Actinomycetes are the potential producers of secondary metabolites of vivid applications; they are isolated from almost all the sources both terrestrial and aquatic habitats. Actinomycetes are a group of Gram-positive bacteria known for their filamentous structure and ability to produce a diverse array of bioactive compounds. These bioactive compounds include antibiotics, antifungals, antivirals, anticancer agents, immunosuppressants, and enzymes. Actinomycetes have been a major source of these bioactive compounds and have played a significant role in the development of many therapeutic drugs. Actinomycetes, which are isolated from practically all sources in both terrestrial and aquatic ecosystems, have the potential to create secondary metabolites with diverse uses. A class of Gram-positive bacteria called actinomycetes is distinguished by its filamentous structure and capacity to manufacture a wide range of bioactive substances. Antibiotics, antifungals, antivirals, cancer preventatives, immunosuppressants, and enzymes are a few examples of these bioactive substances. These bioactive substances have primarily come from actinomycetes, which have also contributed significantly to the creation of several medicinal medications. However, actinomycetes isolation and cultivation can be challenging due to their slow growth rate and complex nutritional requirements. In order to isolate and cultivate actinomycetes, several pre-treatment methods and media can be employed.
Actinomycetes, Streptomyces, Secondary Metabolites, Pre-treatment, International Streptomyces Project
Microbial diversity is a promising area in harnessing potential secondary metabolites that are of vivid applications in the field of synthetic biology since it offers innumerable novel genes and biochemical pathways to depend for antibiotics, enzymes, and other valuable compounds.1 The prominence in harnessing drugs from microorganisms went popular after Penicillin was discovered from Penicillium notatum by Alexander Fleming.2
Actinomycetes are predominantly free-living microorganisms found in a variety of environments. The soil is the most important reservoir for Actinomycetes and also represents the most active zone of interaction between Actinomycetes and the other life forms.3 Streptomycetes are a significant population of Actinomycetes, among all other soil dwelling Actinomycetes species. The genus Streptomyces are undisputed genus isolated in every study which are extraordinarily significant in the synthesis of antibiotics.4
They are studied extensively since they are the potential producers of secondary metabolites which are of great market value yet these organisms are being screened to study other bio-active compounds. Actinomycetes are Gram-positive, filamentous, heterotrophic, saprophytic in nature which play a significant role in organic matter degradation,5 and have high G+C content. They show filamentous structure which can be branched to form a stable mycelium. The mycelium is not always intact or stable and may break into rod or coccus shaped fragments. When grown, the actinomycetes branch profusely forming a network of mycelia, both on the surface of the agar and underneath as well. Surface mycelia growing on the substrate is called aerial mycelia and mycelia submerged beneath substrate is called substrate mycelia.6 Filamentous actinomycetes are known to produce two-thirds of all known antibiotics which fall under categories including, antifungal, antitumor and immunosuppressive agents.7
In addition to being of Actinomycetes origin, which accounts for almost two-thirds of all antibiotics used in clinical practice, Actinomycetes derived natural compounds are also used in the synthesis of several anti-cancer, immunosuppressive, anti-parasitic, and anti-viral agents.8 Streptomyces alone represents around 5% of all known bacterial species within the enormous and diverse branch of microorganisms known as Actinomycetes. They are often categorized as either free or reliant upon the host. Aquatic and terrestrial environments are also potential habitats for these free species. Many diverse eukaryotes, including plants, insects, sea life, and even mammalian and human tissues like the skin, lungs and gastrointestinal system, harbor these microorganisms.9,10
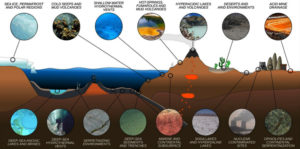
The threat of emerging antibiotic resistant organisms is challenging in today’s world. The quest for discovering novel microorganisms producing secondary metabolites which prove effective against antibiotic resistance pathogens is need of hour.8 In order to meet this challenge, it is necessary to develop novel approaches to search for new molecules. One is the “Renaissance of Antibacterial Discovery from Actinomycetes”. The search for exclusive ecological niches as well as new isolation methods for new genera/species of Actinomycetes can lead to the identification of new gene pools and thus new products (Figure 1).9
Occurrence
Terrestrial environment
Actinomycetes are saprophytes that may be found in a variety of natural environments, such as soil, lakes, oceans, plants, and animals (Figure 2). Novel Actinomycetes continue to flourish in soil. Primary ecological parameters including nutrition, aeration, pH, temperature, salinity, moisture, and organic matter content have a significant impact on the propagation of Actinomycetes thriving in soil and other substrates. Terrestrial Actinomycetes have a wide spectrum of intriguing antibacterial properties.11
Figure 2. Pictorial representation of cross-section of Earth’s crust depicting diversity of actinomycetes.16
Environmental factors like pH, soil type, organic carbon and humus content play a vital role in the distribution and micro-environment of rare actinomycetes. The studies made on distribution of uncommon actinomycetes in native soils of Japan yielded the discovery of Microbispora and Streptosporangium which were known to be more significant in humus-rich acidic soils, similarly Saccharomonospora species were found in abundant in humus-poor alkaline soil environments.10 Other actinomycetes, such as Dactylosporangium and Microtetraspora, Saccharomonospora, and Micromonospora, were commonly isolated from hilly forest soils, level-land forest soils, and pasture soils.12
Dessert soil is regarded as a particularly harsh terrestrial habitat where only a few organisms proliferate, particularly Actinomycetes, frequently use Microcoleus as a food supply. The isolation of actinomycetes from sub-tropical desert soils of Rajasthan, India, proved to be the potential producers of antibacterials against multi-drug resistant and vancomycin-resistant Staphylococcus aureus.13
Soils from the Deccan trap of Karnataka are of interest for many researchers since many studies regarding actinomycetes are revolving around this unique plateau. Rare Actinomycetes isolates of the species Micromonospora, Nonomuraea, Kribbella, Lechevalieria, Saccharotherix, and regularly occurring Streptomyces were isolated from Deccan limestone quarries of Deccan traps, Gulbarga, Karnataka were demonstrated whose antibiotics were proven effective against pathogenic Bacillus subtilis and Fusarium solani.14
Similar studies were carried the successive year regarding the biosynthesis of L-glutaminase, by actinomycetes from the limestone quarries of the Deccan trap. Additionally, soils from agricultural areas, soil from Crab Mountain, and soil from Salim Ali Bird Sanctuary’s mangrove were studied through their cultural characteristics, more study was done on the microscopic observations, and then biochemical observations were conducted. The studies confirmed the strain obtained was Streptomyces rochei isolated from limestone quarry which was confirmed based on 16S rDNA sequencing which was the potential producer of L-glutaminase.11
The largely unexplored acidophilic and obligate acidophilic actinomycetes proliferating in tropical and subtropical red soils of China, pH lying between 4 and 6 were isolated. Red soils shows the characteristic features of low organic carbon content and high iron oxides along with other inorganic metals like Ca, Mg, P, and K. Pure cultures obtained from screening of red soil showed numerous colonies notably belonging to Streptomycetaceae, Pseudonocardiaceae, and Streptosporangiaceae which are the three families of Actinomycetes. Macrolides, polyethers, diketopiperazines, and siderophores accounted for the majority of the secondary metabolites isolated from these isolates.15
The studies on the phylogenetic diversity and physiological heterogeneity among the culturable actinomycetes of the alpine regions of Qinghai-Tibetan Plateau paved the culturing of actinomycetes characterized from near-root soils of various plant groups, which spread in two geographically diverse alpine habitats. This region’s isolates are closely linked to Streptomyces sp. and have antibiotic activity against E. coli, Staphylococcus aureus, Candida albicans, and Pseudomonas aeruginosa.16
The saprophytic actinomycetes dwelling in the rich organic carbon containing soils of forests are drawing attention for researchers to again explore their antimicrobial properties. Actinomycetes strains primarily belonging to the genus Streptomyces, followed by Nocardia and Kribella, were isolated from the Pobitora Wildlife Sanctuary and Kaziranga National Park in Assam, India, which is part of the Indo-Burma mega-biodiversity hotspot. According to the study, the novel strain showed greater similarity between S. malaysiensis which exhibited growth inhibitory properties in methicillin resistant Staphylococcus aureus and Candida albicans.12
The antibacterial properties of actinomycetes from the garden soils of Nasr City, Cairo, Eygpt, yielded the potential secondary metabolites from the strains that proved effective against Escherichia coli, Bacillus megatarium, Pseudomonas aeruginosa and Klebsiella oxytoca.17 Some Streptomyces isolates from the Pindari glacier region of the Indian Himalaya shown significant antifungal activity.18 These findings show that a wide range of Actinomycetes may survive in alpine environments, and the majority of them may produce bioactive compounds. Furthermore, Actinomycetes have been identified as key contributors in the biogeochemical cycles due to their diverse physiology and metabolic flexibility.19
Isolation of actinomycetes from soil as source for studying antimicrobial activity is one of the interesting areas for the researchers, the antimicrobial activity of the soil isolates against Staphylococcus aureus, Escherichia coli and Pseudomonas aeuginosa is well studied. The 6 isolates out of 15 proved effective inhibitory properties, among the three test species, S. aureus showed the maximum inhibitory zone.20
Similarly, several antibiotic-producing actinomycetes strains were isolated from sandy soil that lacked vegetation (bare soil). The discrete isolates obtained after incubation were further streaked on to the selective media in order to obtain pure cultures. The inhibitory growth characteristics of test organisms, Escherichia coli, Pseudomonas aeruginosa, Bacillus subtilis, and Staphylococcus epidermis, were studied cumulatively. The study demonstrated that antibiotic yield was low.4
The antimicrobial activity of actinomycetes from rhizosphere soils of Sugar cane from Piyungan, Bantul, Yogyakarta was processed and treated. On plating numerous colonies appeared which were potent in inhibiting the growth of Gram-positive Staphylococcus aureus, on the other hand E. coli was unaffected. Hence the study proved the isolation of narrow spectrum antibiotic showing inhibitory properties in Gram-positive organisms.21
Actinomycetes isolated from alluvial soils in Punjab were shown to have antibacterial and antifungal properties, as well as the ability to limit growth in a variety of Gram-positive and Gram-negative bacteria, including fungi. TLC examination of the antimicrobial component revealed that it was either alcohol, phenol, or steroid.22
Aquatic environment
Freshwater
Filamentous actinomycetes like Actinoplanes, Micromonospora, Rhodococcus, Streptomyces, and Thermoactinomyces are exclusively found in freshwater, they propagate through the motile zoospores. The zoospores of these freshwater actinomycetes are motile and motility is aided by tufts of flagella that exhibit chemotaxis and frequently require an external source of energy.23 Rifamycin, an anticancer metabolite from a Salinispora strain, marinomycins from Marinophilus sp., abyssomicin-C from Verrucosispora sp., and marinopyrroles from Streptomyces sp. are just a few of the novel bioactive compounds discovered from aquatic actinomycetes.24
Marine
Earth’s surface covers almost two thirds of the ocean surface, the variations in location, temperature, and salinity contribute far higher phylogenetic variety than the terrestrial environment. The ocean is home to between 50 – 80% of all life on Earth. Because of the great genetic and biological variety of marine creatures, the seas provide a unique and plentiful supply of bioactive chemicals for the pharmaceutical industry.25 Microorganisms stick to the sandy ocean floor since the dead and decaying matter of the ocean accumulates here. The sea bed is 10,000 times greater populated than that of the open ocean. The microorganisms here mainly depend on the sea floor feed on sediment. The microbial count beneath the seabed is no astonishing irrespective of unavailability of light and nutrients.26
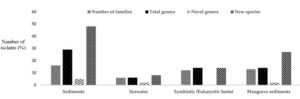
Marine habitats are aquatic environments that have salty water. The salt concentration of a marine environment can range from low to high (e.g., oligohaline to hypersaline) and can be extremely changeable or relatively consistent through time.27 Actinomycetes thriving in marine ecosystems are called marine actinomycetes which often associate with a wide range of aquatic species, including sponges, corals, and echinoderms, as well as vertebrates like pufferfish. These interactions may promote distinct chemical ecologies, regulating secondary metabolic pathway evolution. Marine actinomycetes, in addition to interacting with other species, may thrive in both planktonic and biofilm habitats, even though the majority of strains have been identified from sediments (Figure 3).28
