ISSN: 0973-7510
E-ISSN: 2581-690X
Benzene is an omnipresent aromatic hydrocarbon of the environment and the pollution caused by it is a matter of great public concern. The study was designed to isolate, characterize and identify aerobic bacterial strains capable of benzene degradation from petroleum-contaminated soils of Kolkata, India. Three strains, designated as MKB1b, MKB2a and MKB2d were found to be able to degrade benzene, as the only source of carbon and energy. All the isolated strains had an optimal growth pH of 8.0, and grew best at 37°C to 40°C. Based on their molecular (16S rDNA sequence) characterization, all of the three bacterial strains were phylogenetically similar to the genus Escherichia. Strains MKB2a and MKB2d were identified as Escherichia coli, whereas Strain MKB1b was identified as Escherichia fergusonii; all three sequences were submitted to GenBank bearing accession numbers MK970556, MK970574, MK970557 respectively. This study is the first evidence of isolation, characterization and identification of aerobic benzene-degrading Escherichia spp. from petroleum-contaminated sites of an urban landscape in India. All of these isolated strains may be considered as potent candidates for the bioremediation of urban environment polluted with petroleum products.
Escherichia, aerobic biodegradation, aromatic hydrocarbon, petroleum-contaminated soil.
Benzene (formula C6H6, MW 78); a hydrophobic, volatile, colourless, highly flammable, aromatic hydrocarbon; is a preeminent component of crude as well as refined petroleum and most (98%) of it is commercially used as a resource for petrochemical and chemical industries, petroleum refining industries, and for manufacturing synthetic rubbers, tyres, gums, lubricants, pharmaceuticals, pesticides and other agricultural chemicals, plastics, dyes, polymers, resins, synthetic fibres and consumer products such as marking pens, glues, adhesives, thinners, shoe-polish, perfumes, after shave lotions and paints1,2. Benzene is a major component of petrol, diesel, gasoline and other automobile fuels. After glucosyl residues, benzene ring is globally regarded as the most extensively distributed chemical structure3.
Benzene enters into the human systems by cutaneous exposure or by inhalation4. The human population is exposed to benzene because of its presence in the proximity of benzene utilizing industries and gas stations, and in areas of high vehicular traffic. Cigarette smokers are also directly exposed to benzene5,6. The relation of benzene exposure and the concomitant risk of cancer formation in human tissues and organs have been well-documented7. The toxic effects of benzene have also been related to developing hematopoietic disorders like acute myeloid leukemia, lymphocytic leukemia and non-Hodgkin’s lymphoma8,9, primarily caused by chromosomal alterations attributing to malignant transformation of the genome9,10. It has been also suggested that a number of epigenetic alterations, mostly DNA methylation, may also play a role in formation of tumor12.
Remediation of groundwater contaminated with benzene and other aromatic hydrocarbons has been proven to be an arduous task, due to the fact that these compounds are recalcitrant, water-insoluble and have the ability of diffusing rapidly once they are introduced into the aquifer system13. Mineralization of benzene has been a matter of priority for nearly three decades and microbial bioremediation of the same has been reported under aerobic conditions by genera, Nitrosomonas14, Pseudomonas15,16,17, Acinetobacter18, Lysinibacillus19 as well as in the presence of other electron acceptors by genera, Geobacter20, Dechloromonas21, Desulfobacterium22. Studies on bacterial mineralization of aromatic compounds like benzene have been focused, and microorganisms capable of benzene biodegradation under aerobic conditions have been isolated from industrial areas, and areas adjoining oil fields and oil-refineries22–24. Exploration of aromatic hydrocarbon degrading microbial diversity in and around urban habitats in India as potential sources of bioremediation is severely lacking.
This study has been designed to explore the microbial diversity capable of benzene degradation, in pure culture, as the sole carbon and energy source in petroleum-contaminated sites located in Kolkata, India. Characterization of the microbes as well as assessment of environmental conditions favouring bioremediation of benzene were also studied.
Sample collection
Six gas stations situated in Kolkata, West Bengal, India (22.57° N, 88.36° E) were used as sites for collection of petroleum-mixed soil samples. Around 100 gm of soil from each site was collected in labeled, sterile glass containers from a depth of around 10 cm from top soil-surface and were aseptically transferred to the laboratory for further analysis. Microbiological investigations of all the samples were initiated strictly within 12 hours of collection.
