ISSN: 0973-7510
E-ISSN: 2581-690X
The population rise and industrialization has resulted in the uncontrolled and untreated discharges of various toxic chemicals from different industries and these toxic chemicals cause’s very serious pollution problems. Microbial degradation of toxic pollutants is one of the important way to remove the environmentally harmful compounds. The microorganisms metabolize or enzymatically transform the chemicals to less toxic metabolites. The present research emphasized on the biodegradation of mono-aromatic pollutants like Benzene, Toluene, Xylene and Phenol (BTXP) by a formulated microbial consortium constituted by Alcaligenes sp d2, Enterobacter aerogenes, Raoultella sp and Bacillus megaterium. Statistical design tools have been used for the optimization of different parameters which influencing BTXP biodegradation. The influencing parameters pH, concentration of BTXP and inoculum formulation was identified using Plackett-Burman design and was fine- tuned with Response Surface Methodology. The results specified that pH 6.25 and 250µl of 5% BTXP with the consortium formulated by mixing equal proportions ie, 3 ml of all the four bacterial isolates with OD 1,were found to be optimum for biodegradation. The degradation efficiency of the formulated consortium at its optimized condition was analyses by Fourier Transform Infrared Spectroscopy (FT/IR). About 96% of BTXP was degraded from the medium at the optimized condition and the spectral changes in FT/IR also recommended the effective removal of toxic chemicals from the medium.
Biodegradation, Fourier Transform Infrared Spectroscopy, Microbial consortium, Plackett-Burman design, Response Surface Methodology.
Accelerated industrialization produces increased hazardous waste products. Benzene, toluene, ethyl benzene, xylene and phenol together known as BTEXP with almost the same chemical structure are the second most abundant family of organic pollutants released as mixture in the polluted environment. BTEXP are classified as environmental priority pollutants1,2. BTXP compounds are used as solvents and starting materials by different industries and produced on large scale3-6. Exposure to BTXP is a global environmental problem now a day’s. These toxic xenobiotics can produce neurological disorders and haematological effects, which may finally lead to aplastic anaemia and acute myelogenous leukemia. Pollution with these mono-aromatic hydrocarbons has been increasing and thus needs urgent remediation methodologies to remove these toxic compounds from our ecosystem. Biodegradation is the most attractive and public acceptable way to remove toxic pollutants7. Generally BTXP is encountered as mixture. Microbial consortium is more effective than single microorganism in the degradation of this organic mixture and the polluted sites are rich sources of metabolically active microorganisms8.
The successful eco application of organic mixture biodegradation such as BTXP demands the optimization of medium and growth conditions of the microorganisms. So the optimization is of prime important in the development of a bioprocess using appropriate formulated consortium. The number of conditions or parameters during biodegradation is optimized so far in a one-factor at a time approach, which is extremely time consuming. The statistically designed experiments provide a fast and effective way to face such difficulties9. Very few studies are done on the optimization of various parameters leading to biodegradation of BTXP mixture using microbial consortia. Achievement of appropriate microbial consortia for biodegradation is still in the infant stage due to lack of optimized conditions and studies. To overcome the current difficulties, the present study makes use of efficient statistical tools currently available. Statistical tools like Plackett – Burman10 and Response Surface Methodology (RSM) are more reliable and could reduce the errors in defining the effects of various conditions during the constructions of microbial consortia. A study reported that 71.82% and 79.53% removal of PAHs and crude oil were found at the optimum condition. The different parameters were optimized through Response Surface Methodology11. Optimization of parameters through response surface methodology reported diesel oil removal as 56.568%12.
In the present study, an attempt has been made for modelling and optimizing the BTXP biodegradation process. The study was designed to optimize various factors like pH, dose of inoculums, temperature, concentration of pollutants for the biodegradation of BTXP by a formulated bacterial consortium of –Alcaligenes sp d2, Enterobacter aerogenes SBS1, Raoultella sp SBS2 and Bacillus megaterium SBS3.
