ISSN: 0973-7510
E-ISSN: 2581-690X
Pseudomonas aeruginosa, a Gram-negative bacterium, presents a substantial challenge in healthcare due to its adaptability and resistance. This study delves into its genotypic characteristics, focusing on ESBL and MBL genes. The prevalence of P. aeruginosa in nosocomial infections, the research aims to decipher resistance mechanisms crucial for tailored interventions. The study includes 170 non-repetitive clinical samples with protocols. Antibiotic susceptibility testing reveals diverse resistance patterns. Molecular detection of ESBL and MBL genes involves DNA isolation, PCR amplification, and gel electrophoresis. The study examined 170 P. aeruginosa samples, revealing gender-specific variations with 65.91% male and 34.09% female isolates. Antimicrobial testing displayed resistance in Ceftazidime (59%) and Ciprofloxacin (48%), while Ticarcillin-clavulanic acid showed promising sensitivity (58%). Molecular identification unveiled diverse resistance genes across sample types, emphasizing genetic complexity. The study underscores the urgency for targeted therapeutic interventions and novel antimicrobial strategies against P. aeruginosa infections. As antimicrobial resistance complexities persist, this research guides efforts toward a profound understanding of clinical interventions and strategic antimicrobial management.
Pseudomonas aeruginosa, ESBL, Multidrug Resistance (MDR), Antimicrobial Resistance
In the realm of infectious diseases, the relentless evolution of microbial pathogens poses a constant challenge to healthcare systems worldwide.1 Among these adversaries, P. aeruginosa stands out as a formidable foe, demonstrating remarkable adaptability and resilience. This ubiquitous Gram-negative bacterium is recognized as a significant contributor to both nosocomial and community-acquired infections, exerting its influence across a spectrum of clinical settings. In the pursuit of understanding and combating the threat posed by P. aeruginosa, a pivotal study has been undertaken to unravel the genotypic characteristics of Extended-Spectrum β-Lactamase (ESBL) and Metallo-β-Lactamase (MBL) genes in this microorganism.2,3 The complex and diverse nature of P. aeruginosa, a genus known for its ability to thrive in challenging environments, particularly within healthcare facilities. Nosocomial infections, in particular, are frequently attributed to P. aeruginosa, presenting a formidable challenge due to its intrinsic resistance to various antibiotics and the capacity to develop multidrug resistance (MDR).4,5,6 This study delves into the genotypic landscape of ESBL and MBL genes in β-Lactamase-producing P. aeruginosa strains isolated from a myriad of clinical samples. The primary aim of this research is to decipher the genetic underpinnings of resistance mechanisms exhibited by P. aeruginosa, with a specific focus on ESBL and MBL genes.7 Extended-spectrum β-Lactamases, enzymes capable of hydrolyzing a broad range of β-lactam antibiotics, and Metallo-β-Lactamases, known for their ability to confer resistance to carbapenems, are pivotal players in the intricate web of antimicrobial resistance. Understanding the prevalence and distribution of these genes is crucial not only for unravelling the genetic architecture of resistance in P. aeruginosa but also for informing strategies to mitigate the impact of these infections. The present study aimed to address the current issue of multidrug resistance in P. aeruginosa clinical isolates and illuminates the diverse mechanisms, namely ESBL and MBL genes, contributing to this phenomenon. The implications of these findings extend beyond the laboratory, emphasizing the urgent need for targeted therapeutic interventions and the development of novel antimicrobial strategies to counteract the growing threat posed by P. aeruginosa infections. As healthcare providers and researchers grapple with the complexities of antimicrobial resistance, this study serves as a beacon, guiding efforts towards a more profound understanding of the genetic determinants fueling the adaptability of P. aeruginosa in clinical settings.