Figure 3. Percentage distribution of marine hosts for Actinomycetes based on 16S rRNA gene sequences.31
The Marine ecosystem harbors largest biodiversity on earth and is one of the untapped environments especially for isolating and harnessing the numerous applications of Actinomycetes. The marine water bodies are relatively highly diverse in its microbial flora than compared to any other natural ecosystems. Marine actinomycetes propagating in the marine ecosystems must have undergone evolution in adapting themselves to different pH, high pressure, high acidic conditions, temperature variations upto 8 – 100°C surrounding mid-ocean ridge at the mid-ocean ridges the diversity of culturable and unculturable (rare) actinomycetes associated with marine sources are illustrated in Figure 4.29,30 The first marine Actinomycete species, Rhodococcus marinonascene greatly supported the un-explored biodiversity of Actinomycetes in the oceans. Dietzia, Rhodococcus, Streptomyces, Salinispora, Marinophilus, Solwaraspora, Salinibacterium, Aeromicrobium marinum, Williamsiamaris and Verrucosispora are the most prevalent actinomycetes native to oceanic waters.23
Collection of the samples
Soil is the end product of breakdown and weathering of rock. All soils consist of a combination of mineral and organic particles matter, water and air. The combination of these elements define the properties of the soil: nature, structure, porosity, chemistry and color. Texture and structure are the physical properties, hence soil is also greatly defined by its chemical properties. The percentage of sand, silt, and clay in the soil is referred to as its composition.33
The soil provides a favorable environment for microorganisms and is home to a diverse array of microorganisms, including bacteria, fungus, algae, viruses, and protozoa. Soil mineral components, which are formed as a result of rock weathering and the degradative metabolic activities of soil microorganisms, influence the physical structure, aeration, water holding capacity, and nutrient availability (Figure 5). The study of organisms in soil, their roles, and how they impact soil qualities is known as soil microbiology.34
The selection of environmental samples can have a significant impact on the effectiveness of isolating significant quantities of a certain Actinomycetes. It is better to get soil samples from untouched areas, such as desert, saline and alkaline soils, and ancient forests soils deep from the lithosphere where the microorganismssurvive.36
Oceanic actinomycetes are extensively dispersed, and several natural compounds have been derived from them. With the use of a sampler, sediment from the deep ocean is gathered; the samples are then placed in sterile glass bottles and stored at 4°C.37
Collection of soil samples
Actinomycetes are primary source of soil, they are saprophytic in nature and are exclusively found in the terrestrial habitat. The choice of source of actinomycete would be soil, which maybe agricultural soil, radiation exposed soil, since the actinomycetes that live in it may be new species with novel physiological adaptation mechanisms that produce bioactive chemicals.38,39 Soils from various regions of Belgaum, Karnataka,40 also soil samples acquired from the Thar Desert in India were studied for the isolation of actinomycetes. The Thar desert isolates was discovered to produce yellow color pigment with antibacterial activity.6 The gardens soils of Nasr City, Cairo, Egypt, were collected after digging the site for nearly 5cm deep and were collected in clean polythene bags. The samples were taken to the laboratory and kept at 4°C for further examination.17
Samples from the waste disposal sites at different depths were collected from standard soil collection methods in Ethiopia, further the soil sample was sieved and isolation was carried on dilution methods.41
Actinomycetes were isolated from several soils of varied habitats from 5-25 cm deep in sterile plastic bags for the isolation of microorganisms and transferred aseptically in polyethylene bags to prevent moisture losses during transportation. These environments included Ad-Dawadmi, Saudi Arabia, rhizosphere soils.42 Soil samples were gathered and investigated from several niche habitats such as a field, near plant surface, mining area, Kaketto dam, Kamojiya poultry farm, and well in the Madhya Pradesh district of Sheopur, India. To collect the samples, a sterile polyvinyl corer was put into the sediments around 6-12 inches deep in the earth. These samples were placed in sterile plastic bags, firmly sealed, and promptly transferred to the laboratory. Prior to isolation, these soil samples were air-dried for 3-4 hours at 45°C, crushed, and sieved.43 Soil samples were collected in sterile plastic bags from the 10cm top layer of soil at five random places in the saltran valley of Taza, Morocco, and transferred to the laboratory.44
A study gave insights on the isolation of actinomycetes from six diverse ecosystems in India, including plant rhizosphere soils, agricultural soil, hospital surroundings, river mud, and conserved parts of forest soils. After around 3.0 cm of soil was removed from the surface, samples were collected up to a depth of 20 cm. The soil samples were collected, packed, and stored in polyethylene in a refrigerator.45
The soils collected from the depth of 5 cm were stored in clean polythene bags and dried overnight at 28°C to remove bacterial growth and minimize water content. 1 g of dry soil was taken from each sample and combined with 9 mL of sterile water, vortexed for 5 minutes. The suspension was heated at 50°C for 10 minutes before being vortexed for 5 minutes. The supernatant was serially diluted in 10-2 – 10-4 sterile water.21
Pretreatment methods for soil samples
Actinomycetes must first undergo pretreatment because they grow more slowly than other bacteria and fungus. Pretreatment regimens often choose target actinomycetes by preventing or getting rid of undesirable microorganisms. A wide range of chemical and physical pretreatments have been practiced in selective isolation of actinomycetes (Table 1). Wet incubation with radiation, glycerol treatment, air dry, dry heat, centrifugation, cellulose infiltration, and baiting followed by drying are all physical treatments for actinomycetes.46 Because actinomycete spores are more resistant to desiccation than most bacteria, most undesired Gram-negative bacteria may be entirely removed by simply leaving soil, sediment, lichen, and fecal samples to air dry at room temperature.36
Table (1):
Various Pre-treatment Methods36
Pretreatment Methods |
Targeted Actinomycetes |
Ref. |
|---|---|---|
An hour of air-dried soil heated at 120°C |
Streptosporangium sp Microbispora sp. |
47 |
Soil that has been air-dried and heated to 100°C for 15 minutes |
Actinomadura sp. |
48 |
Water or soil suspensions heated for 10 minutes at 45°C or 50°C |
Streptomyces sp. |
49 |
Water or soil suspensions heated for 30 minutes at 60°C |
Micromonospora sp. |
50 |
An hour of air-dried soil heated at 120°C |
Dactylosporangium Streptosporangium sp. |
51 |
For a week, air-dried soil was heated to 28°C |
Herbidospora cretea |
49 |
For an hour, a soil suspension was heated at 110°C. |
Microtetraspora glauca |
52 |
Treatment of soil sample
There has been increased efforts observed in isolating actinomycetes from various different sources in isolating novel actinomycete strain. The soil samples collected were kept for drying at room temperature followed by continues stirring in an open shaker (Remi) at 100rpm for 5 days results in the formation of a layer of mycelia along the walls of conical flasks which confirms the presence of organisms.53 Drying the soil samples in clean petri dishes at 55°C for ~15 min reduces the unwanted growth of already existing Gram negative bacteria.17
The obtained soil sample was weighed 1g in 100 mL of physiological water (NaCl 8.5 g/L), and the combination was incubated for 30 minutes at 28°C with 200 rpm shaking. The mixtures were allowed to settle before being prepared in successive dilutions of up to 10-5 with sterile physiological water and rapidly agitated with the vortex. An aliquot of 0.1 ml of each dilution between 10-2 and 10-5 was placed on actinomycetes isolation agar and starch casein agar medium. To avoid bacterial and fungal contamination, rifampicin (2.5 µg/mL) and amphotericin B (75 µg/mL) are added to each medium. Plates were checked 48, 72, and 96 hours after incubation at 28 and 37°C.54
To suspend 10 g of each soil sample, 100 ml of sterile physiologic water (0.9% NaCl in distilled water) was used. For 60 minutes, a 50°C heat treatment was performed in an agitated water bath. The samples were serially diluted 10 times in sterile 0.9% saline. Following that, dilutions were plated in triplicate on CSA and incubated at 28°C for 4 to 6 weeks.44
The collected soil samples from several Kala Patthar locations, Mount Everest region, Nepal were dry soil samples (each weighing 4–5 g) were taken at a depth of 4–5 cm, put in clean polyethylene bags, and well mixed with the bag’s already-added 1 gram of CaCO3. The samples were then further dried for roughly three weeks at room temperature. After serial dilution soil samples was spread evenly on starch casein agar plates with cycloheximide and nystatin (each at a concentration of 50 µg/mL of media), actinomycetes were isolated using the spread plate method.1
According to studies, the heating of air-dried soil samples to temperatures of 120°C or 100°C decreased the amount of filamentous bacteria and Streptomycetes present on isolation plates and allowed for the selective separation of a number of uncommon actinomycetes species.55
The river sediments of Krishna and Godavari rivers were collected to isolate actinomycetes. The soil samples were collected and kept in poly bags that had been UV and alcohol sterilized. By using the pour plate technique, soil samples were serially diluted up to a 10-6 concentration, and 1 mL from each dilution was plated on various isolation medium, including starch Casein agar, albumin media, and YMA media, which included the antifungal drug Nystatin 50 µg/mL. The plates were incubated for 7–14 days at various temperatures between 18 and 28°C42
The area of interest in isolating actinomycetes was from waste disposal rhizosphere soil which were collected from different depths 5, 8 and 11 cm. Further was soil was sieved, 1 g of soil sample was transferred from each sample to numerous test tubes containing 10 mL physiological saline (NaCl, 8.5 g/L), and the test tubes were thoroughly agitated with a vortex mixer. The soil samples were sieved using a 250 m pore size sieve, and 1 g of soil was transferred to several test tubes containing 10 mL physiological saline (NaCl, 8.5 g/L), and the test tubes were well shaken using a vortex mixer to form a stock solution, which was then serially diluted before plating on starch casein and oat meal agar plates containing amoxicillin (20 µg/mL) and cyclohexamide (25 µg/mL).41
The various rhizosphere soil samples from medicinal plants at a depth of 10 cm in polythene bags. The soil samples underwent pretreatment to lower the percentage of microorganisms other than actinomycetes. The soil samples were dried for 5 to 10 minutes at a temperature of 50 to 60°C. Further serial dilution is performed before plating.56 Isolation of actinomycetes from Western ghats was carried out, the soil samples were taken at a depth of 15 cm. These soil samples were sieved, crushed, and air dried for 34 hours at 45°C before being used for isolation using the serial dilution technique Agar (AIA) medium with nalidixic acid (100 mg/L) and ketoconazole (30 mg/L) was disseminated with aliquots (0.1 mL) of dilutions 10-2, 10-3, 10-4, and 10-5 and incubated at 30°C for 7–10 days.57
Isolation of the actinomycetes from cave soil was conducted where the upper 5cm of the layer of soil sample was collected at 10 different locations and brought to the laboratory in a sterile polythene bags. Soils were air-dried at room temperature for 7 to 10 days, the samples were grind with mortar and pestle and pH was determined using glass electrode pH meter.58 The collected soil samples 10cm below the surface were treated prior to the isolation by exposing to air and sieved, the obtained soil sample was subjected at 30°C in an hot air oven for a week, 40°C exposure of the samples was done in order to reduce the proportion of bacteria and other actinomycetes while preserving the spores of actinomycetes.59
A different pretreatment involved mixing1 g of air-dried soil with 10 mL of sterile 5 mM phosphate buffer pH 7.2, (i) peptone (6%), and (ii) yeast extract (6%), followed by a heat shock at 50°C for 10 minutes at 200 rpm in an incubator shaker. Inhibiting the growth of Gram-negative bacteria and other unrelated soil bacteria while actinomycetes spores germinate. After autoclave sterilization, the buffer was stored at 4°C in a Schott Duran container (500 mL). The samples were allowed to air dry at room temperature for one week. After allowing the samples to air dry for a week, they were isolated and counted using the serial dilution and spread plate method. Gram staining and lactophenol blue staining were used for microscopic investigation, and spore chain morphology was analyzed using a coverslip culture method and a light microscope on actinomycetes isolation agar. To reduce bacterial and fungal development, nalidixic acid 100 mg/L and actidione 20 mg/L were added, respectively. The plates were then incubated at 30°C for 10 days.42
