 |
NS2B/NS3 |
ARDP0006 (CID 3378440) |
His51, Asp75, Ser135 |
X: 0.643 Y: -6.045 Z: 13.947 |
X: 17.381 Y: 23.658 Z: 43.862 |
33,34 |
NS5 |
SAH (CID 439155) |
Glu37, His441, Cys446, Cys449, Asn492, Gly604, Gly607, Asp663, Asp664, His714, Cys728, Trp823 |
X: 23.702 Y: 58.239 Z: 12.627 |
X: 49.338 Y: 36.839 Z: 34.549 |
35 |
Molecular dynamics analysis
Molecular dynamics was explored to know the stability of ligand-protein interactions.29 This simulation conducted using CABS-flex 2.0 version webserver (http://biocomp.chem.uw.edu.pl/CABSflex2/index) with protein rigidity (1.0), protein restraints (ss2 3 3.8 8.0), global c-alpha restraints weight (1.0), cycle number (50), cycle between trajectory (50), temperature range (1.4), and RNG seed (227) parameter. To maintain the stability, root mean square fluctuation (RMSF) results would be demonstrated with maximum distance of 1-3 Å.30
The optimal criteria of phytochemical compounds utilized as pharmaceutical compounds must meet the pharmacological and pharmacokinetics criteria before. Based on the drug similarity, there were 6 compounds matched the criteria consisting dodecanoic acid (lauric acid) (PubChem ID3893), atroviridin (PubChem ID11267348), naringenin (PubChem ID932), kaempherol (PubChem ID5280863), quercetin (PubChem ID5280343), and gentisein (1,3,7-trihydroxyxanthone) (PubChem ID5281635) (Table 2).
Table (2):
The results of the analysis of drug-likeness analysis
Compound (PubChem ID) |
Lipinski |
Ghose |
Veber |
Egan |
Muegge |
BA score |
GI abs |
Status |
|---|---|---|---|---|---|---|---|---|
Absorbic acid (54670067) |
Yes (0) |
No (2) |
Yes (0) |
Yes (0) |
No (1) |
.56 |
High |
× |
Citric acid (311) |
Yes (0) |
No (2) |
Yes (0) |
No (1) |
No (1) |
.56 |
Low |
× |
Malic acid (525) |
Yes (0) |
No (4) |
Yes (0) |
Yes (0) |
No (2) |
.56 |
High |
× |
Succinic acid (1110) |
Yes (0) |
No (3) |
Yes (0) |
Yes (0) |
No (2) |
.85 |
High |
× |
Tartaric acid (875) |
Yes (0) |
No (4) |
Yes (0) |
Yes (0) |
No (2) |
.56 |
Low |
× |
Hydroxycitric acid (123908) |
Yes (0) |
No (3) |
No (1) |
No (1) |
No (2) |
.11 |
Low |
× |
Pentadecanoic acid (13849) |
Yes (0) |
Yes (0) |
Yes (0) |
No (1) |
No (1) |
.85 |
High |
× |
Nonadecanoic acid (12591) |
Yes (1) |
No (1) |
Yes (0) |
Yes (0) |
No (2) |
.85 |
High |
× |
Dodecanoic acid (3893) |
Yes (0) |
Yes (0) |
Yes (0) |
Yes (0) |
Yes (0) |
.85 |
High |
√ |
Atroviridin (11267348) |
Yes (0) |
Yes (0) |
Yes (0) |
Yes (0) |
Yes (0) |
.55 |
High |
√ |
Atrovirisidone (10342405) |
Yes (0) |
Yes (0) |
Yes (0) |
Yes (0) |
No (1) |
.55 |
High |
× |
Naringenin (932) |
Yes (0) |
Yes (0) |
Yes (0) |
Yes (0) |
Yes (0) |
.55 |
High |
√ |
Morelloflavone (5464454) |
No (3) |
No (2) |
No (1) |