Sample collection
The present descriptive study was conducted at a tertiary care institute in Meenakshi Medical College Hospital and Research Institute with prior approval from the Institutional Ethical Committee (Ref: IEC/09/MMCHRI/2021). The sample size comprised 170 patients, from new patients providing samples to the Department of Microbiology, with proper concern the clinical isolates of P. aeruginosa were collected. Informed written consent was obtained from all patients after a thorough explanation of the study, and the research proceeded following the ethical guidelines. Patients meeting the predefined inclusion criteria, indicative of suspected cases of reinfection of the genitourinary tract, skin and soft tissue, and central nervous system, were selected for the study. The study’s exclusion criteria such as patients who did not satisfy the predetermined criteria for probable cases of reinfection in the genitourinary tract, skin, soft tissue, and central nervous system. Furthermore, those who were unable to give informed written consent or had a previous involvement in the study were not included. The inclusion and exclusion criteria were rigorously adhered to during the patient selection process. All participants provided their informed and written consent before enrollment. Subsequently, pus, wound swabs, and sputum samples were transported to the Central Research Laboratory, Meenakshi Medical College Hospital and Research Institute, Kanchipuram for the study of genetic characterization of AMR and identification of P. aeruginosa. Identification includes Gram staining to observe the morphology of Gram-negative bacilli. All collected samples, excluding blood, were streaked onto the surfaces of different media such as nutrient agar, blood agar, and MacConkey agar plates. These plates were meticulously labelled, incubated at 37°C for 24-48 hours, and scrutinized for signs of growth and colony appearance characteristic of P. aeruginosa. Blood samples, after inoculation into Brain Heart Infusion broth (BHI broth), underwent subculturing on various media and were observed for subsequent growth. For the identification of Pseudomonas species, standard microbiological techniques were used such as conventional techniques like Gram stain staining, evaluation of colony morphology, and biochemical assays. The entire methodology was executed in accordance with established ethical standards and stringent adherence to the inclusion and exclusion criteria.3,2
Antibiotic susceptibility testing
Antibiotic susceptibility testing was conducted on P. aeruginosa isolates using the disc diffusion method, following the CLSI 2020 guideline. Fifteen antibiotic discs, including Imipenem, meropenem, cefotaxime, ceftazidime, piperacillin, ciprofloxacin, and gentamycin, were employed to determine resistance patterns. The disc diffusion assay involved placing these discs on cultured plates, According to the CLSI guidelines, The disc was procured from Himedia Pvt Ltd., The test results were determined such as zones of inhibition was measured to ascertain susceptibility or resistance. Additionally, the micro-broth dilution method assessed colistin susceptibility. Varying concentrations of colistin were introduced to bacterial isolates, determining the Minimum Inhibitory Concentration (MIC).8 This combined approach offers a comprehensive insight into P. aeruginosa‘s antibiotic susceptibility, crucial for informed therapeutic decisions.9
Detection of ESBL genes
DNA isolation was conducted by transferring 1 ml of a 24-hour bacterial broth culture into sterile Eppendorf microcentrifuge tubes, followed by centrifugation at 5000 rpm for 5 min at 4°C. Pellets were dissolved in 300 µl of tris-ethylenediaminetetraacetic acid (EDTA) buffer, incubated at 65°C for 5 min, and treated with isopropanol. After centrifugation, resulting pellets were resuspended in TE buffer with mRNAase A, followed by prokinase K treatment. Phenol-chloroform extraction and subsequent steps were performed to obtain purified DNA, which was confirmed using a spectrophotometer.3,4 PCR was conducted in 50 µl volumes with template DNA, dNTPs, gene-specific primers (TEM, SHV, IMP), and Taq polymerase. DNA amplification occurred in a master cycler under defined cycling parameters (Table 1). PCR products were analyzed on a 1% agarose gel containing ethidium bromide, and gel images were captured under ultraviolet light using a gel documentation system.