Isolation of bacterial strains
Alcaligenes sp d2, a phenol degrading microorganism available in the culture collection center of School of Biosciences, Mahatma Gandhi University, was used as one of the primary member during biodegradation studies. The other isolates were collected from soil sample through enrichment technique. The soil sample was collected from detergent contaminated area. The soil enrichment was initiated by adding 0.136 mM benzene, 0.109 mM toluene, 0.093 mM xylene and 0.122 mM phenol to 1 g of soil in 100 ml Mineral Salt Medium (MSM). The enrichment progressed up to a maximum growth limiting substrate concentration of 0.545 mM benzene, 0.437 mM toluene, 0.372 mM xylene and 0.489 mM phenol and the soil sample was kept on a shaker at 150 rpm at room temperature up to five days. The isolates which could able to grow at the maximum growth limiting substrate concentration were selected13.
Identification of bacterial strains
The isolates screened through soil enrichment method were identified by performing Gram’s staining and biochemical reactions. The identity of the bacterial isolates was confirmed by 16S rDNA sequence analysis using the forward primer sequence (5´-AGA GTT TGA TCM TGG CTC-3´) and the reverse sequence (5´-AAG GAG GTG WTC CAR CC-3´). The final concentration of the reagents was 1 mM MgCl2, 200 µM dNTP, 100 pmol primers and 50 ng DNA. Polymerase Chain Reaction (PCR) was carried out in MycyclerTM (Bio-Rad, USA) with the following PCR Cycle: one cycle at 94oC for 2 min, followed by 35 cycles at 94oC for 1 min, 55oC for 1 min, 72oC for 2 min, followed by final 2 min incubation at 72oC and the PCR products were sequenced at Scigenome labs, Pvt Ltd, Cochin, Kerala13.
Microbial Consortia formulation
One loopful of each of the selected cultures was individually inoculated to 50 ml nutrient broth containing 50µl phenol, benzene, xylene and toluene and the flasks were incubated over night at room temperature at 150 rpm. From the culture the cells were harvested by centrifugation. The pellets were collected and suspended in physiological saline (0.85% NaCl) to obtain the inoculum of 1.OD concentration. The microbial consortium for biodegradation studies was formulated by mixing equal proportions ie, 3 ml of each culture.
Biodegradation studies
A defined Mineral Salt-BTXP (MS-BTXP) medium with the compositions (g/l): KH2PO4 -1, (NH4)2SO4 -1, MgSO4.7H2O – 0.5 and CaCl2 – 0.01 was used as the sample for the biodegradation studies. Benzene, Xylene, Toluene and phenol (99% purity) was purchased from Himedia Pvt. Ltd, India. Different concentrations of 5% BTXP were added to the basal medium. 100µl and 250µl of 5% BTXP was added to basal medium in Plackett-Burman design trials. The substrate concentration as per Response Surface Methodology was 6,6.25,6.5,6.75 and 7. The biodegradation was carried out for a time of 24 hrs and 48 hrs at pH between 6 and 7. All the conditions for the degradation studies were according to Plackett-Burman design (Table 1, 2 and 4). The cells were removed by centrifugation at 10,000 rpm for 10 minutes. The collected supernatant was subjected to solvent extraction with diethyl ether and subjected to FT/IR.
Table (1):
The selected parameters and their value applied in the Plackett-Burman model for process optimization.
| Factors | Parameters | Units | Values | |
|---|---|---|---|---|
| Lowest | Highest | |||
| A | 5% BTXP | µl | 100 | 250 |
| B | Alcaligenes sp d2 | ml | 1 | 3 |
| C | Enterobacter aerogenes | ml | 1 | 3 |
| D | Raoultella sp | ml | 1 | 3 |
| E | Bacillus megaterium | ml | 1 | 3 |
| F | Alcaligenes + Raoultella + Bacillus | ml (each) | 0.33 | 1 |
| G | Enterobacter + Raoultella + Bacillus | ml (each) | 0.33 | 1 |
| H | Incubation Time | hours | 24 | 48 |
| J | pH | 6 | 7 | |
| K | Incubation Temperature | oC | 30 | 35 |
| L | Distilled water | ml | 99 | 100 |
Table (2):
The selected parameters and their value applied in the Response surface methodology-optimal design model for process optimization.
Name |
Unit |
Lowest |
Highest |
|---|---|---|---|
pH |
6 (6,6.25,6.50,6.75,7) |
7 |
|
5% BTXP |
µl |
100 (100,150,175,200,250) |
250 |
Inoculum |
ml |
ABC (ABCD,ACD,ABC,BCD,BDA) |
ABCDa |
Table (3):
Morphological and biochemical characteristics of isolates bacterial strains.