Varieties of physicochemical approaches were employed to kill common bacteria in soil samples from various geographic locations. One gram of each soil sample was suspended in 10 mL of normal saline and split into aliquots for physicochemical treatment. One aliquot of the soil sample was heat treated at 120°C for 1 hour and the other at 30°C for 30 minutes with 1.5% phenol. The soil samples were then stirred and allowed to stand for 30 minutes before being serially diluted and 0.1mL of each “dilution” was plated onto nutrient agar (NA), Actinomycete Isolation Agar, yeast malt glucose agar (M6), antibiotic assay agar, starch casein agar, and Czapek Dox agar.45
The soil samples collected was weighed 20 g and were air-dried. The dried soil sample was then pounded into a fine powder in a pestle that had been disinfected with ethanol on the surface. The soil samples were then dry-heated for 1 hour at 50°C to lower the amount of other bacteria and enable for the preferred isolation of actinomycetes. One gram of soil was added to 100mL of water and incubated for one hour in an orbital shaker at 200 rpm with 30°C temperature control. After leaving the mixtures to settle, the spore suspensions were diluted up to 10-5. With a sterile L-shaped glass rod, 1mL of each dilution was collected and dispersed equally over the surface of starch casein agar (SCA), then incubated at 30°C and colonies were inspected for appearance. The streak-plate technique was used to obtain purified actinomycete isolates, and the pure cultures were kept at 4°C on SCA slants.22
Collection of marine sample
Isolation of novel marine actinomycetes requires knowledge and expertise of actinomycetes taxonomy, physiology, and environmental parameters, such as pH, culture temperature, oxygen, nutritional needs, etc.32
Soil sediments from the Bay of Bengal shore near Chennai, Tamil Nadu, India, were chosen and dried for a week in sterile conditions.60 Soil samples from the saltpan of Ennore were collected in sterile polythene bags and preserved at 4°C for further study.61 South Andaman Island of India, noted for mangrove vegetation offered four marine sediment samples which were taken from the Bay of Bengal coast and the Andaman Sea coast. The samples were collected on the spot in sterile plastic containers, transferred to the laboratory, and stored for further analysis.62
Treatment of marine samples
The marine soil sediments were completely dried before plating on CSPY-ME medium in aged sterile sea water followed by serial dilution. Further, fluconazole (50 µg/mL) was dispensed in media to inhibit the fungal growth.60 The importance of salt concentration in isolating marine actinomycetes was studied, the absence of NaCl in the culture medium yielded little or no growth, on the other hand isolates were able to grow in 3% NaCl, some isolates were able to grow in 10% NaCl. Further, the study concluded that none of the organisms grew on the media containing salt concentration upto 14%. Therefore it can be concluded that the terrestrial actinomycete forms have eventually evolved in salt conditions.63
The importance of dilution (1:1000) of the marine water and sediment samples to obtain the isolated colonies prior to plating. The sediments were heat treated for 30 minutes at 60°C to limit the growth of weedy or vegetative forms, allowing only spore-forming actinomycetes to proliferate. A calcium carbonate-enriched treatment was also used to suppress the growth of vegetative forms.64 The central portions of the marine sediments were transferred to the laboratory in the ice containing cold box. 10 grams of sediment samples were placed in laminar air flow for 10 to 12 hours followed by heat treatment at 41°C for 10, 20 and 60 days prior to isolation100 ml of sterile natural seawater was used to suspend ten grams of each sediment sample and heat-treated sample. Rotary shaking flasks at 150 rpm for 30 minutes produced a suspension, from which 1:10, 1:100, 1:1000, and 1:10000 dilutions were created. 200 µL of each suspension were applied to the surface of various agar medium made with natural seawater, and the plates were cultured at 28°C for two to three weeks.65
Placing these sediment samples for about 60 minutes, in a 50°C water bath prevented the growth of unwanted bacterial flora. Prior to injection onto the isolation plates, all the treated samples were diluted (up to 10-6) with 0.5% saline aseptically.66
Studies proved that the marine sediments was the ideal source in the selective isolation of actinomycetes, it therefore yielded good number of isolates than the plant tissues of mangroves vegetation. They concluded that the organisms needed seawater for their growth.67
Soil samples were collected from coastal Andhra Pradesh (AP) areas at a depth of 10 – 15 cm in a sterile polythene bag and maintained at 4°C until brought to the laboratory usage for the isolation of marine actinomycetes from salt pans. Actinomycetes were isolated and cultivated using selective media such as actinomycetes isolation agar (AIA), Kuster’s agars, and Starch Caesin agar. After being diluted to a concentration of 10-7, one millilitre of the serially diluted soil samples was put to medium. Cyclohexamide (100 µg/mL) was added to all of these medium to avoid fungal infection. The inoculated plates were incubated at room temperature for seven days.68
Studies provided the insights of performingthe pour plate technique from the soil samples (1 gm) which were serially diluted and plated on Starch Caesin agar (SCA). For the production of the medium, filtered seawater was employed. The plates were incubated at 30°C for 7 to 10 days, after which the colonies were streaked using the quadrant streaking method.61
Multiple pretreatment procedures and three selective media were utilized in a study to identify physiologically active actinomycetes from mangrove sediments in India’s Andaman and Nicobar Islands. Sediment samples were collected from four distinct mangrove areas and processed using the dry heat technique. The antibacterial activity of the 42 isolates against harmful bacteria was tested in two distinct mediums. Antibacterial metabolite synthesis was found in 22 of the strains examined. Staphylococcus aureus, Bacillus subtilis, Salmonella typhi, and Klebsiella pneumoniae were used as test microbes. Among all these strains, the identified Streptomyces spp. showed the highest activity against all pathogens used in both media.62
The studies on marine actinomycetes were made by taking a sample from the coastal area of Chennai (Tamil Nadu). 34 strains were isolated and 10 of them were examined as potential marine actinomycetes by crossing them with 5 fish-pathogenic bacteria. The extract was tested against pathogenic fish bacteria by disc diffusion method, and the ethyl acetate extract exhibited a good zone of inhibition with a diameter of 6-15 mm. Streptomyces spp. was found and noted as the potent actinomycete strain in the study.69
Cultivation techniques
Microbiology has made great strides through the discovery and improvement of culture medium. Louis Pasteur pioneered the first artificial liquid culture medium in 1860. Bacterial growth had previously been witnessed on common things such as food. These findings emphasized the significance of bacteria’s natural habitat and nutritional requirements in the designing of culture medium for their isolation. The culture medium is essentially composed of basic components (water and nutrients) to which various growth factors necessary to each bacteria are added for its growth.70
Nutrients are required for microorganism’s growth and play a significant role in the proper culturing of microorganisms in the laboratory as well as their development in the natural environment. Nutrients needed include those that provide energy, carbon, and other materials essential to life. Cell-specific nutrients are employed for growth, propagation and are mainly based on their cellular and metabolic processes. Carbon, nitrogen, sulfur, phosphorus, potassium, magnesium, calcium, oxygen, iron, and other trace minerals are common nutrients required by all living creatures. Essential nutrients are the nutrients that the body need at all times. There are two types of important nutrients: macronutrients and micronutrients. Macronutrients are required in vast quantities; micronutrients, on the other hand, are normally required in lesser quantities and are frequently trace elements.71
Actinomycetes cultivation is significant because these microorganisms have received impeccable attention for their relevance as a source of secondary metabolites that are useful to humans. Therefore, discovering the existence of these microbes in every different habitat illuminates the development of systematic approach for isolating and identifying its pure cultures. The optimization of nutritional conditions for maximum growth and production of desired metabolites has provided valuable insights about the regulatory element factors of metabolism in actinomycetes.72
The ability of actinomycetes to synthesis secondary metabolites is greatly conditioned by the availability of nutrition and during cultivation. The nutrient availability factor and physical parameters can be changed accordingly to extract greater or lesser yield of secondary metabolites; hence, the cultivable environment for an isolate is considered very crucial. In order to maximize productivity and minimize costs, media composition is crucial. As a result, creating a suitable fermentation medium is essential for the generation of secondary metabolites. Besides, the production of bioactive metabolites is also significantly influenced by a number of culture conditions, including pH, incubation time, and temperature.73
Isolation and characterization of actinomycetes is done on selective media by performing serial dilution and plating techniques on Starch Casein Agar, Glucose Yeast Agar, Actinomycetes Isolation Agar, Coal Vitamin Agar supplemented with considerable amounts of antibacterial and antifungal agents.74,75 Humic Acid Vitamin Agar with varied pH was formulated to isolate the rare actinomycetes of mountain soil where in humic acid serves as a source of carbon and nitrogen for the growth of microorganisms.76
Incubation temperature required for the optimal growth of actinomycetes culture is well studied to be 30°C for weeks.17,44 Antibiotic producing actinomycetes isolated from sugarcane rhizosphere soil required a temperature of 28°C for 2 weeks.21 Similarly, isolation of thermophilic actinomycetes requires high temperatures of about 55°C incubated for a period of 7 days in a moist chamber.77
The ideal pH parameters for the optimal growth of actinomycetes would be 7 to 7.5. A study mentioned that the majority of soil actinomycetes thrive between the pH ranges of 5.0 to 9.0 and are regarded as neutrophils.17 However isolation and culturing of moderate acidophilic actinomycetes and obligate acidophilic actinomycetes require pH 6.0 and pH 4.0 respectively.15
Several of the media have been suggested in International Streptomyces Project78,79 that have shown to be beneficial in characterization Actinomycetes strains. Different ISP media were used to study morphology and aerial and substrate mycelium.78 Tryptone yeast extract agar (ISP 1), yeast extract-malt extract agar (ISP 2), oatmeal agar (ISP 3), inorganic salts-starch agar (ISP 4), glycerol-asparagine agar (ISP 5), peptone yeast iron agar (ISP 6), and Tyrosine agar (ISP 7) were the most often used medium for growth studies. The Actinomycetes cultures were inoculated in the agar plates prepared for the above mentioned media. The inoculated plates were allowed to incubate for 14 days at 37°C. The color of the aerial mycelial mass, the reverse colony color and color of soluble pigments, if any, were recorded.80
For the selective isolation of marine Actinomycetes, six different ISP series medium such as Starch Casein Agar (SCA), Nutrient Agar, and Actinomycetes Isolation Agar (AIA), were used. The samples were isolated on Starch Casein Agar (SCA), ISP-4, and ISP-7, which yielded a large number of morphotypes from the samples. The samples collected during monsoon yielded highest number of morphotypes on ISP-5 and ISP-7. For that of samples collected during post-monsoon seasons showed almost equal number of colonies on Nutrient Agar, ISP-2, ISP-4, ISP-5, and ISP-7.81
The isolation and cultivation on different media such as ISP2 (yeast extract-malt extract agar), ISP3 (oatmeal agar), ISP4 (inorganic salts-starch agar), ISP5 (glycerol-asparagine agar), nutrient agar, and Bennett medium to study the pigmentation of the aerial mycelium and the structure of sporophores which is denoted as an important criteria for classification was made.82
ISP 1 tryptone yeast extract broth
Tryptone Yeast Extract Broth (ISP Medium No.1) is indicated as a general purpose enrichment medium for microorganisms that are not particularly fastidious. This medium has been developed in accordance with the International Streptomyces Project. Tryptone and yeast extract supply nitrogenous chemicals, carbon, sulfur, Vitamin B complex, and trace elements, all of which are required for bacterial metabolism.78
ISP 2 yeast malt agar
For the isolation and culture of yeast, mold, and other acidic microorganisms, Yeast Malt Agar (YM Agar) is recommended. Fungistatic agents, such as sodium diphenylpropionate, are added to YM agar to suppress mold growth and therefore allow yeast enumeration from mixed populations.78
Actinomycetes from the Algerian Sahara Biotope were isolated on ISP 2 medium supplemented with 40 µg/mL actidione to prevent the growth of eukaryotic microbes. Furthermore, the isolated colonies were kept on the same agar slants at 4°C for future studies.82
ISP 3 Oat meal agar
According to the International Streptomyces Project, ISP medium number 3 is suggested for the culture and characterization of Streptomyces species.78 The medium is often referred to as oatmeal agar. Oats provide the necessary nutrients for the growth of Streptomyces. The trace salt solution consisting of iron sulphate, manganese chloride and zinc sulphate supplies important electrolytes and minerals.