Table (1):
ESBL gene primer and cycle procedures
| Gene Type | Sequence | Product Size (bp) | Procedure |
|---|---|---|---|
| TEM | 5′-ATAAAATTCTTGAAGAC-3′ | 1,075 | 5 min at 94°C and 32 cycles of amplification consisting of 30 s at 95°C, 1 min at 54°C, and 2 min at 72°C, with 5 min at 72°C for the final extension |
| 5′-TTACCAATGCTTAATCA-3′ | |||
| SHV | 5′-TGGTTATGCGTTATATTCGCC-3′ | 867 | |
| 5′-GCTTAGCGTTGCCAGTGCT-3′ | |||
| IMP | 5’-CTACCGCAGCAGAGTCTTTGC-3′ | 432 | 95°C for 5 min, followed by 30 cycles of denaturation at 95°C for 1 minute, annealing at 50°C for 1 minute and extension at 68°C for 1 minute |
| 5’-GAACAACCAGTTTTGCCTTACC-3′ | |||
| VIM | 5’-CAAGTCCGTTAGCCCATTCC-3’ | 539 | |
| 5’-GGCACAACCACCGTATAGCAC-3’ |
The results of the study examining the demographic distribution and gender disparities in age groups among P. aeruginosa isolated cases were summarized in Table 2. A total of 170 samples were analyzed, and the data is presented based on the sample type and gender. In the Pus category, there were 47 cases among males and 37 cases among females, resulting in a total of 84 cases. For Sputum, the male cases were 36, female cases were 8, with a total of 44 cases. In the Urine category, 1 case was identified in males, while females accounted for 33 cases, totaling 34 cases. The Blood category revealed 7 male cases and 1 female case, totaling 8 cases. Overall, the study encompassed 91 male cases, 79 female cases, and a total of 170 cases. These findings provide valuable insights into the gender-specific variations in P. aeruginosa isolates across different sample types, contributing to a more nuanced understanding of the demographic characteristics of these infections and facilitating targeted intervention strategies and treatment approaches. The gender-based distribution of P. aeruginosa isolates across diverse clinical samples were tabulated in Table 2. Analyzing a total of 88 isolates, the data is organized according to the type of clinical sample and the gender of the individuals. In the Pus category, 28 (31.82%) isolates were identified in males and 15 in females (17.05%), accounting for a total of 43 isolates. For Sputum, 20 isolates were found in males (22.73%), and 4 in females (4.55%), with a cumulative total of 24 isolates (27.27%). The Urine category revealed 7 isolates in males (7.95%), and 10 in females (11.36%), totaling 17 isolates (19.32%). In the Blood category, 3 isolates were identified in males (3.41%), and 1 in females (1.14%), summing up to 4 isolates (4.55%). In totality, 58 isolates were from males (65.91% of the overall distribution), 30 from females (34.09%), resulting in 88 isolates overall. These findings not only shed light on the prevalence of P. aeruginosa isolates within specific clinical samples but also emphasize gender-specific patterns, crucial for tailoring effective clinical interventions and devising targeted treatment strategies.
Table (2):
Demographic Distribution and Gender Disparities Among P. aeruginosa with Isolate
| S. No. | Type of sample | Number of samples | Total Number of sample | Number of P. aeruginosa isolates collected | |
|---|---|---|---|---|---|
| Male | Female | ||||
| 1. | Pus | 47 | 37 | 84 | 43(48.86%) |
| 2. | Sputum | 36 | 8 | 44 | 24(27.27%) |
| 3. | Urine | 1 | 33 | 34 | 17(19.32%) |
| 4. | Blood | 7 | 1 | 8 | 4(4.55%) |
| Total | 91 | 79 | 170 | 88 | |
Antimicrobial susceptibility testing
Table 3 outlines the results of antimicrobial susceptibility testing for P. aeruginosa clinical isolates, employing both disc diffusion assay and Minimum Inhibitory Concentration (MIC). For Ceftazidime, the disc diffusion assay revealed resistance of 59%, intermediate (6%), and sensitivity (23%), while MIC results showed resistance (55%), intermediate (2%), and sensitivity (31%). Ciprofloxacin demonstrated a better susceptibility profile, with resistance (48%), intermediate (1%), and sensitivity (39%) in the disc diffusion assay, and MIC results showed resistance (38%), intermediate (4%), and sensitivity (46%). Piperacillin displayed resistance (58%), intermediate (14%), and sensitivity (16%). Ticarcillin-clavulanic acid exhibited resistance (29%), intermediate (1%), and sensitivity (58%). Notably, Polymyxin-B, Cefoxitin, and Nitrofurantoin displayed excellent sensitivity, with 100% sensitive in both disc diffusion and MIC assays.
