ISSN: 0973-7510
E-ISSN: 2581-690X
AcrAB-TolC is a resistance nodulation division type of efflux pump present in Enterobacteriaceae. It non-specifically effluxes antibiotics out of the bacterial cell, thus conferring drug resistance. Increase in the expression of the AcrAB-TolC efflux pump increases resistance to antibiotics. We aimed to study the expression levels of acrA and acrB that encodes AcrAB-TolC efflux pump, to understand efflux pump mediated resistance to carbapenem among clinical isolates of Enterobacteriaceae obtained from various clinical samples. Additionally, co -production of carbapenemase was also detected in the isolates demonstrating efflux pump mediated resistance to carbapenems. A total of 127 carbapenem resistant clinical isolates of Enterobacteriaceae, isolated from a tertiary care hospital were included in the study. An efflux pump inhibition (EPI) assay with reserpine, an efflux pump inhibitor, was performed to screen for isolates exhibiting efflux pump activity. Real Time Reverse Transcriptase qPCR was performed to detect the mRNA over expression levels of acrA and acrB that encodes AcrAB-TolC efflux pump. The control strains K. pneumoniae BAA2146 and E. coli AcrB were used. EPI assay with carbapenem showed that 56 /127(44%) isolates were screen positive indicating efflux pump activity. A total of 12 isolates showed 101 to 107 increase in the expression of both acrA and acrB when compared with the controls indicating a strong efflux pump activity, in addition to producing carbapenemase. The study highlights the role of efflux pump AcrAB-TolC in conferring resistance to carbapenem among clinical isolates of Enterobacteriaceae.
AcrAB-TolC, Reserpine, Carbapenem Resistance, Clinical Isolates of Enterobacteriaceae, Real Time RT qPCR
Enterobacteriaceae are a large family of Gram negative bacilli that often cause a variety of clinical infections – urinary tract infections, wound infections, sepsis, respiratory infections, etc.1 Infections caused by Enterobacteriaceae often require treatment with antibiotics. The efficient antimicrobial agents against this group of bacteria are cephalosporins, carbapenems, aminoglycosides and fluoroquinolones.2,3 Antibiotic resistance (including that related to Enterobacteriaceae) to the commonly used antimicrobials is a growing problem in both developed and developing countries and especially in ICUs.
Resistance to carbapenems is often mediated by enzymes called as carbapenemase and also by non-enzyme mediated methods like over expression of efflux pumps and loss of porin proteins. Efflux pump mediated resistance to carbapenems is one of the few other studied mechanisms of resistance among clinical isolates of Enterobacteriaceae. AcrAB-TolC is a multidrug Resistance Nodulation Division (RND) type of efflux pump, which is present among Enterobacteriaceae. This is made up of three components AcrA – the periplasmic adaptor protein that connects AcrB and TolC, AcrB – the active pump that pushes the substrate from within the bacterial cell and TolC – the outer membrane channel that efflux out the substrate.4,5 Over expression of the AcrA and AcrB, contributes to drug resistance by effective effluxing out of carbapenems, cephalosporins, fluoroquinolones and aminoglycosides. The AcrAB-TolC efflux pump plays an important role in producing multidrug resistance in Gram-negative bacteria since it is not substrate specific.6
Genes for efflux pumps are present on plasmids as well as chromosome. Mutations in the local repressor genes or activation of a regulon regulated by a global transcriptional regulator such as MarA or SoxS of E. coli, results in over expression of efflux pumps.7-10 Since efflux pumps have a broad substrate range, over expression of a pump will confer resistance to more than one class of antibiotics, as well as to some dyes and detergents.