Biochemical Tests |
d2 |
SBS1 |
SBS2 |
SBS3 |
|---|---|---|---|---|
Grams staining |
-ve |
-ve |
-ve |
+ve |
Motility |
motile |
motile |
Non motile |
Motile |
Morphology |
rod |
rod |
rod |
Rod |
Spore staining |
-ve |
-ve |
-ve |
+ve |
Indole |
-ve |
-ve |
+ve |
-ve |
MR |
-ve |
-ve |
-ve |
-ve |
VP |
-ve |
+ve |
+ve |
-ve |
Citrate |
+ve |
+ve |
+ve |
-ve |
Urease |
-ve |
-ve |
+ve |
-ve |
TSI |
-ve |
+ve |
-ve |
+ve |
Gas |
-ve |
+ve |
+ve |
-ve |
Glucose |
-ve |
+ve |
+ve |
+ve |
Lactose |
-ve |
+ve |
+ve |
+ve |
Mannitol |
-ve |
+ve |
+ve |
+ve |
Sucrose |
-ve |
+ve |
-ve |
+ve |
Oxidase |
-ve |
-ve |
-ve |
-ve |
Catalase |
+ve |
+ve |
+ve |
+ve |
Starch hydrolysis |
+ve |
-ve |
+ve |
+ve |
Gelatin hydrolysis |
-ve |
-ve |
-ve |
-ve |
Identification of the isolates |
Alcaligenes sp. d2 |
Enterobacter aerogenes |
Raoultella sp. |
Bacillus megaterium |
Table (4):
The Plackett-Burman design with BTXP degradation as the response.
Sl. No |
A |
B |
C |
D |
E |
F |
G |
H |
J |
K |
L |
OD at 251 nm |
Response (initial OD 3.98) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
1 |
-1 |
-1 |
-1 |
1 |
1 |
-1 |
-1 |
1 |
-1 |
1 |
-1 |
0.499 |
88 |
2 |
1 |
-1 |
-1 |
1 |
-1 |
1 |
-1 |
-1 |
1 |
-1 |
-1 |
0.275 |
93 |
3 |
-1 |
1 |
-1 |
-1 |
-1 |
-1 |
1 |
-1 |
1 |
-1 |
-1 |
0.347 |
91 |
4 |
1 |
-1 |
1 |
1 |
1 |
-1 |
-1 |
-1 |
-1 |
-1 |
1 |
0.239 |
94 |
5 |
1 |
1 |
1 |
1 |
-1 |
-1 |
1 |
1 |
-1 |
1 |
1 |
0.142 |
96 |
6 |
1 |
1 |
-1 |
-1 |
1 |
-1 |
1 |
1 |
1 |
1 |
1 |
0.236 |
94 |
7 |
1 |
1 |
-1 |
1 |
1 |
1 |
1 |
-1 |
1 |
-1 |
-1 |
0.206 |
95 |
8 |
-1 |
-1 |
1 |
1 |
-1 |
1 |
-1 |
1 |
-1 |
1 |
-1 |
0.628 |
84 |
9 |
-1 |
1 |
1 |
-1 |
1 |
1 |
1 |
1 |
1 |
1 |
-1 |
0.191 |
95 |
10 |
-1 |
-1 |
1 |
-1 |
1 |
1 |
-1 |
-1 |
-1 |
-1 |
1 |
0.282 |
93 |
11 |
1 |
1 |
1 |
-1 |
-1 |
-1 |
1 |
-1 |
1 |
-1 |
1 |
0.242 |
94 |
12 |
-1 |
-1 |
-1 |
-1 |
-1 |
1 |
-1 |
1 |
-1 |
1 |
1 |
0.295 |
93 |
aA- 5% BTXP; B-Alcaligenes sp d2; C- Enterobacter aerogenes; D– Raoultella sp; E- Bacillus megaterium; F- Alcaligenes sp d2+Raoultella sp+ Bacillus megaterium; G- Enterobacter aerogenes + Raoultella sp + Bacillus megaterium; H-Incubation Time; J- pH; K-Incubation Temperature; L-Distilled Water
Process Optimization
Design Expert software version 8.0.7.1 was downloaded from the website www.statease.com and used to create the Plackett-Burman and Response Surface Methodology-optimal design matrices for analyzing the data.