Reagents and Culture Media
Microbial culture media mentioned in the experiments were procured from HIMEDIA (India), whereas, aromatic hydrocarbons were procured from Sigma Aldrich (USA)and were of HPLC grades. Pure cultures were obtained, and growth experiments were carried out with sterile nutrient agar and Luria agar medium used after sterilization at 120°C temperature and 105 K Pa pressure, for 15 minutes. Sterile Minimal salt media (MSM), having the composition of Na2HPO4, 3.61 g; (NH4)2SO4, 2g; KH2PO4, 1.75g; MgSO4.7H2O, 0.2g; CaCl2.H2O, 50mg; FeSO4.7H2O, 1mg; ZnSO4. 7H2O, 70µg; CuSO4.5H2O, 50µg; H3BO3, 10µg; MgSO4.5H2O,10µg; MoO3, 10µg; pH 7.0, was employed as a source of essential macro and micro-nutrients for culture of bacterial strains with aromatic hydrocarbons. Benzene was added to MSM, as and when required, after filter sterilization with 0.22µ bacteriological filter.
Isolation, purification and characterization of benzene-degrading strains
From each labeled sample, 5 gm of soil was inoculated in MSM (1L), supplemented with (1% v/v) n-hexadecane and incubated at 37°C temperature, under shaking condition (120 rpm) until the occurrence of visible growth. Inoculum from this MSM-n-hexadecane culture (10-4 to 10-8 dilutions) were then spread in MSM agar (2% w/v), supplemented with (2% v/v) n-hexadecane and once again incubated at 37°C temperature. After 72 hours of incubation, the bacterial isolate / colony was checked for purity by photomicrography with a Zeiss Axio Scope A1 Microscope, as well as spreading them on sterile Luria Agar plates. All the isolated and purified bacterial strains were stored both as nutrient agar stab and 15% glycerol stock at 4°C and –80°C, respectively. Strains capable of utilizing n-hexadecane as the sole carbon source were then screened for their aerobic benzene degradation ability by the following method. Each of the strains was inoculated into 15 ml MSM supplemented with benzene (1% v/v), and incubated at 37°C, at 120 rpm in an orbital shaker for 7 days. Cultures with visible growth were again spread onto MSM agar plates supplemented with benzene (1% v/v) in serially diluted suspensions (10-4 to 10-8 dilutions). Well-isolated colonies were chosen from these plates based on their cultural characteristics like, colour of surface, shape, margin, texture and elevation using single colony isolation procedure. An array of biochemical tests were performed to characterize the strains according to Bergey’s Manual of Systematic Bacteriology26.
Antibiogram test
Antibiogram or antibiotic susceptibility of the isolated strains was executed by the disk diffusion method27 on Mueller-Hinton agar (MHA) plates with antimicrobial susceptibility test disks (HiMedia) of 10 antimicrobial drugs. Bacterial isolates were analyzed for susceptibility, reduced susceptibility, or resistance towards an antimicrobial drug based on the inhibitory zone diameter matching the manufacturer’s interpretive table’s criteria, following the recommendations of the National Committee for Clinical Laboratory Standards28. Escherichia coli ATCC 25922 strain was used for quality control (qc).
Measurement of temperature and pH optima
Temperature and pH optima of the strains were tested to evaluate the culture conditions needed for most effective aerobic bioremediation of benzene. For testing the pH optima, each isolated strain was grown in six tubes each containing 5 ml of sterile nutrient broths (having pH varying from 2 to 9) and 15 x 107 ml-1 cells from overnight culture were added and incubated at 37°C temperature, at 120 rpm in an orbital shaker.
Temperature optima of each isolates were evaluated by inoculating 15 x 107 ml-1 cells from overnight cultures to six tubes each containing 5 ml of sterile nutrient broths (pH 7) and incubated at temperatures of 25°, 30°, 35°, 37°, 40° and 45°C in an orbital shaker at 120 rpm for 12 hours.
In both experiments, cell density of each tube, after 12 hours of incubation, was measured spectrophotometrically at 660 nm by a SYSTRONICS 2202 Double Beam Spectrophotometer.