Even though most of the bacteria carry genes that encode MDR efflux pumps on their chromosomes, some of these efflux pump encoding genes have been mobi¬lized onto plasmids that may have the potential to transfer between bacteria. This situation is disquieting, since it would result in dissemination of multidrug resistance, more so because efflux pumps have a broad substrate profile. A recent study reported an IncH1 plasmid that was isolated from a Citrobacter freundii strain that contained the RND MDR efflux pump and also carried the gene for the beta lactamase NDM.11
In order to understand enterobacterial resistance to carbapenems, it is imperative to comprehend different mechanisms of drug resistance. Enterobacteriaceae that over expresses efflux pumps have been isolated from patients earlier.12 Understanding the mechanism of over expression may help in designing novel therapeutics to prevent the production of efflux-pump proteins. A study from Turkey reported efflux pumps as the causative mechanism for multidrug resistant K. pneumoniae isolates.13 Tsai et al.14 from Taiwan showed that the over expression of AcrAB-TolC by ramR deletion in K. pneumoniae resulted in increased antibiotic resistance and reduced virulence at the same time. There are limited studies that have explored both beta lactamase as well as efflux pump mediated resistance, among clinical isolates of Enterobacteriaceae cultured from subjects in different tertiary care hospitals. Here we have demonstrated AcrAB-TolC efflux pump mediated resistance to carbapenems among clinical isolates of Enterobacteriaceae.15,16
Study Type: Cross Sectional
Inclusion Criteria
Clinical isolates of Enterobacteriaceae obtained from various clinical samples (urine, blood, sputum, body fluids) that were resistant to carbapenem were included in the study.
Study Isolates
A total of 127 carbapenem resistant clinical isolates of Enterobacteriaceae from clinical samples – urine, blood, sputum, body fluids, obtained from patients attending a tertiary care hospital were included in the study. The bacterial isolates were identified by standard microbiological procedures.
Phenotypic Screening of Efflux Pump Mediated Resistance to Carbapenems17
Reserpine, a non-specific efflux pump inhibitor was used to block the activity of efflux pumps involved in conferring antibiotic resistance. An efflux pump inhibition assay (EPI assay) was performed by determining MIC (agar dilution method) of imipenem in the presence and in the absence of reserpine at a fixed concentration of 25µg/mL. A 2 or more fold decrease in the MIC of imipenem in the presence of Reserpine when compared to the MIC of imipenem alone was considered to indicate the activity of efflux pump in conferring drug resistance.
Real Time Reverse Transcriptase qPCR to Confirm the Over Expression of Efflux Pump AcrAB-TolC
Real Time Reverse Transcriptase qPCR (Real Time RTqPCR) (Step One PlusTM Real Time PCR system, Applied BiosystemsTM) was performed to study the mRNA expression levels of acrA and acrB of the AcrAB-TolC efflux pump, that is known to confer resistance to antimicrobial drugs. Representative isolates which were screen positive for efflux pump activity against imipenem by EPI assay were grown overnight in LB broth (Hi Media). Total RNA was extracted using FavorPrep tissue total RNA mini kit and RNA was quantified for purity and concentration using Nanodrop at 260/280 and 260/230 nm. RNA was converted to cDNA by RT-PCR using Thermo scientific Verso cDNA synthesis kit. RT- PCR was performed in a conventional thermal cycler. Real Time RT qPCR was performed with SYBR Premix Ex Taq from Takara Clontech in Step One PlusTM Applied BiosystemTM using Comparative ΔΔCT program. The test was done in triplicates. gapA was used as the housekeeping gene for normalizing the transcription levels of target genes. The mean of the normalized expression (transcription) values were calculated for acrA and acrB for each strain manually as well as by using the Step One PlusTM software. Klebsiella pneumoniae BAA2146 was used as a positive control for studying and comparing the mRNA expression levels of both acrA and acrB among the screen positive isolates. E.coli AcrB, a strain that over expresses acrB, was used as a positive control for studying and comparing the mRNA expression levels of acrB among screen positive isolates.