ISP 4 inorganic salt starch agar
According to the International Streptomyces Project, Inorganic Salt Starch Agar is utilized for the culture and characterization of Streptomyces species. ISP Medium No. 4 is formulated based on the original formula.78
Temperature range for isolated strain development employed inorganic salts-starch agar medium (ISP 4) utilizing a temperature gradient incubator.83
ISP 5 glycerol asparagine agar base
ISP Medium No. 5 (Glycerol Asparagine Agar Base) is recommended according to the International Streptomyces Project for the cultivation of Streptomyces species. This medium provides consistent and reproducible characteristics of Streptomyces. Glycerol serves as the carbon source while asparagine is the amino acid source for the growth of Streptomyces species. The need for trace elements of Streptomyces is covered by the trace salt solution, which contains various salts. Dipotassium phosphate buffers the medium.
The International Streptomyces Project recommends ISP Medium No. 5 (Glycerol Asparagine Agar Base) for culturing of Streptomyces species. This media offers consistent and reproducible characteristics of Streptomyces. For the growth of Streptomyces species, glycerol acts as a carbon source, while asparagine serves as an amino acid source. The trace salt solution, which comprises different salts, meets Streptomyces demand for trace elements. The medium is buffered with dipotassium phosphate.78
The diffusible pigment production assay on isolates of actinomycetes from rhizosphere soils was performed using glycerol asparagine agar medium. The cultures were inoculated on Glycerol asparagine media and incubated at 28±2°C for 14 days, after incubation, the production of color such as yellow-brown, blue, green, red, orange, grey, or violet in complete medium was studied.84
ISP 6 peptone yeast extract iron agar
ISP Medium Number 6 (Iron Yeast Extract Peptone Agar) is recommended by the International Streptomyces Project for the cultivation and maintenance of Streptomyces species. Gastric digestion of animal tissue, protease peptone and yeast extract provide carbon, nitrogen, sulfur, B vitamins and other essential nutrients for growth. Dipotassium hydrogen phosphate gives the medium a good buffering capacity. Ferric ammonium citrate and sodium thiosulfate together serve as the Hydrogen Sulfide Indicator System
The International Streptomyces Project recommends ISP Medium Number 6 (Iron Yeast Extract Peptone Agar) for the culture and maintenance of Streptomyces species. Carbon, nitrogen, sulfur, B vitamins, and other necessary elements are provided via gastric digestion of animal tissue, protease peptone, and yeast extract. The medium’s buffering ability is enhanced by dipotassium hydrogen phosphate. The Hydrogen Sulfide Indicator System is made up of ferric ammonium citrate and sodium thiosulfate.78
ISP 7 tyrosine agar
ISP Medium Number 7 (Tyrosine Agar) is used to differentiate Streptomyces species based on tyrosine utilization. The medium contains L-tyrosine used by Streptomyces species. The clearing zone around the colony indicates tyrosine hydrolysis. Trace elements provide essential growth factors for Streptomyces species.78
Actinomycetes producing pigment, melanin, require a selective media for screening, which is directly an organic media, specifically tyrosine agar medium containing tyrosine in the melanin pigment production test. The isolates of actinomycetes were inoculated on tyrosine agar slants and incubated at 28±2°C. Browning of the organic medium, production of soluble pigments, and colors of vegetative and aerial mycelium in the slants were evaluated after 2-4 days of incubation.84
ISP 8 starch casein agar
The detection of saccharolytic marine bacteria and actinomycetes is recommended using Starch M-Protein Agar. The complex carbohydrate supply in this medium is starch, while the nitrogen source is M-protein powder. Seawater salts provide various ionic sources that make the media ideal for marine microbial flora while also buffering the medium.76
Many researchers find Starch Casein Agar media ideal for the isolation of terrestrial actinomycetes from varied soil types. Actinomycetes from the limestone quarries of the Deccan trap for the production of L-glutaminase employed Starch Casein Agar. Besides, soils from agricultural fields, crab mountain soil and soils of mangrove vegetation from Salim Ali Bird Sanctuary were collected and dried, followed by phenol, heat and calcium carbonate treatment. Further serial dilution was made for the isolation of actinomycetes on SCA media, the plates were allowed for the incubation at 35°C for 3 days. The cultural characteristics were observed and further analysis was made on microscopic characteristics followed by biochemical observations.85
Starch Casein Agar (CSA) medium was used for the isolation of actinomycetes. Further the purified colonies of actinomycetes were maintained on the International Streptomyces Project (ISP-2) medium.44
The isolation of soil actinomycetes from the inner depths of rhizosphere soils is accomplished using starch casein agar. The soils were allowed to dry at room temperature before being diluted from 10-1 to 10-10. The inoculum of 0.1 mL from the aliquots was uniformly spread on a solidified sterile petri dish treated with fluconazole (30 µg/L) to prevent the growth of fungus. The inoculated plates were incubated for 7 to 10 days at 28±2°C. After incubation, the characteristic actinomycetes colonies were successfully retained on SCA medium by sub-culturing at 4°C using the streak plate method. The nature of the pigmentation of the aerial mycelium, spore mass, size, pigmentation on the reverse side and diffused pigments and colony characteristics of the pure actinomycetial isolates were studied on SCA media.84
Colonies on Starch-casein agar was made further the cultures were streaked and preserved on SCA slants. It was cultured at 30°C for 5 days and sealed with paraffin and stored at 4°C.22
ISP 9 carbon utilization agar
ISP Medium Number 9 (Carbon Utilization Agar). Characterize Streptomyces utilizing a carbon source. ISP 9 serves as a base the ideal nutritional and cultural parameters of strain (BT-408) for growth and antibiotic production were determined using Pridham and Gottlieb’s inorganic salts medium. It was then supplemented with various carbon and nitrogen sources to study their impact on growth and antibiotic synthesis.83
Alongside the physiological, biochemical, and cultural characteristics of strain BT-408, which was derived from the sediments of the Bay of Bengal Ocean, the strain was initially plated on SCA medium before being subjected to a carbohydrate utilization test to determine growth on carbon utilization medium (ISP 9) supplemented with 1% carbon sources at 28°C.83
Bennett agar, Tryptic soy agar (TSA), Yeast extract-malt extract agar (GLM), Chitin-vitamins agar, and International Streptomyces Project (ISP2) agar were used to isolate actinomycetes. Cycloheximide (50 µg/mL) and nystatin (30 µg/mL) were also added to inhibit the fungal growth. Also to inhibit Gram negative bacteria, nalidixic acid (30 µg/mL) was added to the growth medium. For actinomycete selective isolation, four distinct media were used: Yeast Glucose Agar (GYA), Starch Casein Agar (SCA), Actinomycete Isolation Agar (AIA), and Vitamin Carbon Agar (CVA), all of which are treated with the antifungal cycloheximide. All incubations were carried out at 28°C.86
Air-dried sediment samples were ground aseptically with a pestle and mortar, mixed thoroughly, and passed through a 2 mm filter to remove gravel and stones. For pretreatment, the samples were stored in a separate glass container for 5 minutes at 55°C, 60 minutes at 55°C, 15 minutes at 70°C and 1 hour at 100°C. Subsequently, 10-fold serial dilutions of the sediment samples were performed with 50% sterile seawater. Approximately 0.1 mL of the serially diluted samples were dispensed onto Kuster agar medium with 80 µg/mL cycloheximide and 75 µg/mL nalidixic acid to the three agar media to minimize the growth of other bacteria and fungi. The plates were incubated at 28 ± 2°C for 28 days. After 5 days, the actinomycete colonies grown in Petri dishes were counted at regular intervals. All morphologically distinct actinomycete colonies were grown on yeast extract and malt extract agar (ISP 2) using the streak plate technique. After growth, actinomycete colonies were maintained on the slants of ISP No. 2 for further studies.62
Starch Casein Agar (SCA), Actinomycetes Isolation Agar (AIA), Mueller Hilton agar, nutrient broth, starch casein broth, glucose phosphate broth, peptone water, simmons citrate agar, triple sugar iron agar, dimethylsulfoxide, nalidixic acid, actidione, and ciprofloxacin were the culture mediaand antimicrobials employed in the investigation.42
Identification and characterization
Identification and characterization of actinomycetes methods including morphology, physiology, and biochemical characters from the isolated actinomycetes under study. The morphological techniques include both macroscopic and microscopic characterizations that are well studied. The Macroscopic characterization include studies of isolates which are primarily distinguished based on the size, shape, color, pigmentation, and consistency of their colonies.17 For the purpose of presumptive identification of the isolates, the morphology of the studied isolates was compared to the Actinomycetes morphology listed in Bergey’s Manual.1
Catalase test, citrate utilization test, oxidase test, starch hydrolysis test, nitrate reduction test, starch hydrolysis test, tween 20 hydrolysis, urea hydrolysis, esculin hydrolysis, gelatin hydrolysis and carbohydrate fermentation tests are among the various biochemical tests carried out. The physiological tests included motility, temperature tolerance, and NaCl resistance.1
The initial categorization of Actinomycetes was mostly based on morphological observation; however, this is insufficient to distinguish between many taxa as some have very similar morphologies but differ in their diagnostic chemical makeup. It has been shown that chemotaxonomic marker studies satisfy the prerequisites for dependable classification techniques that represent evolutionary connections, at least to the genus level. Actinomycetes chemotaxonomy studies the distribution of certain cell membrane compounds, including fatty acids, sugars, polar lipids, menaquinones, and amino acids.87
Prokaryotes are currently classified and identified using polyphasic combinations of genotypic, chemotaxonomic, and phenotypic traits. Actinomycetes are first classified into taxa according to phenotypic characteristics including morphology, growth needs, or pathogenic potential. Subsequently, the physiological and biochemical characteristics of bacteria were also employed in this context. Techniques for DNA-DNA hybridization and chemotaxonomy were thereafter widely employed. The development of DNA amplification and sequencing methods specifically, the sequencing of the 16S rRNA gene constituted a significant advancement in establishing the taxonomic status of prokaryotes, significantly boosted the rate of finding new species, and is now frequently used as the initial stage in identifying novel organisms. The 16S rRNA gene was chosen as the ideal target molecule for analyzing phylogenetic relations because it is found in all bacteria, is functionally consistent, and is made up of both highly conserved and variable sections. Other molecular approaches used in prokaryote categorization include multi-locus sequencing typing (MLST), SDS-PAGE analysis of whole cell soluble proteins, secondary structure and signature nucleotide analysis of variable sections of the 16S rRNA gene, and SDS-PAGE analysis of variable areas of the 16S rRNA gene. However, genomic age said that some genomic parameters have significant promise in the taxonomy of bacteria and archaea as a replacement for the old way of determining G+C content molecular percentage and the laborious DNA-DNA hybridization (DDH) approach.88
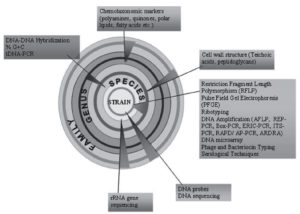