Plackett-Burman design
An efficient tool for process optimization ie, Plackett-Burman design was used to define parameters that significantly influences BTXP biodegradation and to obtain smaller and easily manageable set of parameters. Eleven parameters like, 5% BTXP, different concentrations of inoculums-individual and as mixture, incubation time, incubation temperature and pH were examined with two levels: -1 to denote low level and +1 for high level (Table 1). The percentage reduction of BTXP was treated as the response for each trial and was calculated using the equation,
Control MS medium showed maximum absorption at 251 nm and the reading 3.98 was considered as initial OD.
Response surface methodology-optimal design
The parameters with significant effects on BTXP biodegradation, as recognized by Plackett-Burman design were additionally optimized with Response Surface Methodology (RSM) optimal design. The RSM design consists of 28 experiments in which each parameter were verified at five levels (Table 2) and in multiple combinations with the other parameters. In RSM-optimal design, both numeric and categoric factors were used. pH and BTXP concentration were denoted as numeric and inoculum participation was denoted as categoric factor.
Data analysis
The significance of the model was analyzed through Analysis of variance (ANOVA). Design Expert software version 8.0.7.1 was used for the graphical analysis of the experimental data obtained.
Identification of bacterial strains and formulation of bacterial consortium
The presence of mixture of organic compounds can affect the biodegradation fate by different types of microorganisms. Most of the contaminated sites are polluted with varieties of pollutants. So, biodegradation with single microorganism often results in failure due to many reasons14. These limitations may be overcome by microbial consortium with diverse biodegradation abilities15. Biodegradation of mixed organic pollutants like BTXP by a microbial consortium offers a very promising approach in terms of cost effectiveness and elimination of secondary xenobiotics. Biodegradation of BTXP mixture requires the cooperation of more than one species of microorganisms and bacteria, fungi and algae play key roles in the removal of such compounds16,17. This mixture of contaminants acts as both energy and source of carbon for the growth of microorganisms and thus needs specific conditions for their biodegradation capacities.
Three bacterial strains Enterobacter aerogenes SBS1, Raoultella sp SBS2 and Bacillus megaterium SBS3 were selected as potent BTXP degraders through soil enrichment with benzene, toluene, xylene and phenol. The morphological and biochemical characteristics of all the three newly isolated strains along with Alcaligenes sp d2 were represented in Table 3. The gene sequence of 16S rDNA of SBS1 showed 100% similarity with Enterobacter aerogenes. The gene sequence of 16S rDNA of SBS2 showed 100% similarity with Raoultella sp. and SBS3 showed 99% similarity with Bacillus megaterium. The isolated strains were deposited in the GenBank database under the accession numbers Enterobacter aerogenes SBS1: KC758848, Raoultella sp. SBS2: KC758849 and Bacillus megaterium SBS3: KC758850.
Selection of important parameters with Plackett-Burman design
The biodegradation of BTXP has been shown to be dependent on several environmental and nutritional factors. To get maximum degradation rate, fine tuning and wise blending of these parameters are essential. This can be achieved by optimization procedures. The classical process optimization cannot study all the combinations of parameters and is too laborious. The Plackett – Burman and Response Surface Methodology (RSM) -optimal designs are well established in this field and optimize parameters on a few set of experiments18,19. Plackett-Burman Model was used for the parameter optimization during the effective biodegradation of chloroxylenol20 and crude oil21. The process optimization study by Dutta and Singh22 with Plackett-Burman Model recognized temperature and Na2HPO4 as the most effective factors affecting the petroleum bioremediation by Pseudomonas sp.
Plackett-Burman design is a very useful statistical design tool which helps the investigator to find out the relative importance of various parameters. Therefore, attempt was made to improve the conditions of BTXP biodegradation by simultaneous comparison between two levels of several factors using the design. The information’s in Table 4 showed variations from 84 to 96 % reduction of BTXP. This variation reflects the significance of optimization of parameters to achieve higher biodegradation. The data included in Table 5 has shown that pH (J), concentration of BTXP (A), and inoculum participants (G) along with Alcaligenes sp d2 have statistical significant impact on biodegradation.
Table (5):
Analysis of yields and evaluation of impacts.