The primers used for Real Time RT qPCR of acrA were forward primer-5’CTTAGCCCTAACAGGATGTG3’, reverse primer-5’TTGAAATTACGCTTCAGGAT3’; of acrB were: forward primer-5’CGTACACAGAAAGTGCTCAA3’, reverse primer-5’CGCTTCAACTTTGTTTTCTT3’. Real Time RT qPCR cycling conditions for acrA and acrB were as follows: initial denaturation at 95°C for 15 minutes, cycle denaturation at 94°C for 1 minute, annealing at 52°C for 1 minute, extension at 72°C for 1 minute, melt curve stage (continuous) 95°C for 15 seconds, 60°C for 1 minute, 95°C for 15 seconds.
Ethics
This study was approved by the Institute Ethics Committee of Dr. ALM PG IBMS, University of Madras, Taramani, Chennai. IHEC Approval No. UM/IHEC/01-2014-II, Dated 18-02-2015.
Phenotypic screening of efflux pump mediated resistance to carbapenems detected by efflux pump inhibition assay:
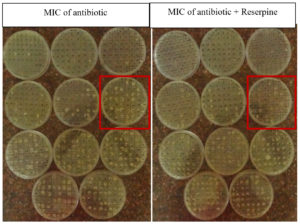
A total of 127 carbapenem resistant clinical isolates of Enterobacteriaceae were phenotypically screened for efflux pump mediated resistance to carbapenem. MIC of imipenem, a carbapenem, was determined for all the 127 isolates by agar dilution method. Concentration of imipenem ranged from 512 µg/mL to 1 µg/mL. MIC of imipenem with reserpine was determined. Isolates showing ≥ 2 fold decrease in MIC of imipenem with reserpine when compared to MIC of imipenem alone was considered to indicate the activity of efflux pumps in conferring resistance to imipenem (Figure 1).
Figure 1. Representative picture of Efflux pump inhibition assay by MIC by agar dilution against Imipenem and Imipenem + Reserpine.
The highlighted boxes indicate the reduced MIC of antibiotic with reserpine when compared to MIC of antibiotic among the tested isolates.
Out of 127 carbapenem resistant clinical isolates of Enterobacteriaceae, 56 (44%) isolates showed ≥ 2 fold decrease in MIC of imipenem with reserpine when compared to MIC of imipenem alone.
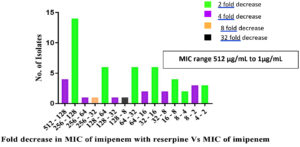
Out of 56 isolates, that showed a decrease in MIC of imipenem with reserpine, 41 isolates (73%) showed a 2 fold decrease in MIC. One isolate showed a 16 fold decrease in MIC, from 128 µg/mL as MIC of imipenem to 8 µg/mL as MIC of imipenem with reserpine. (Table 1) (Figure 2).
Table (1):
Isolates that were screen positive for efflux pump activity detected by efflux pump inhibition assay with imipenem and reserpine.
Fold decrease in MIC of imipenem + Reserpine Vs MIC of imipenem |
No. of isolates (n=56) |
|---|---|
2 fold decrease |
41 (73%) |
4 fold decrease |
13 (23%) |
8 fold decrease |
1 (2%) |
16 fold decrease |
1 (2%) |
Total no. of isolates |
56 |
Figure 2. Isolates showing varying fold decrease in MIC of imipenem with reserpine when compared to MIC of imipenem alone.
Real Time RT qPCR to Study the mRNA Over Expression of acrA and acrB among Clinical Isolates that were Screen Positive for Efflux Pump Mediated Resistance to Carbapenem
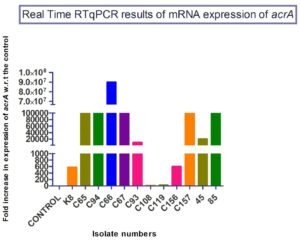
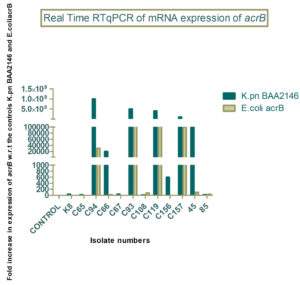
Out of 56 isolates that were screen positive for efflux pump mediated resistance to imipenem, 12 representative isolates were selected for Real Time RTqPCR to study and compare the mRNA expression levels of acrA and acrB with known positive controls.