Polyphasic approach involves the very recent and modern techniques to differentiate between different species of actinomycetes. This method relies on information obtained from genetic methods in addition to morphological and biochemical data. Progress in the polyphasic approach to actinomycetes categorization, including the sequencing of the 16S rRNA gene and the integration of molecular fingerprinting techniques with other molecular markers, polyphasic approach has emerged as vital tool in the field of microbial systematics. The diagrammatic representation of polyphasic characterization of actinomycetes is mentioned in Figure 6.89
Figure 6. Diagrammatic representation of modern polyphasic approach in identifying actinomycetes upto species level.89
Bioactive molecules
Bioactive molecules are the natural products isolated from naturally occurring biological source such as plants, microbes, insects, mammals, etc. These play pivotal role in the therapeutics especially. The word natural here refers to the biological origin which is not synthetic by any means and also does not interfere with the DNA, fatty acids, aminoacids, sugars, which are involved often in the complex metabolism. In other words, naturally occurring secondary metabolites does not have any role to play in the metabolism of an organism. The word product stands for the outputs of the enzyme-mediated chemical reaction in the cell called metabolic pathways. Precisely, the term metabolites are referred instead of products. Hence, the secondary metabolites are those which has no significant role to play in the fundamental processes of life forms.43
Secondary metabolites are tiny compounds that are produced from living organisms but are not required for their growth or development. Most secondary metabolites, on the other hand, are “end products of complex biosynthetic processes” produced by highly structured enzyme systems, such as non-ribosomal peptide synthases (NRPS) and polyketide synthases (PKS), from basic constitutive aspects. Non-ribosomal peptide synthetases are modular enzymes that catalyse the synthesis of key peptide products from a wide range of proteinogenic and non-proteinogenic amino acid substrates.83 Polyketide synthases (PKSs) are multifunctional enzymes that biosynthesize a vast range of natural compounds, many of them are now denoted as antibiotics.90
Microbial secondary metabolites mark low molecular mass, structurally diverse products with a wide range of biological activities such as antimicrobial agents, enzyme inhibitors, antiparasitic and antitumor agents, plant growth stimulators, herbicides, insecticides and anthelmintics. Secondary metabolite synthesis in microorganisms is regulated by processes because it peaks in late growth phase (idiophase), becomes static in logarithmic phase, and diminishes in stationary growth phase. Secondary metabolites synthesized from microorganisms have a particular molecular structure that is not found in chemical libraries, and roughly 40% of microbial metabolites cannot be chemically produced. They are generated by a diverse collection of prokaryotic and eukaryotic organisms, the most persistent and adaptable of which include unicellular bacteria, fungus, and filamentous actinomycetes.91
Microorganisms produce a variety of secondary metabolites with great structural diversity and a wide range of biological activities.92 They produce about two-thirds of all known natural bioactive products, including some of the most commonly used compounds such as anti-cancer, anti-inflammatory, anti-bacterial, anti-fungal, anti-cancer compounds, etc. In fact, it is estimated that around 70% or more of the antibiotics currently in use are produced by organisms of the genus Streptomyces of the Actinomycetes type.93
Actinomycetes are morphologically diverse, aerobic, gram-positive bacteria with a high G+C nucleotide content (>55). They have been considered an abundant source of antibiotics since the discovery of the antibiotic Actinomyces actinomycin by Selman Waksman at Rutgers University in 1940.94 The Actinomycetes group is named after the first described anaerobic species, Actinomyces bovis, which causes actinomycosis, a “radioactive disease,” in cattle. They were originally thought to be an intermediate group between bacteria and fungi, but are now recognized as prokaryotic microorganisms.95 Actinomycetes are one of the most important domain types of bacteria. Surprisingly, the Actinomycetes class includes both a bacterial pathogen (genus Mycobacterium) and industrially important antibiotic-producing microorganisms (genus Streptomyces). Besides, Streptomyces are responsible for two-thirds of all known antibiotics.96
These are used as pharmacologically active compounds (antitumor, anti-inflammatory, neuroactive, antidepressant, anti-Alzheimer, cardioactive, platelet aggregation inhibitors, antioxidants, antihypertensive agents, vasodilators, nerve growth factor, interleukin and endothelin antagonists, estrogens, antiulcers, antiallergy). Antihistamines, anabolic steroids, anesthetics, anticoagulants, hemolytics, hypocholesterolemics, immune actives, immunosuppressants, immunomodulators, immune stimulators), enzymeinhibitors (peptidases, proteinases, glycosidases, amylases, HIV integrases, protein kinases, acetyl-coenzyme A) pesticides and other activities (antiparasitic, antimicrobial, herbicidal, phytotoxic, plant growth regulators, insecticides, nematocides, acaricides, larvicides, anthelmintics). acaricidal antimetabolites, ichthyotoxicants, algaecides, amoebicides, calcium antagonists, chelating agents, siderophores, morphogenic agents, signaling and quorum sensing compounds, radical scavengers, biosurfactants, food additives, microbial hormones and preservatives.97
Studies made on bioactive compounds
Active bio-molecules of actinomycetes and their applications
Actinomycetes secondary metabolites have a wide range of biological effects in addition to antibacterial activity. The manufacture of bioactive chemicals, include more than 10,000 antimicrobial agents used in pharmaceuticals, is carried out by the order Actinomycetales. Numerous biological activity of these compounds, including antibacterial, anticancer, immunosuppressive, and enzyme inhibitory qualities, have been demonstrated via significant research. Actinomycetes generate a variety of bioactive compounds, some of which are listed below.98,99
In 1941, Selman Waksman used the term “antibiotic” as a noun to mean any small chemical substance produced by a bacterium that inhibits the development of other microorganisms. The invention between 1945 and 1955 of penicillin, produced by a fungus, and streptomycin, chloramphenicol and tetracycline, produced by soil bacteria, marked the beginning of the antibiotic era as they are enzyme-catalyzed processes roughly similar to those in Similar to the production of proteins, fatty acids and polysaccharides, the biochemistry of antimicrobial proteins is slightly different and species specific.2
Production of antibiotics
In 1953, discovery of microorganisms that produce terramycin, neomycin, chloramphenicol, and tetracycline revolutionized the invention of new, more potent medications as well as the discovery of chemotherapeutic agents in the field of medicine that which has significantly decreased human suffering. Actinomycetes are known for manufacturing a wide range of antibiotics, including vancomycin, rifamycin, erythromycin, streptomycin, and tetracycline. The treatment of bacterial infections has greatly benefited from these antibiotics (Table 2).98 Two-thirds of microbial antibiotics are synthesized primarily by Actinomycetes species, of which 80% are exclusively produced by Streptomyces.99 These include tetracyclines, anthracyclines (doxorubicin), β-lactam antibiotics (penicillin, streptomycin), insecticides and anthelmintics, macrolides (erythromycin, azithromycin and clarithromycin), aminoglycosides (streptomycin, kanamycin, tobramycin and gentamicin) and aminoglycosides (streptomycin), kanamycin, monobactams, carbapenems and cephalosporins).100
Table (2):
List of applications and antibiotics produced by various Actinomycetes species
Antibiotic compound |
Actinomycetes species |
Source of Actinomycetes sps |
Application |
Ref. |
|---|---|---|---|---|
Abyssomycin |
Verrucosispora spp. |
Sediment sample collected in Japanese sea (289m deep) |
Antibacterial against MRSA (by inhibiting the biosynthesis of pABA pathway) |
101 |
Actinomycins |
Streptomyces anulatus |
Soil |
Antibacterial against different Gram positive and Gram negative organisms |
94 |
Amphomycin |
Streptomyces canus |
Soil |
Antimicrobial against vast range of Gram positive bacteria which includes S. aureus, M tuberculosis and Streptococcus pyogenes. |
102 |
Anthracyclin |
Micromonospora spp. |
Antibacterial |
103 |
|
Antibiotics and Fluorometabolites |
Streptomyces cattley |
Soil |
Antibacterial against both Gram negative and Gram positive bacteria. |
104 |
Aspartocins |
Streptomyces canus |
Soil |
Antibacterial against vast range of Gram positive bacteria which includes S. aureus, M tuberculosis and Streptococcus pyogenes. |
105 |
Avermectin |
Streptomyces avermitilis |
Soil |
Antiparasitic against roundworms, whipworms, filarial worms and scabies mites. |
106 |
Clostomicins |
Micromonospora spp. |
Soil |
Antibacterial against Gram positive bacteria including anaerobes. |
107 |
Chloramphenicol |
S. venezuelae |
Soil |
Antibacterial against Gram positive, Gram negative and anaerobic bacteria. |
107 |
Cycloheximide |
Streptomyces griseus |
Soil |
Antifungal against Candida albicans, Aspergillus fumigatus, Cryptococcus neoformans |
108 |
Erythromycin (Ilotycin) |
Saccharopolyspora erythraea |
Soil |
Antibacterial against S. aureus |
109 |
Gentamicin |
Micromonospora purpurea |
Soil |
Antibacterial effective against Gram negative bacteria including E. coli, P. aeruginosa, K. pneumonia |
110 |
Kanamycin |
Streptomyces kanamyceticus |
Soil |
Effective antibacterial against wide range of both Gram negative and Gram positive organisms. |
111 |
Kasugamycin |
Streptomyces kasugaensis |
Soil from city Nara in Japan |
Bactericidal against P. aeruginosa and Fungicidal against Magnaporthe oryzae (causing rice blast disease). |
112 |
Marinomycin |
Marinispora spp. |
Marine |
Antimicrobial against variety of Gram positive and Gram negative bacteria, as well as some fungi. |
113 |
Netamicin |
Micromonospora spp. |
Soil samples from natal, South Africa. |
Antifungal against Aspergillus, Penicillium and Candida. |
93 |
Oxytetracycline |
Streptomyces rimosus |
Soil |
Antibacterial, wide spectrum antibiotic against all bacteria. |
114 |
Rifamycin |
Streptomyces mediterranei |
Antibacterial |
115 |
|
Streptomycin |
Streptomyces griseus |
Soil |
Antibacterial against Mycobacterium tuberculosis, also effective against E. coli, S. typhi, and Shigella dysenteriae. |
116 |
Tetracycline |
Streptomyces aureofaciens |
Antibacterial effective against Gram negative and Gram positive microorganisms. |
117 |
|
Vancomycin |
Amycolatopsis orientalis |
Soil |
Antibacterial against Gram positive bacteria (cell wall synthesis inhibitor). |
118 |
Production of enzymes
Exsisting literature depicts, different species of actinomycetes produce a wide range of enzymes that can be used in biotechnological and microbiological domains. Actinomycetes are continuously screened based on data obtained from advances made in their genome and protein sequencing. These bacteria are screened for the production of enzymes such as amylases, proteases, chitinases, xylanases, cellulases, and other enzymes (Table 3).11
Table (3):
List of enzymes know to produce by Actinomycetes and their applications
Enzyme |
Industry |
Use |
Streptomyces strains |
Source |
Ref. |
|---|---|---|---|---|---|
Aminoacylase |
Pharmaceuticals |
In the manufacturing of semisynthetic cephalosporin and penicillin |
S. olivaceus |
Soil |
119 |
L-Asparaginase |
Medicine |
In treating of acute lymphoblastic leukemia |
Streptomyces griseus, S. karnatakensis, S. albidoflavus and Nocardia sp. |
Soil |
120 |
Tyrosinase |
Pharmacy |
L-Dopa synthesis |
S. glausescens S.castaneoglobisporus S. castaneoglobisporus |
Soil |
123 124 100 |
Bioherbicides
The bioactive molecules of Actinomycetes offer uses in preventing the growth of unwanted herbs (Table 4).