Table (3):
Antimicrobial susceptibility testing of P. aeruginosa clinical isolates
| Antibiotic | Disc diffusion assay | Minimum Inhibitory Concentration | ||||
|---|---|---|---|---|---|---|
| R | I | S | R | I | S | |
| Ceftazidime | 59 | 6 | 23 | 55 | 2 | 31 |
| Ciprofloxacin | 48 | 1 | 39 | 38 | 4 | 46 |
| Piperacillin | 58 | 14 | 16 | |||
| Ticarcillin-clavulanic acid | 29 | 1 | 58 | |||
| Cefoperazone | 64 | 2 | 22 | |||
| Ceftriaxone | 33 | 5 | 50 | |||
| Cefotaxime | 63 | 18 | 7 | 68 | 5 | 15 |
| Gentamycin | 62 | 2 | 24 | |||
| Ceftazidime-clavulanic acid | 30 | 12 | 46 | 24 | 2 | 62 |
| Amoxiclav | 23 | 1 | 64 | |||
| Piperacillin-tazobactam | 48 | 11 | 29 | |||
| Cefoperazone-sulbactam | 47 | 5 | 36 | |||
| Amikacin | 26 | 62 | ||||
| Tobramycin | 28 | 60 | ||||
| Ofloxacin | 19 | 6 | 63 | |||
| Imipenem | 40 | 10 | 38 | 44 | 2 | 42 |
| Netilmycin | 11 | 1 | 76 | |||
| Polymyxin-B | 0 | 88 | ||||
| Cefoxitin | 0 | 88 | ||||
| Nitrofurantoin | 12 | 76 | ||||
Molecular characterization
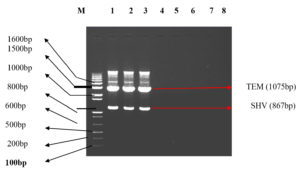
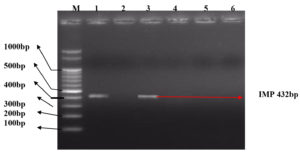
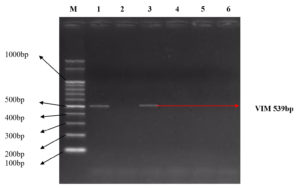
The molecular characterization results revealed the presence of specific resistance genes in P. aeruginosa clinical isolates. In Table 4, Among the 43 Pus samples, the presence of the SHV gene was identified in 5 cases, the TEM gene in 6 cases, and both SHV and TEM genes in 3 cases (Figure 1). Additionally, one sample exhibited the presence of SHV, TEM, and IMP genes. For the IMP gene, 7 cases were identified. In Sputum samples (24), SHV, TEM, and SHV+TEM combinations were found in 4, 1, and 1 case(s), respectively, with 2 cases displaying both SHV and TEM alongside the IMP gene. The IMP gene was present in 2 cases. Among Urine samples (17) (Figure 2), 2 cases showed the SHV gene, 2 cases displayed the TEM gene, and 2 cases exhibited both SHV and TEM genes and 2 VIM positive isolates were found (Figure 3). In the Blood samples (4), 1 case presented the SHV gene. Overall, this table provides a comprehensive overview of P. aeruginosa distribution and associated drug-resistant patterns across various sample types.
