ISSN: 0973-7510
E-ISSN: 2581-690X
Extended spectrum b-lactamases (ESBLs) are one of the major enzymes responsible for antimicrobial drug resistance in bacterial isolates. The objective of this study was to find out the ESBL genes (blaTEM, blaSHV and blaCTX-M) in Enterobacteriaceae. This study was conducted from November 2013 to October 2015. The identification of Enterobacteriaceae isolates & antimicrobial drug resistance was done by conventional standard microbial methods. Further genotypic detection of ESBL was done by multiplex PCR. Results: Among 942 Enterobacteriaceae isolates, 332 (35.24%) isolates was ESBL producers. We observed high prevalence of ESBL enzyme in Klebsiella pneumonia (59.09%). These isolate revealed high resistance to co-trimoxazole, fluoroquinolones and aminoglycosides. Out of 48 randomly selected isolates, ESBL genes were identified in 45 isolates which were found resistant to third generation cephalosporins. Single CTX-M gene noticed in 29 strains of Escherichia coli (E. coli), 02 strains of Klebsiella pneumoniae, and 01 strain of Proteus mirabilis. Occurrence of Combination of the genes e.g., (blaTEM+blaCTX-M), (blaSHV+blaCTX-M), (blaTEM+blaSHV+blaCTX-M) was detected in 08, 01 and 04 isolates respectively. Multiplex PCR in CTX-M carrying isolates revealed the presence of blaCTX-M group-1. The most common blaCTX-M group-1 was observed in all isolates of family Enterobacteriaceae. Antimicrobial drug resistance is a major problem of concern now a day. Studies like this will be helpful to knowing the burden of multi drug resistance as well as formulating antibiotics policy for a particular region.
blaSHV, blaTEM, blaCTX-M group-1, Enterobacteriaceae, Multi drug resistance
The β-lactamases are the groups of enzymes that open up the β-lactam ring by supplementing a water molecule to the β-lactam bond, and this disables the β-lactam antibiotics. A type of β-lactamases enzyme, extended spectrum beta-lactamases (ESBLs), is produced by Enterobacteriaceae which hydrolyze both the oxy-imino beta-lactams and monobactams, but do not have any effect on carbapenems and. cephamycins1,2. The prevalence of ESBLs differs among clinical isolates of various countries.3 Recently prevalence of multiple ESBL genes in a single strain has further complicated the process of detection4. Being plasmid and transposon mediated, it rapidly spread to numerous species of bacteria. In the early 1960s, the first plasmid mediated beta lactamase in gram negative bacteria, TEM-1, was observed5. The next plasmid mediated beta–lactamase discovered was SHV-16. The CTX-M type beta lactamases have become more important and groups in molecular class A β-lactamases, however most of the ESBLs are mutants of the TEM and the SHV enzymes. The CTX-M gene was 1st detected in E. coli strains in Germany and exhibit quickly spreaded to different countries7-11. The CTX-M gene have more than 450 diverse groups depending upon amino acids moiety and distributed into five groups: the CTX-M group 1 CTX-M group 2, CTX-M group 8, CTX-M group 9 and CTX-M group 2512. The CTX-M 15 gene was first detected in New Delhi, India. It refer to the CTX-M group-113. Walsh et al. also described a more prevalence of blaTEM in Indian Enterobacteriaceae and remarkably all were reported as CTX-M group-114. So It becomes essential to find out the prevalence of ESBL producing strains in a geographical area because it can help in judicious use of antibiotics and can guide the empirical therapy. In this consideration this study was planned in, Government Medical College and Dr. Susheela Tiwari Government Hospital, Haldwani to explore ESBL producing Enterobacteriaceae isolates from different clinical samples.
The descriptive study was conducted during November 2013 to October 2015. The clinical Specimens were collected in central laboratory, microbiology section of Dr. Susheela Tiwari Hospital & Government Medical College, Haldwani Uttarakhand. The specimens comprising urine, pus, blood, sputum, body fluids (CSF, pleural fluid, ascetic fluid, and peritoneal fluid), stool and throat swabs were received from outpatients and inpatients. The Enterobacteriaceae members were isolated and identified by standard laboratory protocol15,16. Kirby-Bauer disc diffusion method was utilized for the antimicrobial susceptibility testing as per CLSI guidelines-201217. The antibiotic discs applied for the enterobacteriaceae isolates were Ampicillin (10μg), Cefotaxime (30μg), Ceftriaxone (30μg), Ceftazidime (30μg), Aztreonam (30μg), Gentamicin (10μg), Amikacin (30μg), Ciprofloxacin (5μg), Levofloxacin (5μg), Co-trimoxazole (1.25/23.75μg), Norfloxacin (10μg) and Nitrofurantoin (300μg) both applied for urine samples only. The susceptibility for each antibioticdisc was reported as sensitive, intermediate, or resistant based on the interpretation chart supplied by Hi-Media laboratories Pvt. Ltd – Mumbai.