Variables |
A |
B |
C |
D |
E |
F |
G |
H |
J |
K |
L |
|---|---|---|---|---|---|---|---|---|---|---|---|
ΣH |
567 |
566 |
557 |
551 |
559 |
553 |
566 |
551 |
568 |
551 |
565 |
Σ L |
544 |
545 |
554 |
560 |
552 |
558 |
545 |
560 |
543 |
560 |
546 |
Difference (Σ H – ΣL) |
23 |
21 |
3 |
-9 |
7 |
-5 |
21 |
-9 |
25 |
-9 |
19 |
Mean square (ΣH – ΣL)2 12 |
44.08 |
36.75 |
0.75 |
6.75 |
4.08 |
2.08 |
36.75 |
6.75 |
52.08 |
6.75 |
– |
Error square (Impact factor) |
– |
– |
– |
– |
– |
– |
– |
– |
– |
– |
30.08 |
Mean square Error square |
1.47 |
1.22 |
0.03 |
0.22 |
0.14 |
0.07 |
1.22 |
0.22 |
1.73 |
0.22 |
– |
aA- 5% BTXP; B- Alcaligenes sp d2; C- Enterobacter aerogenes; D- Raoultella sp; E- Bacillus megaterium; F- Alcaligenes sp d2+Raoultella sp+ Bacillus megaterium; G- Enterobacter aerogenes + Raoultella sp + Bacillus megaterium; H-Incubation Time; J-pH; K-Incubation Temperature; L-Distilled Water
Table (6):
Yield obtained for process optimization as per the trials conducted by the response surface methodology-optimal design software analysis.
No |
pH |
5% BTXP µl |
Inoculum ml |
OD at 251 nm |
% reduction in OD (initial OD 3.98) |
|---|---|---|---|---|---|
1. |
7.00 |
250 |
ACD |
0.310 |
92 |
2. |
6.50 |
200 |
BCD |
0.367 |
91 |
3. |
6.50 |
150 |
ABCD |
0.289 |
93 |
4. |
7.00 |
250 |
ABC |
0.319 |
92 |
5. |
6.00 |
100 |
BDA |
0.311 |
92 |
6. |
6.25 |
200 |
BCD |
0.321 |
92 |
7. |
6.00 |
150 |
ABC |
0.428 |
89 |
8. |
6.00 |
250 |
ACD |
0.351 |
91 |
9. |
6.50 |
250 |
ABC |
0.493 |
88 |
10. |
6.75 |
200 |
ABCD |
0.292 |
93 |
11. |
6.00 |
100 |
ABCD |
0.351 |
91 |
12. |
7.00 |
100 |
BCD |
0.410 |
90 |
13. |
7.00 |
250 |
BDA |
0.485 |
88 |
14. |
6.50 |
200 |
BDA |
0.426 |
89 |
15. |
7.00 |
175 |
ABC |
0.398 |
90 |
16. |
6.75 |
175 |
ACD |
0.338 |
92 |
17. |
6.00 |
250 |
BCD |
0.491 |
88 |
18. |
6.75 |
100 |
ABC |
0.400 |
90 |
19. |
6.00 |
250 |
BDA |
0.629 |
84 |
20. |
6.00 |
100 |
BCD |
0.367 |
91 |
21. |
7.00 |
100 |
BDA |
0.507 |
87 |
22. |
6.00 |
250 |
ABC |
0.365 |
91 |
23. |
6.25 |
250 |
ABCD |
0.155 |
96 |
24. |
7.00 |
250 |
ABCD |
0.310 |
92 |
25. |
6.50 |
100 |
ACD |
0.381 |
90 |
26. |
6.00 |
250 |
ABCD |
0.313 |
92 |
27. |
7.00 |
100 |
ACD |
0.421 |
89 |
28. |
7.00 |
250 |
BCD |
0.435 |
89 |
Optimization by Response surface methodology-optimal design
The Plackett-Burman design model does not describe the interactions among different factors. Hence, the interactions among the selected parameters were calculated further by Response Surface Methodology–optimal design. Response Surface Methodology (RSM) is a collection of mathematical and statistical techniques allows the calculation of the optimum levels of different parameters based on a few sets of trials. The one factor at a time optimization approach often leads to misinterpretation of results due to the interactions between various factors23.