Table (4):
List of agriculture important metabolites produced by different Actinomycetes
Bioherbicides |
Biocontrol |
Streptomyces strains |
Source |
Ref. |
|---|---|---|---|---|
Anisomycin |
Effective against Plasmodium falciparum. |
Streptomyces sp. |
Soil |
125 |
Herbicidines and herbimycins |
Wide range of weeds including Monocotyledonous and dicotyledonous weed. |
S. hygroscopicus |
Soil |
126 |
Bialaphos |
Effective in controlling annual and perennial grassy weeds as well as broad-leaved weeds. |
S. viridochromogenes |
Soil |
127 |
Pigments
Microbial pigments are considered safe than that of the synthetic pigments. Microbial pigments produced by the organisms are defensive in nature against harmful environmental conditions. Microbial pigments are non-toxic, non-carcinogenic, and biodegradable, and they have a wide range of applications in industry. These pigments are often blue, violet, red, rose, yellow, green, brown, and black in varied colors that can be dissolved in the medium or retained in the mycelium. These microorganisms may also produce and exude dark pigments known as melanin or melanoid (Table 5).11,128
Table (5):
List of natural pigment produced by Actinomycetes
Pigments |
Streptomyces strain |
Class |
Ref. |
|---|---|---|---|
Actinomycin |
Streptomyces sp. |
Phenoxazinone |
94 |
Melanin |
Streptomyces lavendulae |
129 |
Anti-tumor compounds
Actinomycetes are diverse groups capable of producing distinctive cytotoxic compounds with antitumor activity. Their naturally occurring bioactive chemicals have a wide range of biological actions, including antibacterial and anti-cancer activity (Table 6). For example, doxorubicin, a well-known anticancer drug obtained from the soil-derived Streptomyces peucetius, and marinomycin, an antitumor and antibacterial molecule derived from the marine actinobacter species Marinispora sp.130
Table (6):
List of anti-tumor compounds produced by different Actinomycetes and their applications
Actinomyces strain |
Anti-tumor compounds |
Effective in Treating |
Ref. |
|---|---|---|---|
Micromonospora spp. (Endophytes) |
Anthraquinones |
Epithelial cancers |
131 |
Nocardia asteroides |
Asterobactine |
Breast and Colon cancer |
132 |
Streptomyces spp. |
Borrelidine |
Lung and Breast cancer |
133 |
Micromonospora spp. |
Diazepinomicin |
134 |
|
Actinomadura spp. |
Isotrichodermin (IB-00208) |
Potent inhibitor of tumor cells in vitro |
135 |
Micromonospora spp. |
Megalomicin (LL-E33288 complex) |
136 |
|
Micromonospora spp. |
Lomaiviticins |
Effective against leukemia, lymphoma, breast cancer. |
136 |
Micromonospora spp. |
Lupinacidins |
131 |
|
Thermoactinomyces spp. (Marine) |
Mechercharmycin |
Against variety of cancer cells. |
137 |
Marinospora spp. (Marine) |
Marinomycin |
138 |
|
Salinispora tropica |
Salinosporamide |
139 |
|
Streptomyces peucetius (Soil) |
Doxorubicin (adriamycin) |
Colorectal cancer |
140 |
Anti-viral compounds
The bioactive compounds derived from Actinomycetes are proven effective against virus. Studies of a marine actinomycete strain, Streptomyces kaviengensis, isolated from the coast of New Ireland (Papua New Guinea) enabled to identify a novel metabolite with remarkable antiviral activity tell. The compound, Antimycin A1a, was found to be a derivative of Antimycin A and showed high potency against Western Encephalitis Virus. In addition, previously known antimycin A has demonstrated broad-spectrum activity against a variety of RNA viruses, including members of the families Togaviridae, Flaviviridae, Bunyaviridae, Picornaviridae and Paramyxoviridae (Table 7).141
Table (7):
List of antiviral compounds produced by different Actinomycetes and their applications
Streptomyces species |
Anti-viral compound |
Effective against |
Reference |
|---|---|---|---|
Streptomyces hygroscopicus |
Hygromycin |
142 |
|
Streptomyces spp. |
Panosialins |
HIV,influenza, HSV |
143 |
Streptomyces kaviengensis |
Antimycin A1a |
141 |
Microbial communities comprise an endless and unique array of chemical entities and, therefore, represent a remarkable source for biotechnology and industry. A large number of valuable antibiotics and bioactive metabolites are reported from both terrestrial and aquatic diverse niches. In this sense, the scientific communities have decided to explore new environments for bio-prospecting in search of new biologically active compounds. Actinomycetes population is widespread and its studies are gaining thrust in India and other parts of the world since they produce novel bioactive molecules which can address the emerging problems with respect to environmental concerns, antimicrobial resistance genes in bacteria, agricultural valuable compounds and similar molecules. The culturing of these organisms is quite difficult and optimization of the media fulfils the criteria. Therefore, for effective isolation, physical treatments, chemical treatments or combined treatments alongside employing suitable substrate medium ensures the prolific isolation of Actinomycetes.
ACKNOWLEDGMENTS
The authors would like to express their sincere gratitude to Davangere University, Davangere for its unwavering support and guidance throughout the preparation of this review article.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
This article does not contain any studies on human participants or animals performed by any of the authors.
- Gurung TD, Sherpa C, Agrawal VP, Lekhak B. Isolation and Characterization of Antibacterial Actinomycetes from Soil Samples of Kalapatthar, Mount Everest Region. Nepal J Sci Technol. 1970;10:173-182.
Crossref - Clardy J, Fischbach MA, Currie CR. The natural history of antibiotics. Curr Biol. 2009;19(11):R437-41.
Crossref - Singh R, Dubey AK. Diversity and applications of endophytic Actinomycetes of plants in special and other ecological niches. Front Microbiol. 2018;9:767.
Crossref - Agadagba SK. Isolation of Actinomycetes from Soil. J Microbiol Res.2014(3):136-140.
Crossref - Jose PA, Jebakumar SRD. Phylogenetic appraisal of antagonistic, slow growing actinomycetes isolated from hypersaline inland solar salterns at sambhar salt lake, India. Front Microbiol. 2013;4:1-9.
Crossref - Balagurunathan R, Radhakrishnan M, Somasundaram ST. L-glutaminase producing actinomycetes from marine sediments-selective isolation, semi quantitative assay and characterization of potential strain. Aust J Basic Appl Sci. 2010;4(5):698-705.
- Sapkota A, Thapa A, Budhathoki A, Sainju M, Shrestha P, Aryal S. Isolation, Characterization, and Screening of Antimicrobial-Producing Actinomycetes from Soil Samples. Int J Microbiol. 2020;2716584.
Crossref - Selim MSM, Abdelhamid SA, Mohamed SS. Secondary metabolites and biodiversity of actinomycetes. J Genet Eng Biotechnol. 2021;19(1).
Crossref - Velho-Pereira S, Kamat NM. Actinobacteriological research in India. Indian J Exp Biol. 2013;51(8):573-596.
- Hayakawa M, Ishizawa K, Nonomura H. Distribution of rare actinomycetes in Japanese soils. J Ferment Technol. 1988;66(4):367-373.
Crossref - Mousumi D, Dayanand A. Production and Antioxidant Attribute of L-Glutaminase From Streptomyces Enissocaesilis Dmq-24. 2013;2(3):1-9.
- Sharma P, Thakur D. Antimicrobial biosynthetic potential and diversity of culturable soil Actinomycetes from forest ecosystems of Northeast India. Sci Rep. 2020;10(1):4104.
Crossref - Radhakrishnan M, Suganya S, Balagurunathan R, Kumar V. Preliminary screening for antibacterial and antimycobacterial activity of actinomycetes from less explored ecosystems. World J Microbiol Biotechnol. 2010;26(3):561-566.
Crossref - Quadri SR, Agsar D. Detection of Melanin Producing Thermo-Alkaliphilic Streptomyces from Limestone Quarries of the Deccan Traps. World Journal of Nuclear Science and Technology. 2012;2(2):08-12.
- Guo X, Liu N, Li X, et al. Red soils harbor diverse culturable actinomycetes that are promising sources of novel secondary metabolites. Appl Environ Microbiol. 2015;81(9):3086-3103.
Crossref - Aly MM, Bahamdain LA, Aba SA. Unexplored Extreme Habitats as Sources of Novel and Rare Actinomycetes with Enzyme and Antimicrobial Activities. IOSR J Pharm Biol Sci e-ISSN. 14(6):45-54.
Crossref - Daigham GE, Mahfouz AY. Isolation, characterization, and screening of actinomycetes producing bioactive compounds from Egyptian soil. Egypt Pharmaceut J. 2020;19:381-390.
Crossref - Malviya MK, Pandey A, Trivedi P, Gupta G, Kumar B. Chitinolytic activity of cold tolerant antagonistic species of streptomyces isolated from glacial sites of Indian Himalaya. Curr Microbiol. 2009;59(5):502-508.
Crossref - Shivlata L, Satyanarayana T. Thermophilic and alkaliphilic Actinobacteria: Biology and potential applications. Front Microbiol. 2015;6:1-29.
Crossref - Pandey A, Ali I, Singh Butola K, Chatterji T, Singh V. Isolation and characterization of Actinomycetes from soil and evaluation of antibacterial activities of Actinomycetes against pathogens. Int. J. Appl. Biol. Pharm. 2011;2(4):384-392.
- Aini NN, Sulistyani N. Isolation of Actinomycetes from Sugarcane (Saccharum officinarum) Rhizosphere and the Ability to Produce Antibiotic. Advances in Health Sciences Research. 2019;18:11-16.
Crossref - Rana S, Salam MD. Antimicrobial Potential of Actinomycetes Isolated from Soil Samples of Punjab, India. J Microbiol Exp. 2014;1(2):63-68.
Crossref - Anandan R, Dharumadurai D, Manogaran GP, Anandan R, Dharumadurai D, Manogaran GP. An Introduction to Actinobacteria. In: Actinobacteria – Basics and Biotechnological Applications. IntechOpen. 2016.
Crossref - Gebreyohannes G, Moges F, Sahile S, Raja N. Isolation and characterization of potential antibiotic producing actinomycetes from water and sediments of Lake Tana, Ethiopia. Asian Pac J Trop Biomed. 2013;3(6):426-435.
Crossref - Srinivasan R, Kannappan A, Shi C, Lin X. Marine bacterial secondary metabolites: A treasure house for structurally unique and effective antimicrobial compounds. Mar Drugs. 2021;19(10):530.
Crossref - Gupta C, Prakash D. Nutraceuticals from Microorganisms of Marine Sources. www.intechopen.com
- Dolbeth M, Arenas F. Marine Ecosystems: Types, Their Importance and Main Impacts. In: Leal Filho W, Azul AM, Brandli L, Lange Salvia A, Wall T, eds. Life Below Water. Encyclopedia of the UN Sustainable Development Goals. 2020:1-17:1-17.
Crossref - Jagannathan SV, Manemann EM, Rowe SE, Callender MC, Soto W. Marine Actinomycetes, New Sources of Biotechnological Products. Mar Drugs. 2021;19(7):365.
Crossref - Karuppiah V, Wei S, Zhiyong L. Marine Actinomycetes associated with marine organisms and their potentials in producing pharmaceutical natural products. Appl Microbiol Biotechnol. 2014;98(17):7365-7377.
Crossref - Shivaveerakumar S, Madhusudhan DN , Dayanand A. Catalytic zone, a novel screening approach for the production of extracellular tyrosinase by Streptomyces vinaceusdrappus DSV5. J Pure Appl Microbiol. 2013;7(4):2799-2808.
- Annarita P, Ilaria F, Ida R, Alessia G, Licia L, Barbara N. Microbial Diversity in Extreme Marine Habitats and Their Biomolecules. Microorganisms. 2017;5(2):25.
Crossref - Subramani R, Sipkema D. Marine Rare Actinomycetes: A Promising Source of Structurally Diverse and Unique Novel Natural Products. Mar Drugs. 2019;17(5):249.
Crossref - Nyle C, Ray RW. The Nature and Properties of Soil, 15th Ed. Pearson, Maryland, 2016.