Detection of ESBL
Screening test
All enterobacteriaceae isolates were tested for resistance to aztreonam (30 μg) and the third generation cephalosporins namely cefotaxime (30 μg), ceftriaxone (30 μg) and ceftazidime (30 μg) by the Kirby-Bauer disc diffusion method. This was a part of the routine antimicrobial susceptibility testing. As per 2012 CLSI guidelines, resistance to at least one of the antimicrobials was taken as positive in the screening test for ESBL production17.
Phenotypic confirmatory disc diffusion test (PCDDT)
The ceftazidime (30 μg) disc alone and in combination with clavulanic acid (Ceftazidime + Clavulanic acid, 30/10 μg disc) was applied onto a plate of Mueller Hinton Agar (MHA) for ESBL detection, which was inoculated with the test strain. The culture plates were incubated at 370 C for 16-18 hr and identified more than 5 mm zone of inhibition around the combination disc in comparison to the ceftazidime disc alone was deliberated to be a marker for ESBL production.
E-Test
E-test was applied for further confirmation of ESBL isolates were made by evaluation of minimum inhibitory concentration (MIC). The MICs were evaluated by E-test strips (Hi-Media Laboratories Pvt. Limited, Mumbai). Two ends of The E-test strips were impregnated with cefotaxime (CTX) and cefotaxime + clavulanic acid (CTX+). The E-test strip was placed on the Mueller Hinton Agar (MHA) plate at centre and incubated aerobically at 37°C for 16-18 hours. The interpretation of MIC was considered as the value at the intersection of the growth ellipse with the strip. The ratio of the MIC value of cefotaxime to the MIC value of cefotaxime in combination with clavulanic acid was more than 8 or no zone was observed around cefotaxime but zone was obtained in cefotaxime and clavulanic acid combination was considered as an ESBL producer.
Quality Control
Klebsiella pneumoniae ATCC 700603 strain was used as the positive control whereas ATCC 25922 strain Escherichia coli were used as negative control.
Gene detection in members of family Enterobacteriaceae
Preparation of DNA template
Randomly selected 48 ESBL positive clinical isolates (confirmed by PCDDT) were subcultured on Teepol lactose agar. After overnight incubation, 3-4 discrete colonies of each isolate were inoculated in 100 µl mili Q water and then heated at 95°C for 5 min followed by immediate chilling at 4°C. Primers used for revealing of ESBL genes (TEM, SHV, CTX-M) are shown in Table 1.
Table (1):
ESBL genes (CTX-M, TEM and SHV) Primers.
Target gene |
Primer |
Primer sequence18 |
Amplicon size |
|---|---|---|---|
blaCTX-M |
CTX-MU1 |
5’-ATG TGC AGY ACC AGT AAR GT-3’ |
593bp |
CTX-MU2 |
5’-TGG GTR AAR TAR GTS ACC AGA-3’ |
||
blaTEM |
TEM-F |
5’-KAC AAT AAC CCT GRT AAA TGC-3’ |
936 bp |
TEM-R |
5’-AGT ATA TAT GAG TAA ACT TGG-3’ |
||
blaSHV |
SHV-F |
5’-TTT ATC GGC CYT CAC TCA AGG-3’ |
930 bp |
SHV-R |
5’-GCT GCG GGC CGG ATA ACG-3’ |
Detection of blaCTX-M gene
The blaCTX-M gene areas in all isolates were determined with the primers as depicted by woodford et al.18 Briefly, a total volume of 25 µl reaction mixture was used to amplify the blaCTX-M gene . The polymerase chain reaction (PCR) was performed in MJ-mini Bio-Rad thermal cycler with the cycling conditions. It consists of an early denaturation step at 94°С for 7 minutes and then 35 cycles of denaturation at 94˚С for 50 seconds, annealing at 50°С for 40 seconds and extension for 72°С for 60 seconds. The process was usually concluded with a final extension step at 72°С for 5 minutes. Gel electrophoresis with 2% agarose (Bangalore Genei, India) gel containing ethidium bromide was used to amplify PCR products.