No (1) |
No (3) |
.17 |
Low |
× |
Fukugiside (73157060) |
No (3) |
No (2) |
No (1) |
No (1) |
No (4) |
.17 |
Low |
× |
Kaempherol (5280863) |
Yes (0) |
Yes (0) |
Yes (0) |
Yes (0) |
Yes (0) |
.55 |
High |
√ |
Quercetin (5280343) |
Yes (0) |
Yes (0) |
Yes (0) |
Yes (0) |
Yes (0) |
.55 |
High |
√ |
Garcinol (5281560) |
Yes (1) |
No (4) |
Yes (0) |
No (1) |
No (2) |
.56 |
Low |
× |
Isogarcinol (11135781) |
Yes (1) |
No (4) |
Yes (0) |
No (1) |
No (2) |
.56 |
Low |
× |
α-humulene (5281520) |
Yes (1) |
Yes (0) |
Yes (0) |
Yes (0) |
No (1) |
.56 |
Low |
× |
(-)-ꞵ-caryophyllene (1742210) |
Yes (0) |
Yes (0) |
Yes (0) |
Yes (0) |
No (1) |
.55 |
High |
× |
4-methylhydroatrovirinone (101249096) |
Yes (0) |
No (1) |
Yes (0) |
Yes (0) |
No (1) |
.55 |
High |
× |
Gentisein (5281635) |
Yes (0) |
Yes (0) |
Yes (0) |
Yes (0) |
Yes (0) |
.55 |
High |
√ |
Stigmasterol (5280794) |
Yes (1) |
No (3) |
Yes (0) |
No (1) |
No (2) |
.55 |
Low |
× |
2,6-dimethoxy-p-benzoquinone (68262) |
Yes (0) |
Yes (0) |
Yes (0) |
Yes (0) |
No (1) |
.56 |
High |
× |
According to toxicity analysis from pkCSM, 6 compounds found match with all categories. Atroviridin and gentisein have may cause mutagenic and carcinogenic activities, whereas no compounds show the inhibitor mechanism toward hERG I and II as well as toxicity against liver. On the other hand, the highest MRTD and LD50 found in kaempferol (0.531 log(mg/kg/day) and 2.449 mol/kg and quercetin (0.499 log(mg/kg/day) and 2.471 mol/kg (Table 3).
Table (3):
Toxicity prediction results using pkCSM
Compound |
AMES toxicity |
MRTD |
hERG I/II inhibitor |
LD50 |
Hepatotoxicity |
|---|---|---|---|---|---|
Dodecanoic acid |
No |
-0.340 |
No/No |
1.511 |
No |
Atroviridin |
Yes |
0.161 |
No/No |
1.918 |
No |
Naringenin |
No |
-0.176 |
No/No |
1.791 |
No |
Kaempherol |
No |
0.531 |
No/No |
2.449 |
No |
Quercetin |
No |
0.499 |
No/No |
2.471 |
No |
Gentisein |
Yes |
0.166 |
No/No |
2.135 |
No |
Molecular docking analysis was utilized to determine the binding affinity of chosen phytochemical compounds from G. atroviridis, selected native ligands, and selected synthetic drugs with E, C, NS2B/NS3, and NS5 DENV proteins. The lowest binding affinity of each ligand is projected to play significant and stable biological roles. The results revealed that atroviridin (-7.3, -8.0, and -7.8 kcal/mol) has the most stable binding affinity compared to the native ligands (-3.0, -3.5, and -3.7 kcal/mol) and synthetic drugs (-6.2, -6.2, and -6.6 kcal/mol) against E, NS2B/NS3, and NS5 proteins. Meanwhile, quercetin demonstrated favourable interaction with C protein (-8.6 kcal/mol) although it was still more positive than ST-148 (-8.9 kcal/mol) as synthetic drug (Table 4).