Table (4):
P. aeruginosa distribution among sample types and Drug-resistant pattern
Gene Name |
Pus (43) |
Sputum (24) |
Urine (17) |
Blood (4) |
Total |
|---|---|---|---|---|---|
SHV |
5 |
4 |
2 |
1 |
12 |
TEM |
6 |
1 |
2 |
9 |
|
SHV+TEM |
3 |
3 |
|||
SHV+TEM+IMP |
1 |
1 |
2 |
||
IMP |
7 |
2 |
2 |
11 |
|
VIM |
3 |
5 |
8 |
||
Total |
25 |
13 |
6 |
1 |
45 |
Figure 1. illustration Shows Lane M Containing Marker 100bp DNA ladder. LANE 1,2,3 TEM (1075 bp) and gene SHV (867 bp). LANES 4,5,7,8 were negative for gene TEM (1075 bp) and gene SHV (867 bp)
Figure 2. illustration shows Lane M containing 100bp DNA ladder. Lane 1,3 shows positive for gene IMP (432bp). Lane 2,4,5,6 show negative for gene IMP (432 bp)
The demographic distribution and gender disparities among P. aeruginosa isolated cases were comprehensively examined in our study, presenting valuable insights into the prevalence and characteristics of these infections. As illustrated in The results presented in Table 2 provide a comprehensive overview of the demographic distribution and gender-specific variations observed across different sample types in the study. Among the 170 samples analyzed, the Pus category revealed 47 male cases and 37 female cases, totaling 84 cases. Similarly, the Sputum category exhibited 36 male cases and 8 female cases, constituting 44 cases. The Urine category demonstrated 1 male case and 33 female cases, resulting in 34 cases. In the Blood category, 7 male cases and 1 female case were identified, summing up to 8 cases. Overall, the study comprised 91 male cases, 79 female cases, and a total of 170 cases, shedding light on the gender-specific prevalence of P. aeruginosa across diverse clinical samples. Furthermore, the study conducted by Nabina Maharjan in 2022 reported the isolation of P. aeruginosa in 68 (6.48%) out of 1049 clinical samples showing growth. Among these isolates, 6 (8.82%) were found to be positive for metallo-beta-lactamase. This additional information underscores the clinical significance of the isolated strains, particularly those exhibiting resistance mechanisms, such as metallo-beta-lactamase, which is crucial for understanding the challenges in effective treatment strategies.5 Table 3 further delves into the gender-based distribution of P. aeruginosa isolates across diverse clinical samples. Among the 88 isolates analyzed, the data is organized according to the clinical sample type and the gender of the individuals. The demographic distribution and gender-specific prevalence of P. aeruginosa isolates across distinct clinical samples revealed significant patterns. Analyzing the Pus category, 28 isolates were identified in males (31.82%), while 15 were found in females (17.05%), contributing to a total of 43 isolates (48.86%). Sputum samples showed 20 isolates in males (22.73%) and 4 in females (4.55%), totaling 24 isolates (27.27%). In the Urine category, 7 isolates in males (7.95%) and 10 in females (11.36%) were identified, resulting in 17 isolates (19.32%). The Blood category exhibited 3 isolates in males (3.41%) and 1 in females (1.14%), summing up to 4 isolates (4.55%). Overall, 58 isolates were from males (65.91%), 30 from females (34.09%), with a total of 88 isolates. Comparing these findings with other scientific articles, the study by Nabina Maharjan in 2022 reported the isolation of P. aeruginosa from 68 (6.48%) out of 1049 clinical samples, with 51.47% from male patients and 48.52% from female patients. Additionally, another study highlighted that pus samples accounted for 27.8% of P. aeruginosa isolates, and the highest rate of resistance was observed against ticarcillin-clavulanic acid (89.7%), emphasizing the clinical relevance of drug-resistant strains.5-7 The detection of various resistance mechanisms, including ESBLs, AmpC, and carbapenemase production, underlines the complex nature of P. aeruginosa infections and the importance of tailoring treatment strategies based on gender-specific and resistance pattern variations. These findings not only shed light on the prevalence of P. aeruginosa isolates within specific clinical samples but also emphasize gender-specific patterns, crucial for tailoring effective clinical interventions and devising targeted treatment strategies.6-9 Antimicrobial susceptibility testing was conducted to