The CTX-M gene groups detection by multiplex PCR
The isolates were expressed with the presence of CTX-M genogroups-1, 2, 8, 9 and 25 by using multiplex PCR with few adjustments.18 The MJ-min bio red thermal cycler was used to perform PCR reaction mixture in 25μl (Table A). The PCR cycling conditions have denaturation step in beginning at 94°С for 5 minute, this is followed by 40 cycles of final denaturation at 94°С for 1 minute, annealing at 52°С for 60 second and extension at 72°С for one minute and with a last extension at 72°C for 10 minutes. Gel electrophoresis with 2% agarose (Bangalore Genei, India) gel containing ethidium bromide was used to screen the amplified product.
Identification of blaTEM and blaSHV genes were determined by PCR as adopted by woodford et al18. The polymerase chain reaction was conducted in a reaction mixture of 25µl. (Table 2) The PCR cycling conditions have initial denaturation step at 94° C for 3 min, then 30 cycles of final denaturation at 94° C for 30 sec, annealing at 64° C for 30 sec, extension at 72° C for 1 min with final extension at 72° C for 10 min.
Table (2):
PCR Reaction mixture for β-lactamase genes.
Target gene |
2X PCR Master Mix |
Forward Primer |
Reverse Primer |
Mili Q |
Total volume |
|---|---|---|---|---|---|
blaCTX-M |
12.5µl |
0.05 µl |
0.05µl |
9.9µl |
25µl |
blaTEM |
12.5 µl |
0.05 µl |
0.05 µl |
9.9 µl |
25 µl |
blaSHV |
12.5 µl |
0.05 µl |
0.05 µl |
9.9 µl |
25 µl |
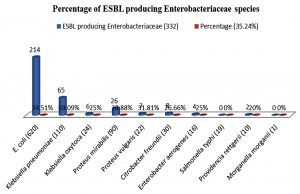
A total of 1346 gram negative bacilli (GNB) were isolated from different, non-repeated clinical specimen received during this period of study. Out of these 1346 GNB isolates, 942 (78.08%) isolates were belongs to Enterobacteriaceae family. Among the ESBL producing Enterobacteriaceae, 332 (35.24%) isolates were detected. Escherichia coli was observed in maximum number (N=214) while higher percentage of ESBL was found in Klebsiella pneumoniae (59.09%) followed by Escherichia coli (34.51%), Proteus vulgaris (31.81%), Proteus mirabilis (28.88%), Citrobacter freundii (26.66%), Klebsiella oxytoca and Enterobacter aerogens (25%) each and 20% Providencia rettgeri (Fig. 1). Maximum numbers of ESBL producing Enterobacteriaceae were found from the urine. (Table 3) Among ESBL producing Enterobacteriaceae strains, more drug resistance were seen to co-trimoxazole (100%) followed by gentamicin (63.85%) and ciprofloxacin (45.83%).
Table (3):
Specimen wise distribution of ESBL and non-ESBL producing Enterobacteriaceae.
| Clinical specimens (Enterobacteriaceae isolates) | ESBL Enterobacteriaceae isolates (%) | Non ESBL Enterobacteriaceae isolates (%) | chi-square value, p value |
|---|---|---|---|
| Urine (N=455) | 190 (41.75%) | 265 (58.24%) | 0.612 (P value <0.001) |
| Pus (N=378) | 123 (32.53%) | 255 (67.46%) | |
| Other samples (N=109) | 19 (17.43%) | 90 (82.56%) |
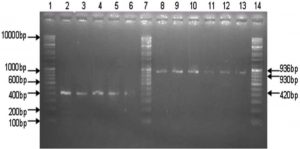
Fig. 2. 2 % Agarose gel is showing positive isolates of CTX-M-group-1 type gene (Lane 2-6) of 420 bp, positive isolates of SHV (lane 8-10) of 930 bp, positive isolates of TEM (lane 11-13) of 936bp, lanes 1,7,14 are showing Genei high range DNA ruler
Table (4):
Comparison of Antibiotic resistance pattern between ESBL producer and non-ESBL producers of family Enterobacteriaceae.