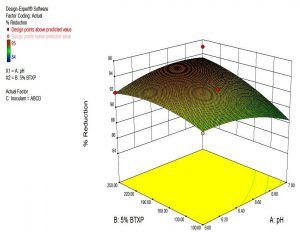
The individual and interactive influence of significant parameters was further optimized by Response Surface Methodology- optimal design. The actual levels of each parameter and the results were indicated in Table 6. The highest % reduction of BTXP of 96 obtained at pH 6.25 and 250µl of 5% BTXP. The data obtained were fitted onto a quadratic model regression equation for the calculation of BTXP biodegradation as a function of pH, concentration of 5% BTXP along with inoculum participants. Table 7 indicated the validity of model by Analysis of variance (ANOVA). The Pred-R2 of 0.2601 was in reasonable agreement with Adj- R2 of 0.3251 and confirmed that the completed experiments were reliable.
Table (7):
ANOVA test for Response surface methodology-optimal design.
Source |
Sum of Squares |
DF |
Mean Square |
F Value |
Prob> F |
Status |
|---|---|---|---|---|---|---|
Model |
63.04 |
6 |
10.51 |
3.17 |
0.0226 |
Significant |
Residual |
69.64 |
21 |
3.32 |
|||
Total |
132.68 |
27 |
R2 =0.2601; Adj-R2 = 0.3251
The data obtained were used to create three dimensional response surface curves to find out the comparative effects of any two variables when the concentration of the third variable maintained at the ABCD level.
The three dimensional response surface curve also showed comparative effects of any two variables, when the concentration of the third variable is maintained at particular level. An idea about the estimated range of the three parameters, which would result in maximum degradation of BTXP, was obtained from the figure. Fig.3 shows the interactive effects of pH and 5% BTXP on biodegradation when the inoculum participation maintained at the ABCD level. The highest % reduction of BTXP of 96 obtained at pH 6.25 and 250 µl of 5% BTXP.
Recently few studies reported the optimization of factors which influences the biodegradation process through Plackett- Burman and Response Surface Methodology. A research work reported that the concentration of microorganisms, phenol concentration, reaction time and the interactions between these factors are the most important factors which influences the phenol degradation process. These influencing factors were identified by Plackett- Burman and Response Surface Methodology24. Response Surface Methodology was used for the optimization of BTEX25 by Pseudomonas putida and maximum removal efficiency of 94, 98, 95 and 98% for benzene, toluene ethylbenzene and xyene was obtained at optimized conditions. The air flow rate had the most significant impact on BTEX biodegradation.
Fourier Transform Infrared spectroscopy (FT/IR) analysis of BTXP biodegradation
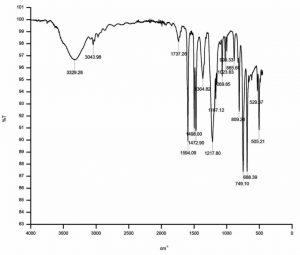
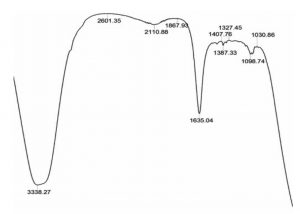
FT/IR analysis of uninoculated BTXP medium-control (Fig.1) showed specific bands representing Benzene, Toluene, Xylene and Phenol. FT/IR analysis of consortium inoculated medium showed the absence of many of the specific bands of BTXP on biodegradation. The structural changes indicated in the representation of C-H stretch, C=C stretch, C-O stretch and C-H bends in the FT/IR analysis spectra indicated the effective biodegradation of BTXP by the formulated consortium in the optimized condition (Fig.2). The three bacterial isolates selected through soil enrichment method along with Alcaligenes sp d2, have the capability to grown at this concentration of BTXP and no inhibitory effects were shown among the strains.
Fig. 1. Fourier transform infrared spectroscopy analysis of the mixture of the compounds Benzene, Toluene, Xylene, and Phenol-Control.
Fig. 2. Fourier transform infrared spectroscopy analysis of 250 µl of 5% Benzene, Toluene, Xylene, and Phenol at a pH of 6.25 after 48 h of degradation with the novel bacterial consortium.
The pH of the mineral salt medium plays an important role in the microbial growth and enzyme activities during biodegradation. A study on BTEX biodegradation26 indicated that a pH 6-8 was optimum for BTEX biodegradation and the degradation was inhibited at pH 5, 9 and 10. A bacterial strain Bacillus cereus ATHH39 was selected as a potent toluene degrading organism and the conditions for better degradation were optimized as pH 6.72, 33.16°C, and toluene concentration of 824.15 mg/l through response surface methodology27.