assess the response of P. aeruginosa clinical isolates to various antibiotics, as detailed in Table 4. The results indicated varying levels of resistance, intermediate, and sensitivity for different antibiotics. Ceftazidime showed resistance of 59%, intermediate (6%), and sensitivity (23%) in the disc diffusion assay, with similar patterns observed in Minimum Inhibitory Concentration (MIC) results. Ciprofloxacin displayed better susceptibility, with 48% resistance, 1% intermediate, and 39% sensitivity in the disc diffusion assay, and comparable MIC results. Polymyxin-B, Cefoxitin, and Nitrofurantoin exhibited 0% resistance and high sensitivity percentages in both disc diffusion and MIC assays, highlighting their effectiveness. Molecular characterization results revealed specific resistance genes in P. aeruginosa clinical isolates. Table 4 illustrates the distribution of these genes across various sample types. Among 43 Pus samples, the SHV gene was identified in 5 cases, the TEM gene in 6 cases, and both SHV and TEM genes in 3 cases. One sample exhibited the presence of SHV, TEM, and IMP genes. For the IMP gene, 7 cases were identified. In Sputum samples (24), SHV, TEM, and SHV+TEM combinations were found in 4, 1, and 1 case(s), respectively, with 2 cases displaying both SHV and TEM alongside the IMP gene. The IMP gene was present in 2 cases. Among Urine samples (17), 2 cases showed the SHV gene, 2 cases displayed the TEM gene, and 2 cases exhibited both SHV and TEM genes. In Blood samples (4), 1 case presented the SHV gene. This comprehensive overview highlights the association of drug-resistant patterns with specific genes in P. aeruginosa across different sample types. Comparing this with Din et al.’s study in 2023,7 they genotypically tested P. aeruginosa isolates for various resistance genes and characterized them for ESBL and MBL production. The study reported the presence of nfxB regulator gene, Mex proteins, Opr proteins, and various β-lactamase genes, emphasizing the complexity of resistance mechanisms in P. aeruginosa. Notably, both studies underscore the importance of molecular characterization in understanding drug resistance patterns.7 Mohammedkheir et al.,9 studied the β-lactamase genes in different organisms, including Pseudomonas aeruginosa, indicates the diversity of resistance mechanisms across bacterial species. It reports the presence of VIM, TEM, SHV, CTX-M-1, CTX-M-9, and CTX-M-8/25 genes in P. aeruginosa, aligning with the findings of the present study.9-12 The co-occurrence of these genes in various clinical isolates emphasizes the need for a comprehensive understanding of resistance mechanisms for effective clinical management. Gender disparities, and antibiotic susceptibility patterns among P. aeruginosa clinical isolates. These findings contribute to a deeper understanding of the epidemiology and characteristics of P. aeruginosa infections, facilitating informed clinical decision-making and the development of targeted therapeutic approaches.10-14
In conclusion, our study sheds light on the critical issue of multidrug resistance (MDR) and diverse resistance mechanisms, such as Extended-Spectrum Beta-Lactamase (ESBL) and Metallo-Beta-Lactamase (MBL), within clinical isolates of Pseudomonas aeruginosa. The escalating prevalence of MDR strains underscores the urgent need for comprehensive intervention strategies. Rigorous antibiotic policies, continuous surveillance of susceptibility patterns, and routine screening for ESBL and MBL production, especially in third-generation cephalosporins, are imperative. Early identification of MBL-producing strains is crucial to prevent treatment failure and reduce morbidity and mortality. Our findings emphasize the global significance of antimicrobial resistance and highlight the importance of collaborative efforts among healthcare professionals, policymakers, and researchers. Implementing proactive measures and remaining vigilant to emerging resistance patterns will be pivotal in addressing this pressing public health concern and developing effective treatment strategies.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
This study was approved by the Institutional Review Board, Meenakshi Medical College Hospital and Research Institute with approval number Ref: IEC/09/MMCHRI/2021.