| Antibiotic | Resistance pattern | Chi-squire (χ2)
|
P value (p˂.001) |
|
|---|---|---|---|---|
| ESBL Enterobacteriaceae | Non ESBL Enterobacteriaceae |
|||
| Gentamicin | 212 (63.85%) | 75 (12.29%) | 269.79 | (p˂.001) |
| Amikacin | 78 (23.49%) | 30 (4.91%) | 73.08 | (p˂.001) |
| Levofloxacin | 113 (34.03%) | 52 (8.52%) | 96.84 | (p˂.001) |
| Ciprofloxacin | 152 (45.83%) | 55(9.01%) | 169.50 | (p˂.001) |
All these ESBL Enterobacteriaceae strains were sensitive to Amikacin (75.61%) and Imipenem (100%). Antibiotic Resistance pattern of ESBLs producing organisms is depicted in Table-4. Total 48 isolates (41 Escherichia coli, 6 Klebsiella pneumoniae and 1 Proteus mirabilis) were randomly selected for detection of CTX-M, TEM and SHV genes by conventional PCR. Overall 45 isolates were positive for CTX-M gene. Single CTX-M gene noticed in 29 strains of E. coli, 02 strains of Klebsiella pneumoniae, and 01 strain of Proteus mirabilis however three isolates of E. coli not shown any ESBL genes. The occurrence of the blaCTX-M gene was higher in E. coli as opposed to k. pneumoniae. Further analyses of these isolates were demonstrating the blaCTX-M for genogroup-1 by multiplex PCR. The CTX-M genogroup-1 was detected in all 45 isolates of Enterobacteriaceae. Occurrence of Combination of these genes e.g., (blaTEM+blaCTX-M), (blaSHV+blaCTX-M), (blaTEM +blaSHV+blaCTX-M) in detected in 13 isolate respectively. (Table-5)
Table (5):
ESBL gene detection in Enterobacteriaceae isolates.
ESBL isolates |
CTX-M |
TEM + CTX-M |
SHV + CTX-M |
CTXM+TEM +SHV |
|---|---|---|---|---|
Escherichia coli (41) |
29 |
7 |
0 |
2 |
Klebsiella pneumoniae (6) |
2 |
1 |
1 |
2 |
Proteus mirabilis (1) |
1 |
0 |
0 |
0 |
Total N=48 |
32(66.66%) |
8 (16.66%) |
1 (2.08%) |
4 (8.33%) |
In this recent time, extended-spectrum ß-lactamases (ESBLs) are main global crisis in the clinical as well as community setting due to the increased use of β -lactams and broad-spectrum cephalosporin’s antibiotics19. The first ESBL was detected in 1983 in Germany Since then, within a short span of 25 years, the ESBLs have become a worldwide problem not only producing nosocomial infections as well as it cause community-acquired infections3. This study observed the prevalence of ESBL producers (35.24%) among the members of family Enterobacteriaceae. It is comparable to the finding of Shashwati et al.20 who has reported 48.27% of ESBL isolates of the Enterobacteriaceae family. Among ESBL producing Enterobacteriaceae the prevalence of E. coli was reported 50.14% followed by Klebsiella pneumoniae 48.27%, Proteus 42.59%, Providencia 37.5%, Enterobacter 33.33%, Citrobacter 18.18%, Morganella 12.5% whereas we noticed Klebsiella pneumoniae was predominant ESBL organisms (59.09%) followed by E. coli (34.51%), Klebsiella oxytoca (25%), Proteus mirabilis (28.88%), Proteus vulgaris (31.81%), Citrobacter freundii (26.66%), Enterobacter aerogenes (25%) and Providencia rettgerii (20%). The high prevalence of Klebsiella pneumoniae may be due to the frequent uses of antibiotic in the tertiary care center and the probability for patient to patient spread is highest which may be important factor. Similar to our study ESBL production in Salmonella species have not been reported by Shashwati et al.20 We have seen a high percentage (41.75%) of ESBL producing Enterobacteriaceae in urine samples similar to Metri et al.21 also detected 39.1% of ESBL producing Enterobacteriaceae in urine sample. We have observed the high resistance in aminoglycosides and flouroquinolones antibiotics e.g., gentamicin, amikacin, levofloxacin and ciprofloxacin 63.85%, 23.49%, 34.03%, 45.83% respectively. Kaur et al.22 also observed similar to the resistance pattern against gentamcin, amikacin, levofloxacin, and ciprofloxacin 57.74%, 34.19%, 54.19% and 83.22% respectively. Imipenem was the most effective drug found in my study against the ESBL isolates. The bla CTX-M group-1 gene was observed in 45 isolates of Enterobacteriaceae. The CTX-M gene was the most prevalent and it was present singly or in the combination with TEM and SHV gene. The TEM and SHV gene not detected individually. The incidence of occurrence of CTX-M gene was higher in E. coli. The Padamini et al.23 reported CTX-M group-1 gene predominant in own study. Shahid et al.24 also observed similar results. Three E. coli strains did not reveal any ESBL gene but they were positive by E strip due to other beta-lactamase gene. The phenotypic confirmatory disc diffusion (PCDDT) method was equally sensitive and cost effective in comparison to the expensive E test. The association between these tests was not significant. The chi square value (χ2) = 0.612, P=0.736 (P value >0.05).