Molecular identification of the isolated strains
Genomic DNA of the strains were isolated as described by Sambrook et. al., 2001 29 with the following modifications. Cells were harvested from 1.5 ml of the overnight Luria Broth culture and resuspended in 567µl TE Buffer (10mM Tris, 1mM EDTA), pH 8.0. After addition of 30µl 10% SDS and incubation for 1 hour at 37 °C, 100µl of 5M NaCl was added. After addition of 80µl of cetyltrimethyl ammonium bromide (CTAB)/NaCl, and after mixing vigorously, the mixture was incubated at 65°C for 10 minutes. After extraction with equal volume of chloroform: isoamyl alcohol (24:1) and another extraction with equal volume of phenol: chloroform: isoamyl alcohol, DNA was precipitated with isopropyl alcohol, washed with chilled 70% ethanol, dried and dissolved in appropriate volume of TE buffer, pH 8.0.
The 16S rRNA gene was amplified by PCR (Perkin-Elmer Applied Biosystems, Weiterstadt, Germany) reaction mixtures, with template DNA (1µg); forward primer 27f (5′-AGAGTTTGATCMTGGCTCAG-3′), reverse primer 1492r (5′-GGTTACCTTGTTACGACTT-3′), both 1µl from 10 pmol/µl working solution; 25µl PCR Master Mix (Thermo Scientific, USA); volume made up to 50µl with sterile nuclease-free water. PCR products were purified by GeneJET PCR Purification Kit (Thermo Scientific, USA), electrophoresed in 1.5% (w/v) agarose gel and were visualized over a transilluminator after ethidium bromide staining. Sequencing of the purified amplicons were done by a paid sequencing facility from Xceleris (Ahmedabad, India).
Phylogenetic analysis of the strains
For the identification and phylogenetic analyses of the strains the sequences obtained were queried in BLASTn software and top ten hits were selected for each strain for further downstream tests. CLUSTALW was used to align these sequences and for annotation purposes. The results were then subjected to phylogenetic analysis using Maximum Likelihood method and Jukes-Cantor model30. Initial trees for the heuristic search were obtained with the Neighbor-Joining method forming a pairwise distance matrix predicted using the Maximum Composite Likelihood (MCL) approach31. Branch lengths of the tree is determined by the number of base substitutions per site. All in silico analyses were done on MEGA X software32. The final sequences were then subjected to test for potential chimeras using DECIPHER chimera check software33 and were then submitted to GenBank.
Benzene biodegradation assay
Aerobic degradation of benzene by the isolated bacterial strains along with their time-course study were done as according to Mukherjee et. al., 201034 with following modifications. In sterile 15 ml Teflon-lined capped tubes, inocula (15×107 cells ml-1 from a 12 hour old culture, washed in sterile PBS, pH 7.0) were added into MSM overlaid with HPLC grade benzene (100µ Mole, added from filter-sterilized stock solutions) ensuring enough head-space for the maintenance of aerobic conditions, and incubated at 37°C temperature, 120 rpm in an orbital shaker-incubator. MSM with 100µM benzene supplemented with heat-killed (by 15 minutes incubation at 70°C) cells of each strains and 15×107 cells ml-1 of each strain in MSM, not supplemented with benzene were used at negative controls (Control 1 and Control 2 respectively). A week-long experiment was set in the manner and cell numbers were determined at a gap of 24 hours by centrifugation of the cultures at 8000 rpm for 10 min, washing cell pellets twice with sterile PBS, resuspension in sterile PBS and direct microscopic count under oil immersion.
After each 24 hours of biodegradation, samples of each tube were extracted with 10 ml of HPLC grade chloroform. Concentrations of benzene in each culture was evaluated by high performance liquid chromatography (HPLC) as described previously21, using a Waters model 600 HPLC system equipped with Waters 2996 Photodiode Array Detector (Milford, MA, USA), at 254 nm. HPLC analyses were done isocratically, at room temperature with a mobile phase of methanol: water 60:40 (v/v) at a flow rate of 1 mL min-1, by an analytical column (Symmetry-C18 5µm, 4.6mm i.d. X 25 cm long, Waters, USA).
Statistical analysis of data
Data values are expressed as [mean values ± standard deviation (S.D.)] from at least three replicates of experiments. Differences among sets were considered significant at (P³ 0.05) level. SPSS 17.0 (SPSS Inc., Chicago, USA) was used to implement statistical analyses and to compare means by Student’s t-test.