Table (4):
Docking analysis results against DENV proteins
| Compound | Binding affinity (kcal/mol) | |||
|---|---|---|---|---|
| E | C | NS2B/NS3 | NS5 | |
| Dodecanoic acid | -4.1 | -4.9 | -4.6 | -4.4 |
| Atroviridin | -7.3 | -8.5 | -8.0 | -7.8 |
| Naringenin | -6.7 | -7.8 | -7.4 | -7.0 |
| Kaempherol | -7.1 | -8.2 | -7.3 | -7.0 |
| Quercetin | -6.8 | -8.6 | -7.6 | -7.2 |
| Gentisein | -6.9 | -7.2 | -7.7 | -6.7 |
| NITD448 | -6.2 | |||
| ST-148 | -8.9 | |||
| ARDP0006 | -6.2 | |||
| SAH | -6.6 | |||
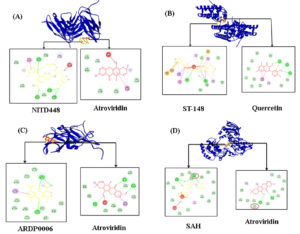
Visualization of ligand-target protein interactions were displayed with different stain of protein and ligand. Protein was labeled yellow and ligand as blue for control and red for selected phytochemical compound. Based on the molecular interaction, atroviridin has less biochemistry interaction against E than the NITD448 as control ligand even though the former one has lower binding affinity value. Both atroviridin and NITD448 do not interact with active sites and have unfavorable bonds that influence the ligand-protein complexes. On the other hand, ligand-capsid protein interactions advantaged ST-148 as control ligand. ST-148 has more interactions than quercetin from G. atrovridis. Quercetin as well as ST-148 does not show interaction toward active sites of capsid protein. Atroviridin has more chemical interaction including the unfavourable donor-donor interaction compared to the ARDP0006 as control. However, neither active sites interaction was formed from complexes against NS2B/NS3 by atroviridin nor ARDP0006. SAH (S-adenocylhomocysteine) as control drug of NS5 has unfavourable bump and donor-donor interactions. Meanwhile, both SAH and atroviridin made up same active site Asp663 of NS5 (Figure 1).
Figure 1. Visualization of docking analysis results against (A) envelop (E), (B) capsid (C), (C) NS2B/NS3, and (D) NS5 proteins. The pink circle indicates the same residue as the active site
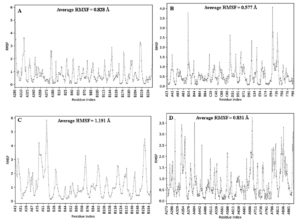
Molecular dynamics results showed that NS2B/NS3-atroviridin has the most flexible result with mean of its RMSF around 1.191 Å. On the other hand, C-quercetin displayed more stable contact with 0.577 for its average RMSF. Apart of that, E-atroviridin has with average RMSF 0.828ֵ and NS5-atroviridin stands with 0.831ֵ. Overall, the selected ligand-protein complexes have dominant RMSF value around 1-3 Å (Figure 2).
DENV protein is made up both structural and non-structural proteins.36 Envelop (E), capsid (C), and membrane proteins are included to the structural ones. Envelop is located outside the virus and contains three ectodomains and transmembrane segment.37 Thus, this protein is in charge to recognize immune cell. DENV via its E protein will attach to several host receptors such as heparin sulphate, b-integrin, and Dendritic Cell-Specific Intercellular adhesion molecule-3-Grabbing Non-integrin (DC-SIGN) or Cluster of Differentiation 2019 (CD209).38 Meanwhile, the capsid protein (C) is involved in crucial functions of multiple processes such as structural maintenance, virus assembly, and viral genome release.7 Recent study showed that C protein shuttles from and to the nucleus of infected cell.8 Moreover, both of the structural proteins can be used as key targets for drug development.6,8,39
Other protein group that discovered in DENV is non-structural protein. This group is classified to NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5 that are transcribed by 11,000 bases genome.40,41 NS2B, NS3, and NS5 are common proteins for classical targets dengue drug development.42 NS2B/NS3 is one of the primary targets of dengue antiviral development in recent years.43 It is trypsin-like serine protease which splits dengue polyprotein into the the separated proteins necessary for viral replication. NS3 plays essential roles for viral growth and maturation post-translation. It is boosted by NS3 that contributed as cofactor for cleavage process.9 Besides, NS5 is the most enormous non-structural protein (±100 kDa) and highly conserved in DENV genome. It has key play in innate immune response impairment during evasion as well as RNA synthesis, capping, and methylation from its N-terminal methyltransferase (MTase) and C-terminal RNA-dependent RNA polymerase (RdRp).10,11,44 This finding raises the possibility of developing antiviral targeting NS2B/NES3 and NS5 as DENV drugs.