The ESBL producing Enterobacteriaceae isolates arising worldwide as described in previous studies in India. Antimicrobial drug resistance is a major global problem of concern now a day. An indiscriminate use of the higher antibiotics and hospitalization are the major risk factors. The present study concludes the existence of CTX-M beta-lactamase gene in singly or various beta-lactamase gene combinations. The occurrence of CTX-M has an harmful effect on public health such as dispersion of hospital-acquired infections to the community25. Studies like this will be helpful to knowing the burden of multi drug resistance as well as formulating antibiotics policy for a particular region.
ACKNOWLEDGMENTS
We are thankful to Dr. Shahid & Mrs. Anuradha Singh, department of Microbiology, Jawaharlal Nehru Medical College & Hospital, Aligarh Muslim University (AMU), U.P. (INDIA) for guidance and support.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
HS drafted the Concepts, manuscript, compiled information from the literature, Clinical studies, and designed the figures and tables. U supervised and reviewed the manuscript, Data Acquisition, Data analysis and Statistical Analysis. VR drafted the manuscript and gathered information from the literature. NN assisted for statistical analysis and Manuscript Editing. S help in data Analysis.
FUNDING
None.
ETHICS STATEMENT
The protocol was approved by the Ethical committee of government medical college, Haldwani, Uttarakhand under the reference number 141-IEC/01/13.
AVAILABILITY OF DATA
All the data pertaining to this study is available with principal investigator, Dr. Hitendra Singh. So it would be produced anytime if it is asked by the editor.
- Ogefere HO, Aigbiremwen PA, Omoregie R. “Extendedspectrum beta lactamase (ESBL)-producing gram-negative isolates from urine and wound specimens in a tertiary health facility in southern Nigeria.” Trop J Pharm Res. TJPR. 2015;14:1089-1094.
Crossref - Mukherjee M, Basu S, Mukherjee SK, Majumder M. Multidrug resistance and extended spectrum beta-lactamase production in uropathogenic E. coli which were isolated from hospitalized patients in Kolkata, India. J Clin Diagn Res. 2013;7:449-453.
Crossref - Bradford PA. Extended-spectrum beta-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin Microbiol Rev. 2001;14:933-951.
Crossref - Samaha-K foury JN, Araj GF. Recent developments in b-lactamases and extended spectrum b-lactamases. BMJ. 2003;327:1209-1213.
Crossref - Paterson DL, Bonomo RA. Extended Spectrum β – Lactamases: a clinical update. Clin Microbiol Rev. 2005;18:657-686.
Crossref - Kaur M, Aggarwal A. Occurrence of the CTX-M, SHV and the TEM Genes among the Extended Spectrum β-Lactamase Producing Isolates of Enterobacteriaceae in a Tertiary Care Hospital of North India. J Clin Diagn Res. JCDR. 2013;7(4):642-645.
Crossref - Bauernfeind A, Stemplinger I, Jungwirth R, Ernst S, Casellas JM. Sequences of beta-lactamase genes encoding CTX-M-1 (MEN-1) and CTX-M-2 and relationship of their amino acid sequences with those of other beta-lactamases. Antimicrob Agents Chemother. 1996;40:509-513.
Crossref - Radice M, Power P, Di Conza J, Gutkind G. Early dissemination of CTX-M derived enzymes in South America. Antimicrob Agents Chemother. 2002;46: 602-604.