Identification of strains and determination of pH and temperature optima
Three bacterial strains were selected for further studies, among 16 isolates capable of utilizing n-hexadecane as sole carbon and energy source, due to their ability of growing in MSM, supplemented with benzene. Results of morphological and biochemical characterization of these three Gram-negative, rod-shaped facultative anaerobic bacterial strains are enlisted in Table 1. MKB1b was found to be the only non-motile bacteria among the three. The three strains exhibited nearly similar biochemical characteristics but differed in their ability in acid production from lactose and sucrose.
Table (1):
Morphological and biochemical characters of the three bacterial isolates obtained from petroleum contaminated soil samples in Kolkata.
| MKB1b | MKB2a | MKB2d | |
|---|---|---|---|
| Morphological Characteristics | |||
| Colony character | Translucent with opaque centre, greyish-white, smooth | Translucent with opaque centre, off-white, smooth, thick | Translucent with opaque centre, colourless, smooth |
| Cell morphology | Rod-shaped | Rod-shaped | Rod-shaped |
| Gram character | – | – | – |
| Motility at 37ºC | + | + | + |
| Spore formation | – | – | – |
| Biochemical characteristics | |||
| Urease | – | – | – |
| DNase | – | – | – |
| Oxidase | – | – | – |
| Voges−Proskauer | – | – | – |
| Gelatinase | – | – | – |
| Methyl Red | + | + | + |
| Indole | + | + | + |
| H2S production | – | – | – |
| Citrate | – | – | – |
| Nitrate reduction | + | + | + |
| Acid formation from | |||
| Lactose | – | + | + |
| Maltose | + | + | + |
| Glucose | + | + | + |
| Mannitol | + | + | + |
| Sucrose | – | – | + |
The three strains were identified and they all belong to the genus Escherichia. Strain MKB2a and MKB2d were identified as E. coli, whereas Strain MKB1b was identified as E. fergusonii. 16S Ribosomal DNA sequences of all three strains were submitted to the GenBank bearing accession numbers MK970556, MK970574, MK970557 respectively. The phylogenetic tree constructed is depicted in Fig. 1. The tree shows the phylogenetic position of the three strains when compared with their nearest neighbours from BLASTn hits. The three strains belong to the same lineage but differs in the strain level as elucidated by the difference in the branch lengths.
Fig. 1. The phylogenetic tree was constructed by using the Maximum Likelihood method and Jukes-Cantor model. The tree with the highest log likelihood (-1546.11) is shown. Initial tree(s) for the heuristic search were obtained by applying the Neighbor-Joining method to a matrix of pairwise distances estimated using the Maximum Composite Likelihood (MCL) approach. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. This analysis involved 30 nucleotide sequences. There was a total of 875 positions in the final dataset. Evolutionary analyses were conducted in MEGA X.
Among the ten antibiotics tested; MKB1b, MKB2a and MKB2d showed resistance to 5, 3 and 3 antibiotics respectively. All the strains showed resistance to Ampicillin (10µg), Erythromycin (15µg) and Vancomycin (30µg) (Table 2).
Table (2):
Antibiogram test results of the isolated Escherichia strains from petroleum contaminated soil samples in Kolkata, using 10 antimicrobial drugs.
Sensitive |
Resistant |
|
|---|---|---|
MKB1b |
S, Of, K, C, G |
A, Co, E, T, Va, |
MKB2a |
S, Of, K, T, C, Co, G |
A, E, Va |
MKB2d |
S, Of, K, T, C, Co, G |
A, E, Va |
A, Ampicillin (10µg); C, Chloramphenicol (30µg); Co, Cotrimoxazole (25µg); E, Erythromycin (15 µg); G, Gentamycin (10 µg); K, Kanamycin (30 µg); Of, Ofloxacin (5 µg); S, Streptomycin (10µg); T, Tetracycline (30 µg); Va, Vancomycin (30 µg)
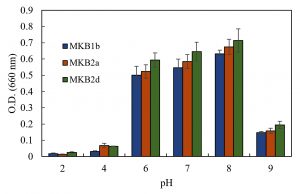
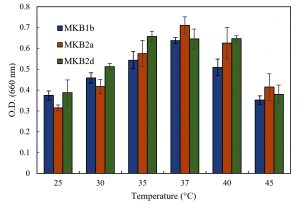
Strains MKB1b, MKB2a and MKB2d showed considerable growth within a range of pH 6.0 to 8.0, having the maximum growth at pH 8.0 (Fig. 2). All the strains exhibited better growth at an alkaline environment (pH 9.0) than in an acidic one (pH 4.0 and 2.0). All the three strains were able to grow at the range of temperature tested, from 25°C to 45°C. Optimum temperature for growth of the strains MKB1b and MKB2a was found to be at 37°C, whereas, MKB2d showed nearly equal growth pattern at both 37°C and 40°C (Fig. 3).