G. atroviridis showed numerous health properties as spices and herbal plant. There are 38 phytochemical compounds found from various parts of this plant based on the prior studies.11 However, limited databases retrieved from PubChem webserver made this study only examine 24 compounds to asses for drug-likeness analysis. Based on the drug-likeness analysis, only 6 phytochemical compounds fulfilled the criteria. In order to analyze the toxicity, the filtered compounds passed with one or no violation of 5 criteria from pkCSM toxicity prediction. After that, selected compounds docked to investigate the interaction stability. The result showed that atroviridin was the most promising pyranoxanthone compounds as anti-DENV inhibits E, NSB/NS3, and NS5 proteins. The binding affinity scores were lower than the selective synthetic drug. Meanwhile, quercetin as a part of flavonoid compounds performed more positive result than ST-148 as the control for C protein. The lowest or the most negative binding affinity is needed to support the stability interaction during the cellular process and has capability as probable inhibitors.45
Atroviridin revealed as the most effective compound inhibitor of E protein compared to drug control. In comparison, atroviridin formed π-donor hydrogen bond with Asn390 from subunit A. There are other chemical interactions including hydrophobic (π-π stacked and π-alkyl) and van der Waals (vdw). Meanwhile, NITD448 demonstrated other chemical interactions including carbon hydrogen and conventional hydrogen bonds that known as the strongest chemical interaction.46 However, the halogen interaction of fluorine and chlorine made this complex satisfied. Halogen can sustain inter- and intramolecular ligand-protein interaction and affect molecular folding but it is weaker than hydrogen bonds. As a result, 20% of drugs that approved by Food and Drug Administration (FDA) were halogenated compounds.47 On the other hand, some of fluoride in biological interactions show that fluoride is highly oxidative that generate of reactive oxygen species, cell necrosis, and apoptosis.48 Because of that, the binding affinity of NITD448 had a higher binding than the atroviridin. Both of the compounds exhibited unfavorable donor-donor interactions and not form any interactions towards active sites (Figure 1). But they share same interaction with Trp391 of subunit A with multiple hydrophobic interactions to help inhibit virion recognition to host cell with aromatic and electron clouds.49
According to the docking analysis, ST-448 displayed the most negative one among other compounds. ST-148 had hydrogen (conventional), hydrophobic (π-σ, π-π T-shaped, amide-π stacked, alkyl, and π-alkyl), electrostatic (π-cation), and unfavorable interactions. There is miscellaneous interaction towards Trp69 from subunit B namely π-sulphur. This interaction provides aromatic compounds that interact with single sulphur atom.50 Sulphur gains specific function in biological activities consists folding stability and intermolecular interaction that larger than expected from vdw contacts.51 Another research reported that S-arene interactions were preferred over O-arene ones due to the non-covalent bonds.52 Meanwhile, quercetin formed only 3 hydrogen (conventional), 3 hydrophobic (amide-π stacked and π-alkyl), an electrostatic (π-anion), and a vdw interactions. Though quercetin did not possess unfavorable bonds, fewer groups of interaction provided more positive or less satisfied binding affinity via molecular docking.20 Overall, neither chemical interaction with active sites from atroviridin nor quercetin interacted more intensively with various amino acid and C protein subunits.
NSB2/NS3 shares co-dependency to activate protease activities in dengue evasion.42 ARDP0006 as control synthetic drug showed higher binding affinity than the atroviridin from G. atroviridis. It made this compound is preferred as anti-DENV drug candidate. Atroviridin has 3 conventional hydrogen (Lys74, Leu85, and Ala164), hydrophobic (both π-sigma and π-alkyl interact to Leu76), and one unfavorable donor-donor interaction (Trp83). Unfavourable interaction may indicate the presence of repulsive forces between ligand and target protein. It will affect to the more positive results after molecular screening.53-55 Next, ARDP0006 demonstrated only 2 kinds of interactions: conventional hydrogen bonds (2 interactions against Lys74 and one interaction against Trp83) and π-sigma interaction (Leu76). Atroviridin as well as ARDP0006 indicated interactions with amino acid residues from subunit B and share same receptor toward Lys74, Leu76, and Trp83 with various bonds. Despite the lack of contacts with catalytic triad and unfavourable bond, atroviridin still revealed best inhibitory activity across the different interactions.56
Based on the molecular docking, atrovridin also developed the most efficient inhibitor for another non-structural protein, NS5. Conventional hydrogen bonds and π-alkyl assist stabilization of ligand-protein complex and triggering inhibitory responds against genome replication and disabling the innate immunity.34 The results of control ligand showed the interaction against active site Asp663 via conventional hydrogen bonds. Besides, it possessed more hydrogen bonds and one hydrophobic interaction. Therefore, unfavourable donor-donor towards Ser710 made the binding affinity and desirable in computational study lower.54
Protein flexibility can be indicated by measuring the amplitude of atomic movements when it was simulated.57 In this analysis, the protein flexibility was represented by RMSF value. Molecular dynamics showed that the most effective inhibitors of selected phytochemical compounds from G. atroviridis are significantly stable based on the predominantly RMSF value that fall between 1-3 Å.58 However, several residue indexes from 4 selected ligand-target protein complexes demonstrated RMSF value >3 Å.