Biodegradation process was analysed by comparing the FT/IR of uninoculated control BTXP mixture with FT/IR of biodegraded BTXP mixture. A phenol degrading bacterium Alcaligenes sp d228 were successful in degrading all the four pollutants in the mixture of BTXP. The newly isolated BTXP degrading strains through soil enrichment, Enterobacter aerogenes SBS1, Raoultella sp SBS2 and Bacillus megaterium SBS3 were selected as the potent degraders of phenol and benzene, toluene and xylene, and phenol and benzene respectively13. The structural changes in the FT/IR spectrum and introduction of vibration in the range 1000-1300 cm-1 and 1625-1750 cm-1 indicated the presence of C=O stretch of ketones, C-O stretch of esters and carboxylic acids.
Biotechnological applications for waste management have need of the development of a mixed biological system for the detoxification of environmental pollutants. The findings concluded that the optimum condition for BTXP biodegradation were pH 6.25 and 250µl of 5% BTXP with the consortium constituted by mixing equal proportions ie, 3 ml of all the four isolates Alcaligenes sp d2, Enterobacter aerogenes, Raoultella sp and Bacillus megaterium with OD 1. The selected variables indicated significant effects on the biodegradation of BTXP. At the optimum condition the degradation study was able to achieve the maximum removal efficiency of 96% for BTXP mixture. This structural change in the FT/IR spectrum during degradation confirms the effective degradation of BTXP present in the MS-BTXP medium. The application of the optimized microbial consortium can serve as a versatile participant for the treatment of Benzene, Toluene, Xylene and Phenol from polluted environments.
ACKNOWLEDGMENTS
None.
FUNDING
None.
ETHICS STATEMENT
The article does not contain any studies with human participants or animals.
AVAILABILITY OF DATA
All datasets generated or analyzed during this study are included in the manuscript.
- Anneser B., Einsiedl F., Meckenstock R.U., Richters L., Wisotzky F., Griebler C. High-resolution monitoring of biogeochemical gradients in a tar-oil contaminated aquifer. Appl. Geochem., 2008; 23(6): 1715–1730.
Crossref - Jo M.S., Rene E.R., Kim S.H., Park H.S. An analysis of synergistic and antagonistic behaviour during BTEX removal in batch system using response surface methodology. J. Hazard Mater., 2008; 152(3): 1276–1284.
Crossref - Nicholson C.A., Fathepure B.Z. 2004. Biodegradation of benzene by halophilic and halotolerant bacteria under aerobic conditions. Appl. Environ. Microbiol., 2004; 70(2): 1222-1225.
Crossref - Mattison R.G., Taki H., Harayama S. The soil flagellate Heteromita globosa accelerates bacterial degradation of alkyl-benzenes through grazing and acetate excretion in batch culture. Microb. Ecol., 2005; 49(1):142-150.
Crossref - Andreoni V., Gianfreda L. Bioremediation and monitoring of aromatic-polluted habitats. Appl. Microbiol. Biotechnol., 2007; 76(2): 287-308.
Crossref - Lin C.W., Chen L.H., I Y.P., Lai C.Y. Microbial communities and biodegradation in lab-scale BTEX-contaminated groundwater remediation using an oxygen-releasing reactive barrier. Bioprocess Biosyst Eng, 2010; 33(3): 383–391.
Crossref - Machnicka A., Suschka J. Activity of selected microorganisms and mixtures in BTX biodegradation. Pol. J. Environ. Stud., 2001; 10(5): 341-346.
- Narancic T., Djokic L., Kenny S.T., O’Connor K.E., Radulovic V., Nikodinovic-Runic J., Vasiljevic B. Metabolic versatility of Gram-positive microbial isolates from contaminated river sediments. J. Hazard Mater., 2012; 215-216: 243–251.
Crossref - Abdel-Fattah Y.R., El-Helow E.R., Ghanem K.M., Lotfy W.A. Application of factorial designs for optimization of avicelase production by a thermophilic Geobacillus isolate. Res. J. Microbiol., 2007; 2(1): 13-23.
Crossref - Plackett R.L, Burman J.P. The design of optimum multifactorial experiments. Biometrika, 1946; 33(4): 305-325.