Fig. 2. Growth of MKB1b, MKB2a and MKB2d in enrichment media at a pH range of 2 to 9, after 12 hours of incubation at 37°C, 120 rpm.
Fig. 3. Growth of MKB1b, MKB2a and MKB2d in enrichment media at six temperatures within a range of 25° to 45°C, after 12 hours of incubation at 120 rpm.
Aerobic biodegradation of Benzene
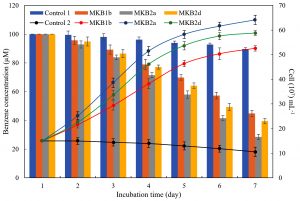
Aerobic benzene biodegradation of strains MKB1b, MKB2a and MKB2d were carried out at their optimum pH and temperature with 100µM benzene and 15×107 cells ml-1 respectively. Depletion of benzene in the heat-killed inoculum (Control 1) was negligible (P < 0.01), and 89.66 ± 0.87µM benzene remained undegraded in the culture, whereas, cell number (10.57×107 ± 1.89 ml-1) in the culture without added benzene did not vary much (P < 0.01) over an incubation period of 7 days (Fig. 4). Strain MKB2a exhibited the most efficient benzene degrading ability of the three, and could degrade benzene up to a final concentration of 28.48 ± 2.17µM after 7 days. In case of the other two strains, 44.82 ± 1.84µM and 39.55 ± 1.87µM of benzene remained unexhausted in the cultures of MKB1b and MKB2d respectively. Bacterial cell density in media, containing benzene as sole carbon and energy source, increased to values of (52.58 ± 0.98), (64.11 ± 1.12) and (58.78 ± 1.88) x 107 cells ml-1 from an initial density of 15×107 cells ml-1 in cultures containing MKB1b, MKB2a, and MKB2d respectively.
Fig. 4. Growth curve and aerobic benzene degradation by MKB1b, MKB2a and MKB2d over a time period of 7 days. Bars and broken lines represent benzene concentration (µM) in culture media and cell number (x 107 ml-1) respectively. MSM added with benzene, inoculated with heat-killed cells and MSM with live cells, but with no added benzene are termed as Control 1 and Control 2 respectively. [n= 3, P ≤ 0.05]
E. coli is the most abundant and successful bacterial flora in the gut ecosystem of warm-blooded animals and is a pathogen of the enteric, urinary, pulmonary, and nervous systems also35. However, this commensal can also thrive in extracorporeal environment36 and has the ability to withstand ample range of physico-chemical conditions, enabling it as a successful survivor in various environments over long periods of time37. E. coli, can also survive and replicate in environments having an abundance of aromatic hydrocarbons; and can actually utilize the aromatic compounds as sources of carbon. On the other hand, E. fergusonii has previously been documented to be able to degrade herbicides like, atrazine and diuron38 and diesel oil39,40.
This is a novel report of aerobic degradation of benzene by E. coli and E. fergusonii strains, isolated from petroleum-contaminated sites in the densely populated city of Kolkata, West Bengal, India. Earlier reports of bacterial species capable of mineralizing benzene and other aromatic hydrocarbons, had been concentrated around oil-field and oil-refineries only41-43. In pure culture, benzene biodegradation have been reported by a number of aerobic bacterial genera like, Geobacter44, Desulfobacterium45, Dechloromonas21 and Bacillus46; and anaerobic genera like, Nitrosomonas 14, Pseudomonas16,17 and Acinetobacter 18. In genus Escherichia, degradation of aromatic compounds like, hydroxyphenyl acetic acid, 3-hydroxyphenylpropionic acid; 3-hydroxycinnamic acid; and phenylacetic acid have been comprehensively studied 47.