NITD448 is DENV fusion inhibitor that might attach to the ג-OG pocket in DENV E protein. But the antiviral effects only were observed during the initial viral entrance.59 Besides, ST-148 proposed to impede viral assembly and release range in DENV-1, -3, and -4. The antivirus potentially beneficial work occurred after post infection and post entry stage.38,60 Following that, ARDP0006 was identified suppress DENV-2 replication via virtual screening and cell culture.61 In addition, SAH in combination with sinefungin, compound 10, and guanosine monophosphate (GMP) failed to block NS5 with good progress due to the cell non-permeability.62 Some of the lack potential of synthetic antivirals above is required to address some of inhibitory functioned compounds towards specific target of DENV both targeting structural and non-structural proteins.38,44 G. atroviridis has the potentials to be the next natural anti-DENV drugs. In silico analysis through molecular docking revealed that atroviridin has the most effective potential against E, NS2B/NS3, and NS5 proteins. Furthermore, in vitro and in vivo analyses still required further to confirm anti-DENV efficiency.
G. atroviridis Griff. ex T. Anders showed anti-DENV properties. Its phytochemical compounds have been discovered as DENV antiviral by inhibiting E, C, NS2B/NS3, and NS5 proteins. Atroviridin has the most negative binding affinity to the E, NS2B/NS3, and NS5 proteins according to the docking analysis. Besides, quercetin showed the second most effective compound by binding to the C protein after the ST-148 potential. Molecular dynamic simulation demonstrated the stable results for those two compounds. Further wet laboratory researches are required to establish the properties efficacy as anti-DENV.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved the final manuscript for publication.
FUNDING
None.
DATA AVAILABILITY
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
ETHICS STATEMENT
Not applicable.
- Messina JP, Brady OJ, Scott TW, et al. Global spread of dengue virus types: mapping the 70 years history. Trends Microbiol. 2014;22(3):138-146.
Crossref - Panraksa P, Ramphan S, Khongwichit S, Smith DR. Activity of andrographolide against dengue virus. Antiviral Res. 2017;139:69-78.
Crossref - Hariono M, Choi SB, Roslim RF, et al. Thioguanine-based DENV-2 NS2B/NS3 protease inhibitors: virtual screening, synthesis, biological evaluation and molecular modelling. PLoS ONE. 2019;14(1):e0210869.
Crossref - Dengue virus type 1 isolate DENV5 polyprotein (POLY) gene complete cds; and sfRNA1 lncRNA gene, partial sequence. https://www.ncbi.nlm.nih.gov/nuccore/?term=denv5, Accessed 28 September, 2023.
- Power CN, Setzer WN. An in-silico investigation of phytochemicals as antiviral agents against dengue fever. Comb Chem High Throughput Screen. 2016;119(7):516-536.
Crossref - Wilder-Smith A, Ooi EE, Horstick O, Wills B. Dengue. Lancet. 2019;393(10169):P350-363.
Crossref - He Y, Wang M, Chen S, Cheng A. The role of capsid in the flaviviral life cycle and perspectives for vaccine development. Vaccine. 2020;38(44):6872-6881.
Crossref - Sallaberry I, Luszczak A, Philipp N, et al. In vivo pair correlation microscopy reveals dengue virus capsid protein nucleocytoplasmic bidirectional movement in mammalian infected cells. Sci Rep. 2021;11(1):24415.
Crossref - Kroneberger T, Serafim MSM, Tondoru AK, Maltarollo VG, Poso A. Ligand accessibility insights to the dengue virus NS3-NS2B protease assessed by long-timescale molecular dynamics simulations. Chem Med Chem. 2021;16(16):2524-2534.