- Sachidanand B, Mitra N, Vinod K, Richa R, Mishra B. Soil as a Huge Laboratory for Microorganisms. Agric Res Technol Open Access J. 2019;22(4):113-138.
Crossref - Fayuan W, Quanlong W, Catharine A. A, Yuhuan S, Shuwu Z. Effects of microplastics on soil properties: Current knowledge and future perspectives. J Hazard Mater. 2022;424(Pt C):127531.
Crossref - Jiang J, Trundle P, Ren J, et al. An Introduction to Actinobacteria. Intech. 2010;34(8):57-67.
Crossref - Kumar V, Singh Bisht G, Gusain O. Terrestrial Actinomycetes from Diverse Locations of Uttarakhand, India: Isolation and Screening for Their Antibacterial Activity. Iran. J. Microbiol. 2013;5(3): 299-308.
- Ma A, Zhang X, Jiang K, et al. Phylogenetic and Physiological Diversity of Cultivable Actinomycetes Isolated From Alpine Habitats on the Qinghai-Tibetan Plateau. Front Microbiol. 2020;11:555351.
Crossref - You KM, Park YK. A new method for the selective isolation of actinomycetes from soil. Biotechnol Tech. 1996;10(7):541-546.
Crossref - Basavaraj KN, Chandrashekhara S, Shamarez AM, Goudanavar PS, Manvi FV. Isolation and morphological characterization of antibiotic producing actinomycetes. Trop J Pharm Res. 2010;9(3):230.
Crossref - Bizuye A, Moges F, Andualem B. Isolation and screening of antibiotic producing actinomycetes from soils in Gondar town, North West Ethiopia. Asian Pacific J Trop Dis. 2013;3(5):375-381.
Crossref - Sheik GB, Maqbul MS, S. GS, S RM. Isolation and Characterization of Actinomycetes From Soil of Ad-Dawadmi, Saudi Arabia and Screening Their Antibacterial Activities. Int J Pharm Pharm Sci. 2017;9(10):276.
Crossref - Chaudhary HS, Yadav J, Shrivastava AR, Singh S, Singh AK, Gopalan N. Antibacterial activity of actinomycetes isolated from different soil samples of Sheopur (A city of central India). J Adv Pharm Technol Res. 2013;4(2):118-123.
Crossref - El Karkouri A, Assou SA, El Hassouni M. Isolation and screening of actinomycetes producing antimicrobial substances from an extreme Moroccan biotope. Pan Afr Med J. 2019;33:329.
Crossref - Singh V, Haque S, Singh H, et al. Isolation, Screening, and Identification of Novel Isolates of Actinomycetes from India for Antimicrobial Applications. Front Microbiol. 2016;7:1921.
Crossref - Shivabai C, Gutte S. Isolation of Actinomycetes From Soil Sample Using Different Pretreatment Methods And Its Comparative Study. Int. J. Res. Anal. Rev. 2019;6(2):697-702
- Hayakawa M, Momose Y, Yamazaki T, Nonomura H. A method for the selective isolation of Microtetraspora glauca and related four-spored actinomycetes from soil. J Appl Bacteriol. 1996;80(4):375-386.
Crossref - Antal N, Fiedler HP, Stackebrandt E, Beil W, Stroch K, Zeeck A. Retymicin, galtamycin B, saquayamycin Z and ribofuranosyllumichrome, novel secondary metabolites from Micromonospora sp. T 6368. I. Taxonomy, fermentation, isolation and biological activities. J Antibiot. 2005;58(2):95-102.
Crossref - Hayakawa M, Iino H, Takeuchi S, Yamazaki T. Application of a method incorporating treatment with Chloramine-T for the selective isolation of Streptosporangiaceae from the soil. J Ferment Bioeng. 1997;84(6):599-602.
Crossref - Matsukawa E, Nakagawa Y, Iimura Y, Hayakawa M. Stimulatory effect of indole-3-acetic acid on aerial mycelium formation and antibiotic production in Streptomyces spp. Actinomycetologica. 2007;21(1):32-39.
Crossref - Takahashi Y, Omura S. Isolation of new actinomycete strains for the screening of new bioactive compounds. J Gen Appl Microbiol. 2003;49(3):141-154.
Crossref - Bredholdt H, Galatenko OA, Engelhardt K, Fjaervik E, Terekhova LP, Zotchev SB. Rare actinomycete bacteria from the shallow water sediments of the Trondheim fjord, Norway: isolation, diversity and biological activity. Environ Microbiol. 2007;9(11):2756-2764.
Crossref - Das M. Exploration of Plastic Degrading Actinomycetes from Deccan Trap of Karnataka. Journal of Innovative Research in Engineering & Technology-ISSN. 2017;(01):1-5.
- Kumar N, Singh R, Mishra S, Singh A, Pachouri UC. Isolation and screening of soil Actinomycetes as source of antibiotics active against bacteria. Int J Microbiol Res. 2010;2(2):12-16.
Crossref - Nonomura H, Ohara Y. Distribution of actinomycetes in soil. VI. A culture method effective for both preferential isolation of Microbispora and Streptosporangium strains in soil (Part I). J Ferment. Technol. 1969;47:463-469.
- Dhawane VP, Zodpe SN. Screening and Isolation of Pigment Producers and Non-pigment Producers Actinomycetes from Rhizosperic Soil Sample. Int J Curr Microbiol App Sci. 2017;6(5):1570-1578.
Crossref - Ganesan P, Reegan AD, David RHA, et al., Antimicrobial activity of some actinomycetes from Western Ghats of Tamil Nadu, India. Alexandria J Med. 2017;53(2):101-110.
Crossref - Niyomvong N, Pathom-aree W, Thamchaipenet A, Duangmal K. Actinomycetes from Tropical Limestone Caves. Chiang Mai J Sci. 2012; 39(3): 373-388
- Ayoib A, Gopinath SCB, Yahya ARM, Zakaria L. Coal-vitamin medium for improved scheme of isolating biosurfactant-producing actinomycetes of rare species from soil samples. Biomass Convers Biorefin. 2023.
Crossref - Kalimuthu K, Kumaravel M, Haldar D, Perumal A. Isolation and Characterization of Marine Actinomycetes – Bioprospecting for Antimicrobial, Antioxidant and Anticancer Properties. Res Sq. 2022.
Crossref - Lakshmipathy D, Krishnan K. A Morphological, Biochemical and Biological Studies of Halophilic Streptomyces sp. Isolated from Saltpan Environment. Am J Infect Dis. 2009;5(3):207-213.
Crossref - Baskaran R, Vijayakumar R, Mohan P. Enrichment method for the isolation of bioactive actinomycetes from mangrove sediments of Andaman Islands, India. Malays. J Microbiol. 2011;7(1):26-32.
Crossref - Ellaiah P, Reddy A. Isolation of actinomycetes from marine sediments off Visakhapatnam, east coast of India. Indian Journal of Geo-Marine Sciences (IJMS). 1987;16(2):134-135.
- Sheikh M, Rathore D, Gohel S, Singh S. Cultivation and characteristics of the Marine Actinobacteria from the Sea water of Alang, Bhavnagar. Indian Journal of Geo-Marine Sciences. 2019;48:1896-1901.
- Kokare CR, Kadam SS, Mahadik KR, Chopade BA. Studies on bioemulsifier production from marine Streptomyces sp. S1. Indian J Biotechnol. 2007;6(1):78-84.
- Vijayakumar R, Muthukumar C, Thajuddin N, Panneerselvam A, Saravanamuthu R. Studies on the diversity of actinomycetes in the Palk Strait region of Bay of Bengal, India. Actinomycetologica. 2007;21(2):59-65.
Crossref - Hong K, Gao AH, Xie QY, et al. Actinomycetes for marine drug discovery isolated from mangrove soils and plants in China. Mar Drugs. 2009;7(1):24-44.
Crossref - Sathish KS, Kokati VBR. In-vitro antimicrobial activity of marine actinobacteria against multidrug resistance Staphylococcus aureus. Asian Pac J Trop Biomed. 2012;2(10):787-792.
Crossref - Velmurugan S, John ST, Nagaraj DS, Ashine TA, Kumaran S, Pugazhvendan SR. Isolation of Actinomycetes from Shrimp Culture Pond and Antagonistic to Pathogenic Vibrio spp. and WSSV. Int J Curr Microbiol App Sci, 2015;4(7): 82-92: 2319-7706. https://www.ijcmas.com/vol-4-7/S.%20Velmurugan,%20et%20al.pdf.
- Bonnet M, Lagier JC, Raoult D, Khelaifia S. Bacterial culture through selective and non-selective conditions: the evolution of culture media in clinical microbiology. New Microbes and New Infections. 2020;34:100622.
Crossref - Santos Junior VD, Nizoli ֹ, Galvan D, et al. Micronutrient requirements and effects on cellular growth of acetic acid bacteria involved in vinegar production. Food Sci Technol. 2022;42:e05121.
Crossref - Srinivasan MC, Laxman RS, Deshpande MV. Physiology and nutritional aspects of actinomycetes: an overview. World J Microbiol Biotechnol. 1991;7(2):171-184.
Crossref - Bundale S, Singh J, Begde D, Nashikkar N, Upadhyay A. Culturable rare actinomycetes from Indian forest soils: Molecular and physicochemical screening for biosynthetic genes. Iran J Microbiol. 2018;10(2):132-142.
- Pridham TG, Gottlieb D. The Utilization of Carbon Compounds by Some Actinomycetales as an Aid for Species Determination. J Bacteriol. 1948;56(1):107-114.
Crossref - Shivaveerakumar S, Madhusudhan DN, Raghavendra H, Agsar D. Screening of streptomyces and process optimization for the production of tyrosinase. J Pure Appl Microbiol. 2014;8(4):3245-3252.
- Hayakawa M, Nonomura H. Humic acid-vitamin agar, a new medium for the selective isolation of soil actinomycetes. J Fermentat Technol. 1987;65(5):501-509.
Crossref - Jiang Y, Li Q, Chen X, Jiang C. Isolation and Cultivation Methods of Actinobacteria. In: Dhanasekaran D, Jiang Y, eds. Actinobacteria – Basics and Biotechnological Applications. InTech. 2016.
Crossref - Shirling EB, Gottlieb D. Methods for characterization of Streptomyces species1. Int J Sys Evol Microbiol. 1966;16(3):313-340.
Crossref - Pridham TG, Anderson P, Foley C, Lindenfelser LA, Hesseltine CW, Benedict RG. A selection of media for maintenance and taxonomic study of Streptomyces. Antibiotics Ann. 1956:947-953.
- Williams ST, Goodfellow M, Alderson G, Wellington EMH, Sneaih PHA, Sackin MJ. Numerical-classification of Streptomyces and related genera. J. Gen. Microbial. 1983;129:1743-1813.
Crossref - Singh Parmar R, Singh C, Saini P, Kumar A. Isolation and Screening of Antimicrobial and Extracellular Pigment Producing Actinomycetes from Chambal Territory of Madhya Pradesh Region, India. International Journal of Advance Research and Innovation. 2016;4(1):138-145.
Crossref - Mohamed H, Miloud B, Zohra F, et al., Isolation and Characterization of Actinobacteria from Algerian Sahara Soils with Antimicrobial Activities. Int J Mol Cell Med.2017;6(2):109-120.
Crossref - Sujatha P, Bapiraju KVVSN, Ramana T. Studies on a new marine streptomycete BT-408 producing polyketide antibiotic SBR-22 effective against methicillin resistant Staphylococcus aureus. Microbiol Res. 2005;160(2):119-126.
Crossref - Padma BJ, Saraswathi K, Arumugam P, Shiny R. A. Isolation, characterization and evaluation of antioxidant activities of secondary metabolites producing actinomycetes of terrestrial origin. Int J Res Med Sci. 2018;6(3):1017.