E. coli and E. fergusonii strains described in this study could degrade benzene as the sole carbon and energy source, in pure culture, up to a concentration of 71.52µM over a period of 7 days under aerobic conditions. In enrichment studies, all of the strains showed maximum proliferation at a temperature range of 37°C to 40°C and in alkaline pH 8.0, making them potential bio-remediators of benzene-contaminated soils. Further insights on the mechanism of benzene mineralization, characterization of the genes involved as well as extensive investigations of urban microbial diversity are warranted to utilize as proficient candidates against aromatic hydrocarbon pollution.
ACKNOWLEDGMENTS
Infrastructural assistance obtained from Central Instrument Centre (CIC) and CIC Imaging, Department of Life Sciences, Presidency University, Kolkata and the Department of Zoology, University of Burdwan is gratefully acknowledged. This communication contains data from a thesis to be submitted for the partial fulfilment of the Degree of Doctor of Philosophy in the Department of Zoology, The University of Burdwan, West Bengal, India.
CONFLICT OF INTEREST
The authors confirm that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
SM performed all the laboratory experiments, analysis of data and manuscript writing. AD was involved in field sampling and molecular phylogenetic studies. NKS and NCS conceived the research idea, oversaw experiments and finalized the manuscript.
FUNDING
The work has been partially funded by the Faculty Research and Development Fund (FRPDF), Presidency University, Kolkata, allocated to SM.
ETHICS STATEMENT
This article does not contain any studies with human participants or animals performed by any of the authors.
AVAILABILITY OF DATA
Data supporting the present study are available from the first and corresponding authors upon justifiable request. 16S rDNA sequences of the bacterial strains are accessible at GenBank (https://www.ncbi.nlm.nih.gov/genbank/) bearing accession numbers MK970556, MK970574 and MK970557.
- Jindrova A, Chocova M, Demnerova K, Brenner V. Bacterial aerobic degradation of benzene, toluene, ehtylbenzene, and xylene. Folia Microbiologica, 2002; 47: 83–93.
Crossref - Smith MT. Advances in Understanding Benzene Health Effects and Susceptibility. Annual Review of Public Health, 2010; 31: 133–148.
Crossref - Song J, Zhao Q, Guo J, Yan N, Chen H, Sheng F, Lin Y, An D. The microbial community responsible for dechlorination and benzene ring opening during anaerobic degradation of 2,4,6 trichlorophenol. Science of The Total Environment, 2019; 651: 1368–1376.
Crossref - Brandt L. Exposure to organic solvents and risk of haematological malignancies. Leuk. Res., 1992; 16: 67–70.
Crossref - Ray MR, Roychoudhury S, Mukherjee S, Lahiri T. Occupational benzene exposure from vehicular sources in India and its effect on hematology, lymphocyte subsets and platelet P-selectin expression. Toxicology and Industrial Health, 2007; 23: 167–175.
Crossref - A.K, M.S.J, E. P, J.J.K.J. Exposure to benzene at work and the risk of leukemia: A systematic review and meta-analysis. Environmental Health: A Global Access Science Source, 2010; 9: 1–8.
Crossref - Falzone L, Marconi A, Loreto C, Franco S, Spandidos DA, Libra M. Occupational exposure to carcinogens: Benzene, pesticides and fibers (Review). Molecular Medicine Reports, 2016; 14: 4467–4474.
Crossref - McHale CM, Zhang L, Smith MT. Current understanding of the mechanism of benzene-induced leukemia in humans: implications for risk assessment. Carcinogenesis, 2012; 33: 240–52.
Crossref - World Heatlh Organization T. Exposure to Benzene: A Major Public Health Concern. Available at http://www.who.int/ipcs/features/benzene.pdf.
- Fenga C, Gangemi S, Costa C. Benzene exposure is associated with epigenetic changes (Review). Molecular Medicine Reports, 2016; 13: 3401–3405.
Crossref - Holeckova B, Piesova E, Sivikova K, Dianovsky J. Chromosomal aberrations in humans induced by benzene. Ann. Agric. Environ. Med., 2004; 11: 175–179.
- Bahadar H, Mostafalou S, Abdollahi M. Current understandings and perspectives on non-cancer health effects of benzene: A global concern. Toxicology and Applied Pharmacology, 2014; 276: 83–94.
Crossref - Meckenstock RU, Boll M, Mouttaki H, Koelschbach JS, Cunha Tarouco P, Weyrauch P, Dong X, Himmelberg AM. Anaerobic degradation of benzene and polycyclic aromatic hydrocarbons. Journal of Molecular Microbiology and Biotechnology, 2016; 26: 92–118.