Crossref - Klema VJ, Ye M, Hindupur A, et al. Deng ue virus nonstructural protein 5 (NS5) assembles into a dimer with a unique methyltransferase and polymerase interface. PLoS Pathog. 2016;12(2):1005451.
Crossref - Petit MJ, Kenaston MW, Pham OH, Nagainis AA, Fishburn AT, Shah PS. Nuclear dengue virus NS5 antagonizes expression of PAF1-dependent immune response genes. PLoS Pathog. 2021;17(11):e1010100.
Crossref - Shahid M, Law D, Azfaralariff A, Mackeen MM, Chong TF, Fazry S. Phytochemicals and biological activities of Garciniaatroviridis: a critical review. Toxics. 2022;10(11):656.
Crossref - Hamidon H, Susanti D, Taher M, Zakaria ZM. Garciniaatroviridis – A review on phytochemicals and pharmacological properties. Marmara Pharm J. 2017;21(1):38-47.
Crossref - Al-Mansoub MA, Asmawi MZ, Murugaiyah V. Effect of extraction solvents and plant parts used on the antihyperlipidemic and antioxidant effects of Garciniaatroviridis: A comparative study. J Sci Food Agric. 2014;94(8):1552-1558.
Crossref - Suwanmanee S, Kitisin T, Luplertlop N. In vitro screening of 10 edible Thai plants for potential antifungal properties. Evid Based Complement Alternat Med. 2014;138587.
Crossref - Mackeen MM, Mooi LY, Amran M, Mat N, Lajis NH, Ali AM. Noncytotoxic and antitumor – promoting activities of garcinia acid esters from Garcinia atroviridis Griff. ex T. Anders (Guttiferae). Evid Based Complement Alternat Med. 2012;892814.
Crossref - Muchtariadi, Nuwarda RF, Ikram EHK, Rahim ASA, Gazzali AM, Wahab HA. Neuraminidase inhibitor of Garcinia atroviridis L. fruits and leaves using partial purification and molecular characterization. Molecules. 2022;27(3):949.
Crossref - Wickasono A, Raihandhany P, Zen TV, et al. Rafflessia and Sapria metabolites using a bioinformatics approach to assess their potential drugs. Philipp J Sci. 2022;151(5):1771-1791.
Crossref - Christina YI, Nafisah W, Atho’illah MF, Rifa’I M, Widodo N, Djati MS. Anti-breast cancer potential activity of Phaleria macrocarpa (Scheff.) Boerl. leaf extract through in silico studies. J Pharm Pharmacog Res. 2021;9(6):824-845.
Crossref - Aini NS, Kharisma VD, Widyananda MH, et al. Bioactive compounds from purslane (Portulacaoleracea L.) and star anise (Illiciumverrum Hook.) as SARS-CoV-2 antiviral agent via dual inhibitor mechanism: in silico approach. Pharmacog J. 2022;14(4):352-357.
Crossref - Widyananda MH, Wicaksono ST, Rahmawati K, et al. A potential anticancer mechanism of finger root (Boesenbergia rotunda) extracts against a breast cancer cell line. Scientifica. 2022;9130252.
Crossref - Fan J, de Lannoy IAM. Pharmacokinets. Biochem Pharmacol. 2014;87(1):93-120.
Crossref - Pires DEV, Blundell TL, Ascher DB. pkCSM: predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures. J Med Chem. 2015;58(9):4066-4072.
Crossref - Nidom CA, Ansori ANM, Nidom AN, Indrasari S, Nidom RV. Curcumin from Curcuma longa L. as Dual Inhibitors Against Indonesian SARS-CoV-2 Isolates: A Molecular Docking Study. Pharmacognosy Journal. 2023; 15(1): 228-232.
Crossref - Yeni Y, Supandi S, Merdekawati F. In silico toxicity prediction of 1-phenyl-1-(quinazolin-4-yl) ethanol compounds by using Toxtree, pkCSM, and preADMET. Pharmaciana. 2022;8(2):205-216.
Crossref - Egbuna C, Patrick-Iwuanyanwu KC, Onyeike EN, Khan J, Alshehri B. FMS-like tyrosine kinase-3 (FLT3) inhibitors with better binding affinity and ADMET properties than sorafenib and gilteritinib against acute myeloid leukemia: in silico studies. J Biomol Struct Dyn. 2022;40(22):12248-12258.