The mRNA expression levels of acrA ranged from 101 to 107. The mRNA expression levels of acrB ranged from 101 to 109. The mRNA expression levels of test isolates were either comparable to the control or highly over expressed when compared to control (Figure 3, Figure 4).
Carbapenem resistance among Enterobacteriaceae is mediated mainly by acquired carbapenemases.18,19 Resistance to carbapenems is also mediated by a combination of non-carbapenemase beta lactamases or ESBLs and reduced drug influx via loss of outer-membrane porins.20 This study demonstrates, efflux pump mediated resistance to carbapenem group of antibiotics observed in clinical isolates of Enterobacteriaceae and also the potential of reserpine to be used along with carbapenem to treat infections caused by strains of carbapenem resistant Enterobacteriaceae showing efflux pump mediated antibiotic resistance.
A study by Adler et al.21 showed that mutations in acrB and acrR activates AcrAB-TolC mediated efflux thereby conferring high level carbapenem resistance in E.coli. Another study showed that over expression of efflux pumps expels carbapenems leading to carbapenem resistance associated with multidrug resistance.22 A study from Taiwan demonstrated that over expression of acrB was observed in 44% of clinical isolates from UTI showing resistance to carbapenem and tigecycline.23 A study from India reported that 35% clinical isolates of E.coli showed efflux pump mediated resistance to carbapenems by efflux pump inhibition assay using CCCP as an efflux pump inhibitor. The same study reported that 52% of the isolates selected for Real Time qRT PCR showed upregulation of acrA.24
In the current study, 127 carbapenem resistant clinical isolates of Enterobacteriaceae were phenotypically screened for efflux pump mediated resistance to carbapenem by efflux pump inhibition assay using reserpine as an efflux pump inhibitor.
Of the 127 isolates, 56 (44%) isolates showed ≥ 2 fold decrease in MIC of imipenem with reserpine when compared to MIC of imipenem alone. This indicates role of an efflux pump inhibitor in mediating resistance to imipenem.
It was interesting to note that one isolate showed a 16 fold decrease in MIC from 128 µg/mL MIC of imipenem to 8 µg/mL MIC of imipenem with reserpine, suggesting the potential use of an efflux pump inhibitor in tandem with an antibiotic to effectively treat infections instead of consorting to higher classes of antibiotics.
Of the 56 isolates that were screen positive for efflux pump mediated resistance to imipenem, 12 representative isolates were selected for Real Time RTqPCR to study the mRNA expression levels of acrA and acrB. All the 12 strains had increased expression levels for acrA that ranged from 101 to 107 and for acrB that ranged from 101 to 109.
Out of 12 isolates, three showed the highest expression of acrA i.e., 107. In addition to the over expression of acrA, these isolates also produced beta lactamases. (a) Isolate no. C65 showed an 8 fold decrease in MIC of imipenem with reserpine, and was also positive for blaNDM. (b) Isolate no. C66 showed a 4 fold decrease in MIC of imipenem with reserpine, and was also positive for blaOXA-48. (c) Isolate no. C67 showed a 4 fold decrease in MIC of imipenem with reserpine, and was also positive for blaNDM and blaOXA-48. All the three isolates were Klebsiella pneumoniae isolated from urine samples of three patients who were above 70 years old suffering from UTI.