Crossref - Hyman MR, Sansome-Smith AW, Shears JH, Wood PM. A kinetic study of benzene oxidation to phenol by whole cells of Nitrosomonas europaea and evidence for the further oxidation of phenol to hydroquinone. Archives of Microbiology, 1985; 143: 302–306.
Crossref - Chang M-K, Voice TC, Criddle CS. Kinetics of competitive inhibition and cometabolism in the biodegradation of benzene, toluene, and p -xylene by two Pseudomonas isolates. Biotechnology and Bioengineering, 1993; 41: 1057–1065.
Crossref - Shirai K. Catechol Production from Benzene through Reaction with Resting and Immobilized Cells of a Mutant Strain of Pseudomonas. Agricultural and Biological Chemistry, 1987; 51: 121–128.
Crossref - Mukherjee S, Bardolui N.K., Karim S, Patnaik V.V., Nandy RK, Bag PK. Isolation and characterization of a monoaromatic hydrocarbon-degrading bacterium, Pseudomonas aeruginosa from crude oil. Journal of Environmental Science and Health – Part A Toxic/Hazardous Substances and Environmental Engineering, 2010.
Crossref - Iqbal A, Arshad M, Hashmi I, Karthikeyan R, Gentry TJ, Schwab AP. Biodegradation of phenol and benzene by endophytic bacterial strains isolated from refinery wastewater-fed Cannabis sativa. Environmental Technology (United Kingdom), 2018; 39: 1705–1714.
Crossref - Sciences B, Science F, Box PO. Isolation and Molecular Identification of New Benzene Degrading Lysinibacillus Strains from Gasoline Contaminated Soil Fawzi I. Irshaid and Jacob H. Jacob., 2016; 8: 34–43.
Crossref - Rooney-Varga JN, Anderson RT, Fraga JL, Ringelberg David, Lovley DR. Microbial Communities Associated with Anaerobic Benzene Degradation in a Petroleum-Contaminated Aquifer. ; 1999: 1-3056–3063. Available at http://aem.asm.org/. Accessed August 11, 2018.
- Coates JD, Chakraborty R, Lack JG, O’Connor SM, Cole KA, Bender KS, Achenbach LA. Anaerobic benzene oxidation coupled to nitrate reduction in pure culture by two strains of Dechloromonas. Nature, 2001; 411: 1039–1043.
Crossref - Da Silva MLB, Alvarez PJJ. Assessment of anaerobic benzene degradation potential using 16S rRNA gene-targeted real-time PCR. 2006.
Crossref - Vogt C, Kleinsteuber S, Richnow HH. Anaerobic benzene degradation by bacteria. Microbial Biotechnology, 2011; 4: 710–724.
Crossref - Meckenstock RU, Boll M, Mouttaki H, Koelschbach JS, Cunha Tarouco P, Weyrauch P, Dong X, Himmelberg AM. Anaerobic degradation of benzene and polycyclic aromatic hydrocarbons. Journal of Molecular Microbiology and Biotechnology, 2016; 26: 92–118.
Crossref - Smith MR. The biodegradation of aromatic hydrocarbons by bacteria. 1990; 191–206.
Crossref - Boone DR, Castenholz RW, Garrity GM. Bergey’s manual of systematic bacteriology. Springer; 2001.
Crossref - Buzon-Duran L, Capita R, Alonso-Calleja C. Antibiotic susceptibility of methicillin-resistant staphylococci (MRS) of food origin: A comparison of agar disc diffusion method and a commercially available miniaturized test. Food Microbiology, 2018; 72: 220–224.
Crossref - Kiehlbauch JA, Hannett GE, Salfinger M, Archinal W, Monserrat C, Carlyn C. Use of the National Committee for Clinical Laboratory Standards guidelines for disk diffusion susceptibility testing in New York state laboratories. Journal of Clinical Microbiology, 2000; 38: 3341–8.
- Sambrook J, Russell DW (David W. Molecular cloning/ : a laboratory manual) . Cold Spring Harbor Laboratory Press; 2001.
- Som A. Theoretical foundation to estimate the relative efficiencies of the Jukes–Cantor+gamma model and the Jukes–Cantor model in obtaining the correct phylogenetic tree. Gene, 2006; 385: 103–110.