INFORMED CONSENT
Written informed consent was obtained from the participants before enrolling in the study.
- Diggle SP, Whiteley M. Microbe Profile: Pseudomonas aeruginosa: opportunistic pathogen and lab rat. Microbiology. 2020;166(1):30-33.
Crossref - Ruppe E. Network for Enhancing Tricycle ESBL Surveillance Efficiency (NETESE) group. Lessons from a global antimicrobial resistance surveillance network. Bull World Health Organ. 2023;101(10):672-678.
Crossref - Sathe N, Beech P, Croft L, Suphioglu C, Kapat A, Athan E. Pseudomonas aeruginosa: Infections and novel treatment approaches “Knowing the enemy” the threat of Pseudomonas aeruginosa P. aeruginosa and exploring novel treatment approaches. Infect Med. 2023:2(3):178-194.
Crossref - Qin S, Xiao W, Zhou C, et al. Pseudomonas aeruginosa: pathogenesis, virulence factors, antibiotic resistance, interaction with host, technology advances and emerging therapeutics. Signal Transduct Target Ther. 2022;7(1):199.
Crossref - Maharjan N. Pseudomonas aeruginosa P. aeruginosa Isolates among Clinical Samples showing Growth in a Tertiary Care Centre: A Descriptive Cross-sectional Study. J Nepal Med Assoc. 2022;60(252):676-680.
Crossref - Mehrotra T, Konar D, Pragasam AK, et al. Antimicrobial resistance heterogeneity among multidrug-resistant Gram-negative pathogens: Phenotypic, genotypic, and proteomic analysis. Proc Natl Acad Sci U S A. 2023;120(33):e2305465120.
Crossref - Din M, Awan MA, Rahman SU, Ali M, Aslam M. Co-existence of blaIMP, blaNDM-1, and blaSHV, genes of Pseudomonas aeruginosa isolated from Quetta: Antimicrobial resistance and clinical significance. Pak J Med Sci. 2023;39(5):1507-1511.
Crossref - Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing (301th ed.). Clinical and Laboratory Standards Institute.2020.
- Mohammedkheir MIA, Gaafar EM, AbdAlla EGE. Assessment of BlaTEM, BlaSHV, and BlaCTX-M genes of antibiotic resistance in Gram-negative bacilli causing urinary tract infections in Khartoum State: a cross-sectional study. BMC Infect Dis. 2024;24(1):141.
Crossref - Gales AC, Stone G, Sahm DF, Wise MG, Utt E. Incidence of ESBLs and carbapenemases among Enterobacterales and carbapenemases in Pseudomonas aeruginosa isolates collected globally: results from ATLAS 2017-2019. J Antimicrob Chemother. 2023;78(7):1606-1615.
Crossref - Mallikarjuna PV, Dhanashree B. Phenotypic and genotypic characterization of clinical Pseudomonas aeruginosa. J Taibah Univ Med Sci. 2022;18(3):480-487.
Crossref - Tenover FC, Nicolau DP, Gill CM. Carbapenemase- producing Pseudomonas aeruginosa – an emerging challenge. Emerg Microbes. 2022;11(1):811-814.
Crossref - Chegene Lorestani R, Shojaeian A, Rostamian M. Phenotypic, genotypic, and metabolic resistance mechanisms of ESKAPE bacteria to chemical disinfectants: a systematic review and meta-analysis. Expert Rev Anti Infect Ther. 2023;21(10):1097-1123.
Crossref - Yang X, Zeng Q, Gou S, et al. Phenotypic heterogeneity unveils a negative correlation between antibiotic resistance and quorum sensing in Pseudomonas aeruginosa clinical isolates. Front Microbiol. 2024;15:1327675.
Crossref
© The Author(s) 2024. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.