Out of 12 isolates, four showed a very high expression of acrB i.e., 108 – 109. In addition to the over expression of acrB, these isolates also produced beta lactamases. (a) Isolate no. C94 from a drain sample of a 50 year old female patient showed a 16 fold decrease in MIC of imipenem with reserpine, and was also positive for blaNDM. (b) Isolate no. C93 from a urine sample showed a 4 fold decrease in MIC of imipenem with reserpine, and was also positive for blaNDM. (c) Isolate no. C119 from a urine sample showed a 4 fold decrease in MIC of imipenem with reserpine, and was also positive for blaOXA-48 and blaKPC. (d) Isolate no. C157 from a urine sample showed a 4 fold decrease in MIC of imipenem with reserpine, and was also positive for blaNDM. All the four isolates (C94, C93, C119 and C157) were E. coli.
Over expression of the efflux pump AcrAB-TolC contributes to antibiotic resistance. If the genes that encode for over expression of the efflux pump are present on plasmids, along with genes that encode for enzyme mediated resistance, then it would result in rampant dissemination of multi-drug resistance that is mediated by multiple mechanisms. However, the presence of genes for AcrAB-TolC on plasmids is not explored in this study.
The present study demonstrates efflux pump AcrAB-TolC mediated resistance to carbapenems among clinical isolates of Enterobacteriaceae. Antibiotic resistance, mediated by over expression of acrA and acrB that encodes the efflux pump AcrAB-TolC in Enterobacteriaceae enhances the non-susceptibility of the bacteria to a range of antibiotics, because of the non-specificity of substrates that could be effluxed out of the efflux pump.
ACKNOWLEDGMENTS
The authors would like to thank Dr. Waheeta Hopper, Head, Department of Biotechnology, School of Bioengineering, Faculty of Engineering & Technology, Kattankulathur Campus, SRM Institute of Science and Technology for providing the control strain E.coli AcrB.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
PS conceptualized, carried out the work, and wrote the manuscript. PK supervised, provided critical feedback and reviewed and edited the manuscript. DM edited the manuscript and provided the strains included in the study. PB provided the strains included in the study. All authors read and approved the final manuscript for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
This study was approved by the Institute Ethics Committee of Dr. ALM PG IBMS, University of Madras, Taramani, Chennai, India with reference number UM/IHEC/01-2014-II.
- Murray PR, Rosenthal KS, Pfaller MA. Medical Microbiology, 5th ed. Philadelphia, Pennsylvania, USA 2005.
- Paterson DL. Resistance in gram-negative bacteria: enterobacteriaceae. Am J Med. 2006;119(6 Suppl 1):S20-S28.
Crossref - Falagas ME, Bliziotis IA. Pandrug-resistant Gram-negative bacteria: the dawn of the post-antibiotic era? Int J Antimicrob Agents. 2007;29(6):630-636.
Crossref - Dinh T, Paulsen IT, Saier Jr MH. A family of extracytoplasmic proteins that allow transport of large molecules across the outer membranes of gram-negative bacteria. J Bacteriol. 1994;176(13):3825-3831.
Crossref - Hayashi K, Nakashima R, Sakurai K, et al. AcrB-AcrA Fusion Proteins That Act as Multidrug Efflux Transporters. J Bacteriol. 2016;198(2):332-342.
Crossref - Perez A, Poza M, Fernandez A, et al. Involvement of the AcrAB-TolC efflux pump in the resistance, fitness, and virulence of Enterobacter cloacae. Antimicrob Agents Chemother. 2012;56(4):2084-2090.
Crossref - Wang H, Dzink-Fox JL, Chen M, Levy SB. Genetic characterization of highly fluoroquinolone-resistant clinical Escherichia coli strains from China: role of acrR mutations. Antimicrob Agents Chemother. 2001;45(5):1515-1521.
Crossref - Webber MA, Piddock LJ. Absence of mutations in marRAB or soxRS in acrB-overexpressing fluoroquinolone-resistant clinical and veterinary isolates of Escherichia coli. Antimicrob Agents Chemother. 2001;45(5):1550-1552.
Crossref - Duval V, Lister IM. MarA, SoxS and Rob of Escherichia coli – Global regulators of multidrug resistance, virulence and stress response. Int J Biotechnol Wellness Ind. 2013;2(3):101-124.