Crossref - Quinteros M, Radice M, Gardella N, Rodriguez MM, Costa N, Korbenfeld D, et al. Extended-spectrum beta lactamases in enterobacteriaceae in Buenos Aires, Argentina, public hospitals. Antimicrob Agents Chemother.2003;47:2864-2867.
Crossref - Kim J, Lim YM, Jeong YS, Seol SY. Occurrence of CTX-M-3, CTX-M-15, CTX-M-14, and CTX-M-9 extended spectrum beta-lactamases in Enterobacteriaceae clinical isolates in Korea. Antimicrob Agents Chemother. 2005;49:1572-1575.
Crossref - Livermore DM, Hawkey PM. CTX-M: changing the face of ESBLs in the UK. J Antimicrob Chemother. 2005;56: 451-454.
Crossref - Ehlers MM, Veldsman C, Makgotlho EP, Dove MG, Hoosen AA, Kock MM. Detection of blaSHV, blaTEM and blaCTX-M antibiotic resistance genes in randomly selected bacterial pathogens from the Steve Biko Academic Hospital. FEMS Immunol Med Microbiol. 2009;56:191-196.
Crossref - Karim A, Poirel L, Nagarjan S, Nordmann P. Plasmid mediated extended-spectrum β-lactamase (CTX-M-3 like) from India and gene association with insertion sequence ISEcp1. FEMS Microbiol Lett. 2001;201: 237-241.
Crossref - Walsh TR, Toleman MA, Jones RN. Comment on: occurrence, prevalence and genetic environment of CTX-M lactamases in Enterobacteriaceae from Indian hospitals. J Antimicrob Chemother. 2007;59:799-800.
Crossref - Collee JG, Fraser AG, Marmian BP, Simmons A, editors. Mackie and McCartney practical medical microbiology. 14th ed. Standard ed. Churchill livingstone, London. 1996. (Reprinted 1999)
- Win WC, ALLEN SD, Janda WM, Koneman EW, Procop GW, Schreckenberger PC, Wood G, edidors. Colour atlas and text book of diagnostic microbiology. 6th ed. Philadelphia: Lippincott Williams.
- Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing. Twenty second informational supplement ed. CLSI document M100-S22. Wayne, PA: CLSI. 2012.
- Woodford N, Fagan EJ, Ellington MJ. Multiplex PCR for rapid detection of genes encoding CTX-M extended-spectrum -lactamases. J Antimicrob Chemother. 2006;57:154-155.
Crossref - Zamani H, Salehzadeh A, Zarrin S. Prevalence of ambler class a extended-spectrum-b-lactamases (ESBLS) among Uropathogenic Escherichia coli strains isolated from Rasht city, Iran . Bioscience Journal. 2017;33(5). 1314-1320.
Crossref - Shashwati N, kiran T, Dhanvijay. Study of extended spectrum β-lactamase producing Enterobacteriaceae and antibiotic coresistance in a tertiary care teaching hospital. J of Nat Science. 2014;5:30-35.
Crossref - Basavaraj C. Metri. The Prevalence of ESBL among Enterobacteriaceae in a Tertiary Care Hospital of North Karnataka, India. J Clin Diagn Res. JCDR. 2011;5: 470-475.
- Kaur M, Aggarwal A. Occurrence of the CTX-M, SHV and the TEM Genes Among the Extended Spectrum b-Lactamase Producing Isolates of Enterobacteriaceae in a Tertiary Care Hospital of North India. J Clin Diagn Res. JCDR. 2013;7:642-645.
Crossref - Baby Padamini S, Appalaraja B, Mani KK. Detection of Enterobactericeaeproducing CTX-M extended spectrum β-lactamases from a tertiary care hospital in South India. Indian J Med Microbiol. 2008;26:163-166.
Crossref - Shahid M, Singh A, Sobia F, et al. bla(CTX-M), bla(TEM), and bla(SHV) in Enterobacteriaceae from North-Indian tertiary hospital: high occurrence of combination genes. Asian Pac J Trop Med. 2011;4:101-105.
Crossref - Woodford N, Ward ME, Kaufmann ME, et al. Community and hospital spread of Escherichia coli producing CTX-M extended-spectrum β-lactamases in the UK. J Antimicrob Chemother. 2004;54:735-743.
Crossref
© The Author(s) 2020. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.