Crossref - Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol., 1987; 4: 406–425.
- Tamura K, Nei M, Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. USA. 2004; 101: 11030–11035.
Crossref - Wright ES, Yilmaz LS, Noguera DR. DECIPHER, a search-based approach to chimera identification for 16S rRNA sequences. Applied and Environmental Microbiology, 2012; 78: 717–725.
Crossref - Mukherjee S, Bardolui NK, Karim S, Patnaik VV, Nandy RK, Bag PK. Isolation and characterization of a monoaromatic hydrocarbon-degrading bacterium, Pseudomonas aeruginosa from crude oil. Journal of Environmental Science and Health – Part A Toxic/Hazardous Substances and Environmental Engineering, 2010; 45.
Crossref - Di E, Ferrandez A, Prieto MA, Garci L, Ferra A. Biodegradation of Aromatic Compounds by Escherichia coli Biodegradation of Aromatic Compounds by Escherichia coli. Society, 2001; 65: 523–569.
Crossref - Jang J, Hur H-G, Sadowsky MJ, Byappanahalli MN, Yan T, Ishii S. Environmental Escherichia coli: ecology and public health implications-a review. Journal of Applied Microbiology, 2017; 123: 570–581.
Crossref - Boyte S, Quaife S, Horswell J, Siggins A. Survival of Escherichia coli in common garden mulches spiked with synthetic greywater. Letters in Applied Microbiology, 2017; 64: 386–391.
Crossref - Moretto JAS, Braz VS, Furlan JPR, Stehling EG. Plasmids associated with heavy metal resistance and herbicide degradation potential in bacterial isolates obtained from two Brazilian regions. Environmental Monitoring and Assessment. 2019; 191: 314.
Crossref - Pasumarthi R, Chandrasekaran S, Mutnuri S. Biodegradation of crude oil by Pseudomonas aeruginosa and Escherichia fergusonii isolated from the Goan coast. Marine Pollution Bulletin, 2013; 76: 276–282.
Crossref - Sriram MI, Gayathiri S, Gnanaselvi U, Jenifer PS, Mohan Raj S, Gurunathan S. Novel lipopeptide biosurfactant produced by hydrocarbon degrading and heavy metal tolerant bacterium Escherichia fergusonii KLU01 as a potential tool for bioremediation. Bioresource Technology, 2011; 102: 9291–9295.
Crossref - Maiti A, Das S, Bhattacharyya N. Bioremediation of High Molecular Weight Polycyclic Aromatic Hydrocarbons by Bacillus thuringiensis Strain NA2. ; 2012. Available at www.worldsciencepublisher.org. Accessed July 29, 2018.
- Mukherjee AK, Bordoloi NK. Biodegradation of benzene, toluene and xylene ( BTX ) in liquid culture and in soil by Bacillus subtilis and Pseudomonas aeruginosa strains and a formulated bacterial consortium. 2012; 3380–3388.
Crossref - Taylor P, Basak SP, Sarkar P, Pal P. Journal of Environmental Science and Health , Part A/ : Toxic / Hazardous Substances and Environmental bacteria from industrial effluent-contaminated soil and Isolation and characterization of phenol utilizing bacteria from industrial effluent-contaminated. 2014;37–41.
- Rooney-Varga JN, Anderson RT, Fraga JL, Ringelberg David, Lovley DR. Microbial Communities Associated with Anaerobic Benzene Degradation in a Petroleum-Contaminated Aquifer. ; 1999: 1-3056–3063. Available at http://aem.asm.org/. Accessed August 6, 2018.
- Jensen A-M, Finster KW, Karlson U. Degradation of carbazole, dibenzothiophene, and dibenzofuran at low temperature by Pseudomonas sp. strain C3211. Environmental Toxicology and Chemistry, 2003; 22: 730–735.
Crossref - Dou J, Ding A, Liu X, Du Y, Deng D, Wang J. Anaerobic benzene biodegradation by a pure bacterial culture of Bacillus cereus under nitrate reducing conditions. Journal of Environmental Sciences, 2010; 22: 709–715.
Crossref - Burlingame R, Chapman PJ. Catabolism of phenylpropionic acid and its 3-hydroxy derivative by Escherichia coli. Journal of Bacteriology, 1983; 155: 113–21.
© The Author(s) 2019. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.