Crossref - Pomposiello PJ, Demple B. Identification of SoxS-regulated genes in Salmonella enterica serovar typhimurium. J Bacteriol. 2000;182(1):23-29.
Crossref - Dolejska M, Villa L, Poirel L, Nordmann P, Carattoli A. Complete sequencing of an IncHI1 plasmid encoding the carbapenemase NDM-1, the ArmA 16S RNA methylase and a resistance-nodulation-cell division/multidrug efflux pump. J Antimicrob Chemother. 2013;68(1):34-39.
Crossref - Davin-Regli A, Pages J-M, Ferrand A. Clinical Status of Efflux Resistance Mechanisms in Gram-Negative Bacteria. Antibiotics. 2021;10(9):1117.
Crossref - Hasdemir UO, Chevalier J, Nordmann P, Pages JM. Detection and prevalence of active drug efflux mechanism in various multidrug-resistant Klebsiella pneumoniae strains from Turkey. J Clin Microbiol. 2004;42(6):2701-2706.
Crossref - Tsai Y, Chang J, Liou C, et al. Regulation of AcrAB-TolC and the roles of this efflux pump on antibiotic resistance and virulence in Klebsiella pneumonia. J Microbiol Immunol Infect. 2015;48(2):S173-S174.
Crossref - Viderman D, Brotfain E, Khamzina Y, Kapanova G, Zhumadilov A, Poddighe D. Bacterial resistance in the intensive care unit of developing countries: Report from a tertiary hospital in Kazakhstan. J Glob Antimicrob Resist. 2019;17:35-38.
Crossref - Lepape A, Jean A, De Waele J, et al. European intensive care physicians’ experience of infections due to antibiotic-resistant bacteria. Antimicrob Resist Infect Control. 2020;9(1):1.
Crossref - Surendranath CR, Ramana BV, Chaudhury A, Kumar AG. Functional study of efflux pump inhibitors in clinical isolates of multidrug resistant Klebsiella pneumonia. Asian J Pharm Clin Res. 2014;7:80-82.
- Nordmann P, Dortet L, Poirel L. Carbapenem resistance in Enterobacteriaceae: here is the storm! Trends Mol Med. 2012;18(5):263-272.
Crossref - Queenan AM, Bush K. Carbapenemases: the versatile beta-lactamases. Clin Microbiol Rev. 2007;20(3):440-458.
Crossref - Lumbreras-Iglesias P, Rodicio MR, Valledor P, Suarez-Zarracina T, Fernandez J. High-Level Carbapenem Resistance among OXA-48-Producing Klebsiellapneumoniae with Functional OmpK36 Alterations: Maintenance of Ceftazidime/Avibactam Susceptibility. Antibiotics. 2021;10(10):1174.
Crossref - Adler M, Anjum M, Andersson DI, Sandegren L. Combinations of mutations in envZ, ftsI, mrdA, acrB and acrR can cause high-level carbapenem resistance in Escherichia coli. J Antimicrob Chemother. 2016;71(5):1188-1198.
Crossref - Meletis G, Exindari M, Vavatsi N, Sofianou D, Diza E. Mechanisms responsible for the emergence of carbapenem resistance in Pseudomonas aeruginosa. Hippokratia. 2012;16(4):303-307. PMCID: PMC3738602
- Chiu SK, Chan MC, Huang LY, et al. Tigecycline resistance among carbapenem-resistant Klebsiella Pneumoniae: Clinical characteristics and expression levels of efflux pump genes. PLoS One. 2017;12(4):e0175140.
Crossref - Chetri S, Bhattacharjee A, Chanda D, Chakravarty A. Transcriptional response of AcrAB-TolC conferring carbapenems resistance within Escherichia coli associated with community acquired infection. Int J Infect Dis. 2016;45(Suppl. 1):84.
Crossref
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.