Crossref - Virupakshappa P.K., Krishnaswamy M.B., Mishra G., Mehkri M.A. Optimization of crude oil and PAHs degradation by Stenotrophomonas rhizophila KX082814 strain through response surface methodology Using Box-Behnken design. Biotechnol. Res. Int., 2016; 2016: 1-13.
Crossref - Olawale O., Oyawale F.A., Adepoju T.F., Aikulolu S., Akinmoladun A.I. Optimisation of Diesel Polluted Soil Using Response Surface Methodology. International Journal of Environmental Protection and Policy, 2015; 3(6): 194-202.
Crossref - Vijayan D., Kochupurackal J., Abraham A., Nair I.C. Microbial consortia formulation for the effective biodegradation of benzene, toluene, xylene and phenol. J. Microbiol. Biotechnol. Food Sci., 2014; 3(6): 457-462.
- Herrero M., Stuckey, D.C. Bioaugmentation and its application in wastewater treatment: a review. Chemosphere, 2015; 140: 119–128.
Crossref - Gurav R., Lyu H.H., Ma J.L., Tang J.C., Liu Q.L., Zhang H.R. Degradation of n-alkanes and PAHs from the heavy crude oil using salt-tolerant bacterial consortia and analysis of their catabolic genes. Environ. Sci. Pollut. Res., 2017; 24: 11392–11403.
Crossref - Schulze S., Tiehm A. Assessment of microbial natural attenuation in groundwater polluted with gasworks residues. Water Sci. Technol., 2004; 50(5):347-353.
Crossref - Nikolova N., Nenov V. BTEX degradation by fungi. Water Sci. Technol., 2005; 51(11): 87-93.
Crossref - Weuster-Botz D. Experimental design for media development: statistical design or global random search? J. Biosci. Bioeng., 2000; 90(5): 473-483.
Crossref - Abdel-Fattah Y.R. Optimization of thermostable lipase production from a thermophylic Geobacillus sp. using Box-Behnken experimental design. Biotechnol. Lett., 2002; 24(14): 1217-1222.
Crossref - Ghanem K.M., Al-Fassi1 F.A., Al-HazmiN.M. Optimization of chloroxylenol degradation by Aspergillus niger using Plackett-Burman design and response surface methodology. Afr. J. Biotechnol., 2012; 11(84): 15040-15048.
- Berekaa M.M. Towards efficient crude oil degradation by Pseudomonas sp. strain-O2: Application of Plackett-Burman design for evaluation of cultivation conditions. Afr. J. Microbiol. Res., 2013; 7(39): 4722-4729.
Crossref - Dutta S., Singh P. The Plackett-Burman model-optimization of significant nutritional parameters for petroleum bioremediation by Pseudomonas sp. Int. J. Adv. Res., 2014; 2(1): 898-902.
- Myers R.H., Montgomery D.C., AndersonCook C.M. Response Surface Methodology: process and product optimization using designed experiments, 4th ed.; John Wiley and Sons, 2016; 856.
- Priyadharshini S.D., Bakthavalsalam A.K. Optimization of phenol degradation by the microalga Chlorella pyrenoidosa using Plackett- Burman and Response Surface Methodology. Bioresour. Technol., 2016; 207: 150-156.
Crossref - El Telib A.E., El- Nass M.H., Acio J.A. Biodegradation of BTEX: Optimization through Response Surface Methodology. American Journal Of Engineering and Applied Science, 2017.
Crossref - You Y.1., Shim J., Cho C.H., Ryu M.H., Shea P.J., Kamala-Kannan S., Chae J.C., Oh B.T. Biodegradation of BTEX mixture by Pseudomonas putida YNS1 isolated from oil contaminated soil. J. Basic Microbiol., 2013; 53(5): 469-475.
Crossref - Heydarnezhad F., Hoodaji M., Shahriarinour M., Tahmourespour A., Shariati S. Optimizing toluene degradation by bacterial strain isolated from oil-polluted soils. Pol. J. Environ. Stud., 2018; 27(2): 655-663.
Crossref - Nair I.C., Shashidhar S. Microbial degradation of phenol by a species of Alcaligenes isolated from a tropical soil. Soil SciT.5, 2004; 3(4): 47- 51.
© The Author(s) 2019. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.