Crossref - Chen G, Seukep AJ, Guo M. Recent advances in molecular docking for the research and discovery of potential marine drugs. Mar Drugs. 2020;18(11):545.
Crossref - Wargasetia TL, Ratnawati H, Widodo N, Widyananda MH. Bioinformatics study of sea cucumber peptides as antibreast cancer through inhibiting the activity of overexpressed protein (EGFR, PI3K, AKT1, and CDK4). Cancer Inform. 2021;20:11769351211031864.
Crossref - Aurora Y, Tarigan IPN, Suryanto NMM, Santosa P, Pricilla V, Parikesit AA. Identification of flavonoids of Kalanchoe pinnata as candidate drugs for COVID-19 gamma-varint treatment. Mal J Fund Appl Sci. 2022;18(6):630-643.
Crossref - Wijaya RM, Hafidzhah MA, Kharisma VD, Ansori ANM, Parikesit AA. COVID-19 in silico drug with Zingiber offcinale natural product compound library targeting the Mpro protein. Makara J Sci. 2021;25(3):162-171.
Crossref - Cockburn JJB, Sanchez MEN, Fretes N, et al. Mechanism of dengue virus broad cross-neutralization by a monoclonal antibody. Struct. 2012;20(2):303-314.
Crossref - Xia H, Xie X, Zou J, et al. A cocrystal structure of dengue capsid protein in complex of inhibitor. Proc Natl Acad Sci USA. 2020;117(30):17992-18001.
Crossref - Erbel P, Schiring N, D’Arcy A, et al. Structural basis for the activation of flaviviral NS3 proteases from dengue and West Nile virus. Nat StructMol Biol. 2006;13: 372-373.
Crossref - Kharisma VD, Probojati RT, Murtadlo AAA, Ansori ANM, Antonius Y, Tamam MB. Revealing potency of bioactive compounds as inhibitor of dengue virus (DENV) NSB2/NS3 protease from sweet potato (Ipomeabatatas L.) leaves. Indian J Forensic Med Toxicol. 2021;15(1):1627-1632.
Crossref - Yap TL, Xu T, Chhen Y-L, et al. Crystal structure of the dengue virus RNA-dependent RNA-polymerase catalytic domain at 1.85-angstrom resolution. J Virol. 2007;81(9):4753.
Crossref - Meng F, Badierah RA, Almehdar HA, Ridwan EM, Kurgan L, Uversky VN. Unstructural biology of the dengue virus proteins. Eur Biochem J. 2015;282(17):3368-3394.
Crossref - Reyes-Ruiz JM, Osuna-Ramos JF, Jesus-Gonzalez LAD, et al. Isolation and characterization of exosomes released from mosquito cells infected with dengue virus. Virus Res. 2019;266:1-14.
Crossref - Lee MF, Wu YS, Poh CL. Molecular mechanisms of antiviral agents against dengue virus. Viruses. 2023;15(3):705.
Crossref - Fadholly A, Ansori ANM, Proboningrat A, et al. Apoptosis of HeLa cells via caspase-3 expression induced by chitosan-based nanoparticle of Annona squamosal leaf extract: in vitro study. Indian J Pharm Edu Res. 2020;54(2):416-421.
Crossref - Rushika P, Richard JK. Structural proteomics of dengue virus. Curr Opin Microbiol. 2008;11(4):369-377.
Crossref - Kharisma VD, Ansori ANM, Nugraha AP. Computational study of ginger (Zingiber officinale) as E6 inhibitor in human papillomavirus type 16 (HPV-16) infection. Biochem Cell Arch. 2020;20(1):3155-3159.
- Wu H, Bock S, Snitko M, et al. Novel dengue virus NS2B/NS3 proteaase inhibitor. Antimicrob Agents Chemother. 2015;59(2):1100-1109.
Crossref - Lim SYM, Chieng JY, Pan Y. Recent insights on anti-dengue virus (DENV) medicinal plants: review on in vitro, in vivo and in silico discoveries. All Life. 2020;14(1):1-33.
Crossref - Obi JO, Gutierrez-Barbosa H, Chua JV, Deredge DJ. Current trends and limitations in dengue antiviral research. Trop Med Infect Dis. 2021;6(4):180.