Crossref - Das M, Agsar D, Shivaveerakumar S. Production of L-glutaminase by Streptomyces rochei Detected from the Lime Stone Quarries of Deccan Trap. J Pure Appl Microbiol. 2013;7(3):2251-2260
- Djebbah FZ, Al-Dhabi NA, Arasu MV, et al. Isolation and characterisation of Streptomyces sp. strain GLD25 with antimicrobial and antioxidant effects from Gueldaman cave (GLD1), Akbou-Algeria. J King Saud Univ. 2022;34(1):101-719.
Crossref - Wang Y, Jiang Y. Chemotaxonomy of Actinobacteria. In: Actinobacteria – Basics and Biotechnological Applications. IntechOpen. 2016.
Crossref - Chen X, Jiang Y, Li Q, Han L, Jiang C. Molecular Phylogenetic Identification of Actinobacteria. In: Dhanasekaran D, Jiang Y, eds. Actinobacteria – Basics and Biotechnological Applications. InTech. 2016.
Crossref - Prakash O, Verma M, Sharma P, et al., Polyphasic approach of bacterial classification – An overview of recent advances. Indian J Microbiol. 2007;47(2):98-108.
Crossref - Nivina A, Yuet KP, Hsu J, Khosla C. Evolution and Diversity of Assembly-Line Polyketide Synthases. Chem Rev. 2019;119(24):12524-12547.
Crossref - Demain AL. Microbial secondary metabolism: a new theoretical frontier for academia, a new opportunity for industry. Ciba Found Symp. 1992;171:3-16.
Crossref - Newman DJ, Cragg GM, Snader KM. Natural products as sources of new drugs over the period. J Nat Prod. 2003;66(7):1022-1037.
Crossref - Berdy J. Bioactive Microbial Metabolites. J Antibiot. 2005;58(1):1-26.
Crossref - Waksman SA, Woodruff HB. Bacteriostatic and Bactericidal Substances Produced by a Soil Actinomyces. Exp Biol Med. 1940;45(2):609-614.
Crossref - Küster E. Taxonomy of soil actinomycetes and related organisms. In Gray TRG and Parkinson D (eds.), The ecology of soil bacteria. Liverpool University Press, Liverpool. 1968: 322-336.
- Okami Y, Hotta K. Search and Discovery of New Antibiotics. In: Goodfellow M, Williams ST, Mordarski M (eds.), Actinomycetes in Biotechnology, Academic Press, London. 1988:33-67.
Crossref - Berdy J. Thoughts and facts about antibiotics: where we are now and where we are heading. J Antibiot (Tokyo) 2012;65:441.
Crossref - Prescott LM, Harley JP, Klein DA. Microbiologie. Bruxelle: De Boek &Larcier; 2007.pp. 805-825
- Pandey B, Ghimire P, Agrawal V. Studies on the antibacterial activity of the Actinomycetes isolated from the Khumbu Region of Nepal. J App Mirobiol. 2004;20.
- Hamdi B, Mohamed H. Streptomyces Secondary Metabolites. In; 2018:99-122.
Crossref - Bister B, Bischoff D, Strobele M, et al. Abyssomicin C – A polycyclic antibiotic from a marine Verrucosispora strain as an inhibitor of the p-aminobenzoic acid/tetrahydrofolate biosynthesis pathway. Angew Chem Int Ed Engl. 2004;43(19):2574-2576.
Crossref - Heinemann B, Kaplan MA, Muir RD, Hooper IR. Amphomycin, a new antibiotic. Antibiot Chemother. 1953;3(12):1239-1242.
- Grein A, Merli S, Spalla C. New Anthracycline Glycosides from Micromonospora I. Description of The Producing Strain. J Antibiot. 1980;33(12):1462-1467.
Crossref - Barbe V, Bouzon M, Mangenot S, et al. Complete genome sequence of Streptomyces cattleya NRRL 8057, a producer of antibiotics and fluorometabolites. J Bacteriol. 2011;193(18):5055-5056.
Crossref - Yang HJ, Huang XZ, Zhang ZL, et al. Two novel amphomycin analogues from Streptomyces canus strain FIM-0916. Nat Prod Res. 2014;28(12):861- 867.
Crossref - Burg RW, Miller BM, Baker EE, et al. Avermectins, new family of potent anthelmintic agents: producing organism and fermentation. Antimicrob Agents Chemother. 1979;15(3):361-367.
Crossref - Omura S, Imamura N, Oiwa R, Kuga H, Iwata R, Masuma R, Iwai Y. Clostomicins. New antibiotics produced by Micromonospora echinospora subsp. armeniaca subsp. nov. I. Production, isolation, and physico-chemical and biological properties. J Antibiot. 1986;39(10):1407-1412.
Crossref - Kominek LA. Biosynthesis of novobiocin by Streptomyces niveus. Antimicrob Agents Chemother. 1972;1(2):123-134.
Crossref - Mc Guire JM, Bunch RL, Anderson RC, et al. Ilotycin, a new antibiotic. Antibiot Chemother (Northfield). 1952;2(6):281-283.
- Weinstein MJ, Luedemann GM, Oden EM, et al. Gentamicin, a new antibiotic complex from Micromonospora. J Med Chem. 1963;6:463-464.
Crossref - Umezawa H, Ueda M, Maeda K, et al. Production and isolation of a new antibiotic: kanamycin. J Antibiot. 1957;10(5):181-188.
- Umezawa H, Okami Y, Hashimoto T, Suhara Y, Hamada M, Takeuchi T. A new antibiotic, kasugsmycin. J Antibiot. 1965;18:101-103.
- Kwon HC, Kauffman CA, Jensen PR, Fenical W. Marinomycins A-D, antitumor-antibiotics of a new structure class from a marine actinomycete of the recently discovered genus “Marinispora.” J Am Chem Soc. 2006;128(5):1622-1632.
Crossref - Rhodes PM. The production of oxytetracycline in chemostat culture. Biotechnol Bioeng. 1984;26(4):382-385.
Crossref - Margalith P, Beretta G. Rifomycin. XI. Taxonomic study on Streptomyces mediterranei nov. sp. Mycopathol Mycol Applic. 1960;13:321-330.
Crossref - Schatz A, Waksman SA. Effect of streptomycin and other antibiotic substances upon Mycobacterium tuberculosis and related organisms. Proc Soc Exp Biol Med. 1944;57(2):244-248.
Crossref - Darken MA, Berenson H, Shirk RJ, Sjolander NO. Production of tetracycline by Streptomyces aureofaciens in synthetic media. Appl Microbiol. 1960;8(1):46-51.
Crossref - Brigham RB, Pittenger RC. Streptomyces orientalis, n. sp., the source of vancomycin. Antibiot Chemother (Northfield). 1956;6(11):642-647.
- Sugie M, Suzuki H. Purification and properties of D-aminoacylase of Streptomyces olivaceus. Agric. Biol. Chem. 1978;42:107–113.
- DeJong P. L-Asparaginase production by Streptomyces griseus. Appl Microbiol. 1972;23(6):1163-1164.
Crossref - Narayana KJP, Kumar KG, Vijayalakshmi M. L-asparaginase production by Streptomyces albidoflavus. Indian J. Microbiol. 2007;48(3): 331-336.
Crossref - Mostafa SA, Ali OA. Identity and lipase productivity of a mesophilic actinomycete isolated from Egyptian soil. Zentralbl Bakteriol. 1979;134(4):316-324.
Crossref - Kupper U, Niedermann DM, Travaglini G, Lerch K. Isolation and characterization of the tyrosinase gene from Neurospora crassa. J Biol Chem. 1989;264(29):17250- 17258.
Crossref - Matoba Y, Kumagai T, Yamamoto A, Yoshitsu H, Sugiyama M. Crystallographic evidence that the dinuclear copper center of tyrosinase is flexible during catalysis. J Biol Chem. 2006;281(13):8981-8990.
Crossref - Sobin, BA, Tanner FW. Anisomycin, a new anti-protozoan antibiotic. J Am Chem Soc. 1954;76(15):4053-4054.
Crossref - Omura S, Iwai Y, Takahashi Y, et al. Herbimycin, a new antibiotic produced by a strain of Streptomyces. J Antibiot. 1979;32(4):255-261.
Crossref - Bayer E, Gugel KH, Hagele K, et al. Stoffwechselprodukte von Mikroorganismen. Phosphinothricin und phosphinothricyl-alanyl-alanin. Helv Chim Acta. 1972;55(1):224-239.
Crossref - Polapally R, Mansani M, Rajkumar K, et al. Melanin pigment of Streptomyces puniceus RHPR9 exhibits antibacterial, antioxidant and anticancer activities. PLoS One. 2022;17(4):e0266676.
Crossref - Mencher JR, Heim AH. Melanin Biosynthesis by Streptomyces lavendulae. J Gen Microbiol. 1962;28(4):665-670.
Crossref - Manivasagan P, Venkatesan J, Sivakumar K, Kim SK. Marine Actinomycetesl metabolites: current status and future perspectives. Microbiol Res. 2013;168(6):311-332.
Crossref - Igarashi Y, Trujillo ME, Martinez-Molina E, et al. Antitumor anthraquinones from an endophytic actinomycete Micromonospora lupini sp. nov. Bioorg Med Chem Lett. 2007;17(13):3702-3705.
Crossref - Nemoto A, Hoshino Y, Yazawa K, et al. Asterobactin, a new siderophore group antibiotic from Nocardia asteroides. J Antibiot. 2002;55(6):593-597.
Crossref - Vino S, Lokesh KR. Borrelidin: a promising anticancer agent from Streptomyces species. Adv Biotech. 2008;6:22-26.
- Charan RD, Schlingmann G, Janso J, Bernan V, Feng X, Carter GT. Diazepinomicin, a new antimicrobial alkaloid from a marine Micromonospora sp. J Nat Prod. 2004;67(8):1431-1433.
Crossref - Malet-Cascon L, Romero F, Espliego-Vazquez F, Gravalos D, Fernandez-Puentes JL. IB-00208, a new cytotoxic polycyclic xanthone produced by a marine-derived Actinomadura. I. Isolation of the strain, taxonomy and biological activites. J Antibiot. 2003;56(3):219 -225.
Crossref - Hirsch AM, Valdes M. Micromonospora: an important microbe for biomedicine and potentially for biocontrol and biofuels. Soil Biol Biochem. 2009;42(4):536-542.
Crossref - Kanoh K, Matsuo Y, Adachi K, Imagawa H, Nishizawa M, Shizuri Y. Mechercharmycins A and B, cytotoxic substances from marine derived Thermoactinomyces sp. YM3-251. J Antibiot. 2005(4);58:289-292.
Crossref - Bull AT, Stach JEM. Marine actinobacteria: new opportunities for natural product search and discovery. Trends Microbiol. 2007;15(11):491-499.
Crossref - Williams DH, Williamson MP, Butcher DW, Hammond SJ. Detailed binding sites of the antibiotics vancomycin and ristocetin A: determination of intermolecular distances in antibiotic/substrate complexes by use of the time-dependent NOE. J Am Chem Soc. 1983;105(5):1332-1339.
Crossref - Arcamone F, Cassinelli G, Fantini G, et al. Adriamycin, 14-hydroxydaunomycin, a new antitumor antibiotic from S. peucetius var. caesius. 1969;11(6):1101-1110.
Crossref - Raveh A, Delekta PC, Dobry CJ, et al. Discovery of potent broad spectrum antivirals derived from marine Actinomycetes. PLoS One. 2013;8(12):e82318.
Crossref - Gonzalez A, Jimenez A, Vazquez D, Davies J, Schindler D. Studies on the mode of action of hygromycin B, an inhibitor of translocation in eukaryotes. Biochim Biophys Acta. 1978;521(2):459-469.
Crossref - Aoyagi T, Yagisawa M, Kumagai M, Hamada M, Okami Y. An enzyme inhibitor, panosialin, produced by Streptomyces. I. Biological activity, isolation and characterization of panosialin. J Antibiot. 1971;24(12):860-869.
Crossref
© The Author(s) 2024. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.