Crossref - Wahyuni DK, Wacharasindhu S, Bankeeree W, et al. Molecular simulation of compounds from n-hexane fraction of Sonchus arvensis L. leaves as SARS-CoV-2 antiviral through inhibitor activity targeting strategic viral protein. J Pharm Pharmacog Res. 2022;10(6):1126-1138.
Crossref - Prahasanti C, Nugraha AP, Kharisma VD, et al. A bioinformatics approach of hydroxyapatite and polymethylmethacrylate composite exploration as dental implant biomaterial. J Pharm Pharmacog Res. 2021;9(5):746-754.
Crossref - Suarez-Castro A, Valle-Sanchez M, Cortes-Garcia CJ, Chacon- Garcia L. Molecular docking in halogen bonding. In: Molecular Docking. Intech Open. 2018;2018.99-114.
Crossref - Barbier O, Arreola-Mendoza L, Razo LMD. Molecular mechanisms of fluoride toxicity. Chem Biol Interact. 2010;188(2):319-333.
Crossref - Verma D, Mitra D, Paul M, et al. Potential inhibitors of SARS-CoV-2 (COVID-19) proteins PLPro&Mpro/3CLpro: molecular docking and simulation studies of three pertinent medicinal plant natural components. Curr Res Pharmacol Drug Discov. 2021;2:100038.
Crossref - Antonius Y, Ongko J, Hardjo PH. Identification of potential activity of volatile compounds derived from pogostemon Cablin benth as antiviral of SARS-CoV-2. Int J App Pharm. 2023; 15(1):93-97
- Beno BR, Yeung K-S, Bartberger MD, Pennington LD, Meanwell NA. A survey of the role of noncovalent sulfur interactions in drug design. J Med Chem. 2015;58(11):4383-4438.
Crossref - Tungary E, Ongko J, Sukweenadhi J. Molecular docking of active compounds from traditional medicinal plants as ACE-2 protein (1R4L) inhibitor in searching for COVID-19 drug. Res J Pharm Technol. 2022; 15(9): 4235-4240
- Wang YZ, Wu AX. π-π interaction in aromatic supramolecular system. Chinese J Org Chem. 2008;28(6):997-1011.
- Yang CY, Phillips JG, Stuckey JA, et al. Buried hydrogen bond interactions contribute to the high potency of complement factor D inhibitors. ACS Med Chem Lett. 2016;7(12):743.
Crossref - Odhar HA, Hashim AF, Humadi SS. Molecular docking analysis and dynamics simulation of salbutamol with the monoamine oxidase B (MAO-B) enzyme. Bioinformation. 2022;18(3):304-309.
Crossref - Padmi H, Kharisma VD, Ansori ANM, et al. Macroalgae bioactive compounds for the potential antiviral of SARS-CoV-2: an In silico study. J Pure Appl Microbiol. 2022;16(2):1018-1027.
Crossref - Kharisma VD, Agatha A, Ansori ANM, Widyananda MH, Rizky WC, Dings TGA, Derkho M, Lykasova I, Antonius Y, Rosadi I, Zainul R. Herbal combination from Moringa oleifera Lam. and Curcuma longa L. as SARS-CoV-2 antiviral via dual inhibitor pathway: A viroinformatics approach. J Pharm Pharmacogn Res. 2022; 10(1): 138-146
- Parikesit AA, Nurdiansyah R. Natural product repurposing of the H5N1-based lead compounds for the fit inhibitors against 3C-like protease of SARS-CoV-2. J Pharm Pharmacogn Res. 2021;9(5):730-745.
Crossref - Poh MK, Yip A, Zhang S, et al. A small molecule fusion inhibitor of dengue virus. Antiviral Res. 2009;84(3):260-266.
Crossref - Scatturo P, Trist IML, Paul D, et al. Characterization of the mode of action of a potent dengue virus capsid inhibitor. J Virol. 2014;88(19):11540-11555.
Crossref - Tomlinson SM, Watowich SJ. Anthracene-based inhibitors of dengue virus NS2B-NS3 protease. Antiviral Res. 2011;89(2):127-135.
Crossref - Lim SP, Noble CG, Shi P-Y. The dengue virus NS5 protein as a target for drug discovery. Antiviral Res. 2015;119:57-67.
Crossref
© The Author(s) 2023. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.